Abstract
Recombineering allows DNA cloned in Escherichia coli to be modified via lambda (λ) Red-mediated homologous recombination, obviating the need for restriction enzymes and DNA ligases to modify DNA. Here, we describe the construction of three new recombineering strains (SW102, SW105 and SW106) that allow bacterial artificial chromosomes (BACs) to be modified using galK positive/negative selection. This two-step selection procedure allows DNA to be modified without introducing an unwanted selectable marker at the modification site. All three strains contain an otherwise complete galactose operon, except for a precise deletion of the galK gene, and a defective temperature-sensitive λ prophage that makes recombineering possible. SW105 and SW106 cells in addition carry l-arabinose-inducible Cre or Flp genes, respectively. The galK function can be selected both for and against. This feature greatly reduces the background seen in other negative-selection schemes, and galK selection is considerably more efficient than other related selection methods published. We also show how galK selection can be used to rapidly introduce point mutations, deletions and loxP sites into BAC DNA and thus facilitate functional studies of SNP and/or disease-causing point mutations, the identification of long-range regulatory elements and the construction of conditional targeting vectors.
INTRODUCTION
Bacterial artificial chromosomes (BACs) have become the DNA of choice for genomic sequencing due to their high stability and large insert size (100–300 kb) (1). BACs are also being used more and more for making transgenic mice, since, in many cases, all of the important regulatory sequences required for normal gene expression can be found on a single BAC (2,3). Many laboratories also use BACs as the starting point for making gene-targeting constructs for manipulating mouse genes using ES cell technology (knock-outs, knock-ins and conditional targeting using Cre/loxP) (4,5).
Recombineering (recombination-mediated genetic engineering) makes it possible to modify BAC DNA via homologous recombination [reviewed in (6,7)]. Recombineering is made possible through the use of three λ Red-encoded genes: exo, bet and gam. exo encodes a 5′–3′ exonuclease that produces 3′ overhangs from introduced double-stranded DNA targeting cassettes (dsDNA). bet encodes a pairing protein that binds to the 3′ overhangs and mediates its annealing and homologous recombination with complementary DNA present on the BAC. At the same time, gam encodes an inhibitor of the Escherichia coli RecBCD exonuclease and thereby protects the linear DNA-targeting cassette from degradation by RecBCD. λ Red (or the corresponding RecE and RecT genes of the prophage Rac) can be expressed from a multicopy plasmid using an inducible promoter (8,9). Alternatively, these genes can be expressed from a stably integrated defective λ prophage, where exo, bet and gam are controlled by the strong phage promoter pL, under stringent control of the temperature-sensitive repressor, cI857 (10,11). In the prophage system, exo, bet and gam are not expressed when the bacteria are kept at 32°C. By shifting the bacteria to 42°C for as little as 15 min, the genes are rapidly induced to very high levels and homologous recombination is very efficient.
Using recombineering, one can easily subclone a genomic fragment from a BAC by gap repair, either for use as a transgene directly or for subsequent manipulation to make a gene-targeting construct. The introduction of selectable markers into a BAC is also very easy using recombineering. However, a major limitation to the usefulness of BACs is the ease and efficiency with which one can make subtle and ‘seamless’ mutations like point mutations and clean deletions or introduce in-frame fusions of cDNAs or epitope tags without leaving at the same time a selectable marker or a loxP/Frt site at the modification site. A handful of methods for making such mutations, all based on homologous recombination in E.coli, have been developed. One method is RecA dependent and relies on the use of a shuttle vector and two recombination steps: integration followed by the resolution of the co-integrate (2,12). This method requires considerable up-front effort using traditional cloning with restriction enzymes and ligation. A simpler and more widely used method is based on positive/negative selection using e.g. a sacB–neo fusion gene (8). neo (kanamycin) resistance is used for positive selection while sucrose toxicity resulting from sacB expression is used for negative selection. A major drawback of this selection system is that spontaneous point mutations in the sacB portion of the sacB-neo fusion gene can occur without influencing the bacteria's ability to grow on kanamycin. These sacB mutants significantly increase the background after negative selection. A related method is based on counterselection using a recognition site for a rare restriction enzyme like I-SceI (13). For this method to work efficiently, the I-SceI restriction enzyme has to be induced in trans, and since there is no selection for maintaining the correct recognition sequence for this enzyme, the background due to point mutation or deletion of the restriction site is high. A method for BAC modification without selection has also been developed (14). Although relatively efficient, this method relies on a PCR-based screening of the resulting colonies to identify the desired clones. Since there is no selection step, the number of manipulations made possible by this selection procedure is more limited.
Here, we report the development of a novel and highly efficient galK-based positive/negative selection system for the manipulation of BACs. The E.coli galactose operon consists of four genes: galE, galT, galK and galM, which are necessary for growth and utilization of galactose as the only carbon source. The galK gene product, galactokinase, catalyzes the first step in the galactose degradation pathway, phosphorylating galactose to galactose-1-phosphate. Galactokinase also efficiently catalyzes the phosphorylation of a galactose analog, 2-deoxy-galactose (DOG). The product of this reaction cannot be further metabolized, leading to a toxic build-up of 2-deoxy-galactose-1-phosphate (15). Thus, both positive and negative selection can be conferred by galK. Because galK is used for both selection steps, background following negative selection is reduced and no colony screening is necessary. The small size of the galK cassette (around 1200 bp plus homology arms) also makes it easier to amplify by PCR and to introduce into bacteria using electroporation.
MATERIALS AND METHODS
General recombineering and galK selection
Recombineering was performed as follows: 500 μl of an overnight culture was diluted in 25 ml Luria–Bertani (LB) medium with or without chloramphenicol selection (12.5 μg/ml) in a 50 ml baffled conical flask and grown at 32°C in a shaking waterbath to an OD600 of 0.6. Then, 10 ml was transferred to another baffled 50 ml conical flask and heat-shocked at 42°C for exactly 15 min in a shaking waterbath. The remaining culture was left at 32°C as the uninduced control. After 15 min the two samples, induced and uninduced, were briefly cooled in an ice/waterbath slurry and then transferred to two 15 ml Falcon tubes (BD Biosciences) and pelleted using 5000 r.p.m. (eppendorf centrifuge 5804R, rotor A-4-44) at 0°C for 5 min. The supernatant was poured off and the pellet was resuspended in 1 ml ice-cold ddH2O by gently swirling the tubes in an ice/waterbath slurry. Subsequently, 9 ml ice-cold ddH2O was added and the samples pelleted again. This step was repeated once more. After the second washing and centrifugation step, the supernatant was removed, and the pellet (∼50 μl each) was kept on ice until electroporated with PCR product, gel-purified fragment or double-stranded oligo. An aliquot of 25 μl was used for each electroporation in a 0.1 cm cuvette (BioRad) at 25 μF, 1.75 kV and 200 Ω. After electroporation the bacteria were recovered in 1 ml LB (15 ml Falcon tube) for 1 h in a 32°C shaking waterbath. For the counterselection step (see below), the bacteria were recovered in 10 ml LB in a 50 ml baffled conical flask and incubated for 4.5 h in a 32°C shaking waterbath. After the recovery period, the bacteria were washed twice in 1× M9 salts as follows: 1 ml culture was pelleted in an eppendorf tube at 13 200 r.p.m. for 15 s and the supernatant was removed with a pipette. The pellet was resuspended in 1 ml of 1× M9 salts, and pelleted again. This washing step was repeated once more. After the second wash, the supernatant was removed and the pellet was resuspended in 1 ml of 1× M9 salts before plating serial dilutions (100 μl, 100 μl each of 1:10, 1:100 and 1:1000 dilutions) on minimal medium plates (see below). Washing in M9 salts is necessary to remove any rich media from the bacteria prior to selection on minimal medium. The uninduced samples routinely had a higher degree of lysis/bacterial death after electroporation, so the uninduced samples were diluted in 0.25–0.75 ml 1× M9 salts in the final step to make up for the difference. Detailed protocols for recombineering can also be found at our website (http://recombineering.ncifcrf.gov).
Minimal media and indicator plates
Minimal media and the indicator plates were prepared using standard methods (16). We added supplements as indicated.
Washing solution: 1× M9 medium.
Gal positive selection: M63 + agar (15 g/l; Difco, BD Biosciences) + d-galactose (0.2%; Sigma) + d-biotin (1 mg/l; Sigma) + l-leucine (45 mg/l; Sigma) and ± chloramphenicol (12.5 μg/ml; Sigma).
Gal counterselection: M63 + agar + glycerol (0.2%; Fischer) + d-biotin (1 mg/l) + l-leucine (45 mg/l) + DOG (0.2%; Ferro Pfanstiehl) and ± chloramphenicol (12.5 μg/ml).
Gal indicator plates: MacConkey agar (Difco, BD Biosciences) + d-galactose (1%) and ± chloramphenicol (12.5 μg/ml). Plates were prepared using manufacturer's instructions.
Bacterial strains
The strains used in this paper are listed in Table 1. SW101 was derived from DY380 by a homologous recombinational exchange of the mutated gal operon leader sequence with a 441 bp PCR product (Expand High-Fidelity PCR System; Roche Applied Science) containing the wild-type gal operon leader sequence, made using W3110 (17) bacteria as the template. The primers used to amplify the wild-type leader were (Integrated DNA Technologies, Inc.): F 5′-CGACGCATGCAGGCATGAA-3′ and R 5′-AGTGGATCACGGTGTCGATA-3′ and the PCR conditions were: 94°C for 15 s, 60°C for 30 s and 72°C for 30 s, for 35 cycles. The PCR product was gel purified (GFX kit; Amersham Biosciences) and eluted in 50 μl ddH2O. An aliquot of 2.5 μl was used in the recombineering experiment as described above and the bacteria were plated on M63 minimal medium + galactose + leucine + biotin. The plates were incubated at 32°C for 2–3 days. A few of the many resulting colonies were streaked for single colonies on indicator plates and a single dark red Gal+ colony was used in the next step. SW102 was derived from SW101 in the following way: two homology arms flanking the galK open reading frame (ORF) were PCR amplified using SW101 bacteria as template and the following primers (Integrated DNA Technologies, Inc.); recognition sites for restriction enzymes are underlined: 5′arm F: 5′-AAATAACTCGAGCAGCTGCACGCGCACTTT-3′; 5′arm R: 5′-AAATAAGAATTCTTCTTACACTCCGGATTCGC-3′; 3′arm F: 5′-AAATAAGAATTCTGTAAACCATCACAAGGAGCAG-3′; 3′arm R: 5′-AATAAAGCGGCCGCCAGCTGGTTAACGCCCTGA-3′. PCR conditions were 94°C for 15 s, 60°C for 30 s and 72°C for 30 s, for 35 cycles. The 5′ homology arm (171 bp) was digested with XhoI and EcoRI, and the 3′ arm (335 bp) with EcoRI and NotI. The digested PCR products were gel purified and triple-ligated into a vector digested with XhoI and NotI. The targeting cassette was then released from the backbone using XhoI and NotI digestion, followed by gel purification. The cassette was eluted in 50 μl ddH2O, and 2.5 μl was used for recombineering as described above. After 4.5 h of outgrowth in LB media and two washes in 1× M9 salts, serial dilutions of the bacteria were plated on minimal plates containing glycerol as carbon source, leucine and biotin, plus DOG for selection against galK. The plates were incubated for 3 days at 32°C. A few of the many resulting colonies were streaked for single white/colorless (Gal−) colonies on indicator plates followed by PCR analysis and sequencing to check for correct deletion of the galK gene. A single, verified colony was expanded and a glycerol stock was made and used for initiation of all subsequent experiments. SW103 and SW104 were derived from EL250 and EL350, respectively, using the same method as for the derivation of SW101. SW105 and SW106 were derived from SW103 and SW104, respectively, using the same method as for the derivation of SW102 from SW101. SW105 expresses Flp under the control of an arabinose-inducible promoter and SW106 expresses Cre under the same promoter.
Table 1.
Recombineering reagents used in this work
| Genotype | Phenotype | Reference | |
|---|---|---|---|
| Strains | |||
| W3110 | Gal+ | (17) | |
| DH10B | mcrA Δ(mrr-hsdRMS-mcrBC) ΔlacX74 deoR endA1 araD139 Δ(ara, leu) 7697 rpsL recA1 nupG φ80dlacZΔM15 galU galK | Gal− | (18) |
| DY380 | DH10B [λc1857 (cro-bioA)<>Tet] galK+gal490 | Gal− | (11) |
| SW101 | DY380 gal+ | Gal+ | This work |
| SW102 | SW101 ΔgalK | Gal− | This work |
| EL250 | DY380 (cro-bioA)<>araC-PBADFlpe | Gal− | (11) |
| EL350 | DY380 (cro-bioA)<>araC-PBADCre | Gal− | (11) |
| SW103 | EL250 gal+ | Gal+ | This work |
| SW104 | EL350 gal+ | Gal+ | This work |
| SW105 | SW103 ΔgalK | Gal− | This work |
| SW106 | SW104 ΔgalK | Gal− | This work |
| Selection cassette | |||
| pgalK | galK wild-type gene driven by the em 7 promoter | This work | |
<> indicates the result of a homologous recombination event. Gal+: the ability to use galactose as the sole carbon source. Gal−: resistance to DOG and lack of the ability to grow on galactose as the sole carbon source.
Construction of pgalK
Two rounds of PCR were performed to construct a galK ORF driven by the prokaryotic em7 promoter. The first round was performed using galK ORF 1st F and galK ORF R primers using W3110 bacteria (17) as template. The resulting PCR product was then used as template for a second round of PCR using galK ORF 2nd F and galK ORF R primers. The product from the second round of PCR was gel purified and digested with EcoRI and BamHI and cloned into EcoRI and BamHI digested pBluescript SK−. pgalK was verified by sequencing [the plasmid sequence is available from our website (http://recombineering.ncifcrf.gov)].The primer sequences were (restriction sites and the galK ATG are underlined and the em7 promoter is in italics): galK ORF 1st F: 5′-CCCAGGAGGCAGATCATGAGTCTGAAAGAAAAAACACAATCTCTGT-3′; galK ORF 2nd F: AATAAAGAATTCCTGTTGACAATTAATCATCGGCATAGTATATCGGCATAGTATAATACGACAAGGTGAGGAACTAAACCCAGGAGGCAGATCATG; galK ORF R: AATAAAGGATCCTCAGCACTGTCCTGCTCCTT-3′. PCR conditions for both rounds were 94°C for 15 s, 60°C for 30 s and 72°C for 1.5 min, for 30 cycles. Primers were from Invitrogen.
PCR amplification of the galK targeting cassette
Sequences of all galK primers (Invitrogen) used in the experiments described in this paper are given below. em7-galK was PCR amplified using 1 ng pgalK as template and the following conditions: 94°C for 15 s, 60°C for 30 s and 72°C for 1 min, for 30 cycles. After completion of PCR, 2 μl DpnI was added to each 25 μl reaction and incubated for 2 h at 37°C to remove any plasmid template. The DpnI-digested reaction mix was run on a 1% agarose gel over night, and the PCR product was purified and eluted in 50 μl ddH2O. An aliquot of 2.5 μl was used for each experiment. For making the Nras G12D substitution the following primers were used for the first step, introducing galK. Homology to Nras sequence is in italics and the sequence recognizing em7-galK is underlined: Nras galK F: 5′-TTTTTGCTGGTGTGAAATGACTGAGTACAAACTGGTGGTGGTTGGAGCAGCCTGTTGACAATTAATCATCCGCA-3′; Nras galK R: 5′-CAAAGTGGTTCTGGATTAGCTGGATCGTCAAGGCGCTTTTCCCAACACCATCAGCACTGTCCTGCTCCTT-3′. For making 50, 75 and 100 kb deletions in the RP23-341F12 BAC, the following primers were used (homology to BAC sequence is in italics and the sequence recognizing em7-galK is underlined): galK F 341F12 start: 5′-ACTCCCACTGGAAGCTTTTTACAAAACATGTGTTGCTGACATGTTGACAGCCTGTTGACAATTAATCATCGGCA3′; galK R 341F12 50 kb: 5′-ACCCAAACCAAACAACATCCAAACCAAAAACACAGACAAAACCAAATATGTCAGCACTGTCCTGCTCCTT-3′; galK R 341F12 75 kb: 5′-ACACTAAGCCAAACTCCTTGCCTGGGCTATTTCTCTTTGTTTTTCCAAATTCAGCACTGTCCTGCTCCTT-3′; galK R 341F12 100 kb: 5′-TATGTGTCTGTGTGTGTATGTACAGTTCTTTGTTTTTGTTTTTTTTCTTTTCAGCACTGTCCTGCTCCTT-3′. For insertion of a loxP511 site in the RP23-341F12 BAC, the following primers were used for the first step (introduction of galK). Homology to BAC sequence is in italics and the sequence recognizing em7-galK is underlined: 95 kb loxP511 galK F: 5′-GGACAGAGGCGTGACAGACGTGTGAGCTCCGTGGACAACTCTCCCCGAAGCCTGTTGACAATTAATCATCGGCA-3′; 95 kb loxP511 galK R: 5′-GACTCTGAGCAGCAACGGCTGAGCCTCACTTGAGAGGGTCCCTGAGTCAC TCAGCACTGTCCTGCTCCTT-3′.
Oligos for recombineering
Oligos used to replace the galK-targeting cassette were obtained from Invitrogen. dsDNA was used and the oligos (sense and antisense) annealed in vitro: 10 μg of each oligo (sense and antisense) was mixed in an eppendorf tube in a total volume of 100 μl of 1× PCR buffer (Expand High Fidelity PCR kit, Roche Applied Science) and boiled for 5 min, allowed to cool to room temperature for 30 min, ethanol precipitated and resuspended in 100 μl ddH2O to a final concentration of 200 ng/μl double-stranded oligo. An aliquot of 1 μl (200 ng) was used in the recombineering experiments. To introduce the G12D (G<>A) point mutation in the Nras BAC CITB 50J2, the following oligos were used for the second step (the introduced adenosine/thymidine base pair is underlined, the flanking sequences are homologous to the Nras BAC sequence): G12D S: 5′-TTTTTGCTGGTGTGAAATGACTGAGTACAAACTGGTGGTGGTTGGAGCAGATGGTGTTGGGAAAAGCGCCTTGACGATCCAGCTAATCCAGAACCACTTT-3′; G12D AS: 5′-AAAGTGGTTCTGGATTAGCTGGATCGTCAAGGCGCTTTTCCCAACACCATCTGCTCCAACCACCACCAGTTTGTACTCAGTCATTTCACACCAGCAAAAA-3′. To introduce a loxP511 site in the RP23-341F12 BAC, the following oligos were used for the second step (loxP511 is underlined, the flanking sequences are homologous to the BAC sequence around position 95 kb): 95 kb loxP511 S: 5′-ACGTGTGAGCTCCGTGGACAACTCTCCCCGAAGATAACTTCGTATAGTATACATTATACGAAGTTATGTGACTCAGGGACCCTCTCAAGTGAGGCTCAGC-3′; 95 kb loxP511 AS: 5′-GCTGAGCCTCACTTGAGAGGGTCCCTGAGTCACATAACTTCGTATAATGTATACTATACGAAGTTATCTTCGGGGAGAGTTGTCCACGGAGCTCACACGT-3′.
Verification of positive recombinants
In the G12D Nras experiment, the selected Gal− clones were analyzed by SpeI digestion of BAC miniprep DNA using unmodified CITB 50J2 BAC DNA as a control. Clones without rearrangements were analyzed by PCR using 1 μl BAC miniprep DNA as the template. The PCR products were gel purified and sequenced using the same primers as were used for PCR. Primers flanking the targeted mutation were: Nras test F: 5′-CACTCATCTGCAAGGAATGCT-3′; Nras test R: 5′-CCTCAGTAAGCACGAACTTGT-3′. PCR conditions were 94°C for 15 s, 60°C for 30 s and 72°C for 30 s, for 30 cycles. Modifications of the RP23-341F12 BAC (50, 75 and 100 kb deletions and the introduction of a loxP511 site) were tested by SpeI restriction analysis of BAC miniprep DNA and compared with unmodified 341F12 BAC DNA. In the loxP511 experiment, clones 3, 5 and 6 were further tested for correct insertion of the loxP511 site by transforming 1 μl of BAC miniprep DNA into electrocompetent and arabinose-induced EL350 cells (11) and plating on LB plates with chloramphenicol. Two colonies from each starting clone were tested by SpeI digestion of BAC miniprep DNA for the 95 kb Cre-mediated deletion. Finally, the Cre-recombined clones were tested by PCR with one primer mapping to the end of the pBACe3.6 BAC backbone and the other mapping to a position 95 kb away on the wild-type BAC. The primers (Invitrogen) used for this analysis were: 95 kb loxP511 check F: 5′-GCGGATGAATGGCAGAAATTC-3′; 95 kb LoxP511 check R: 5′-TTTGCCAGACTGGTGCCTAA-3′. PCR conditions were 94°C for 15 s, 60°C for 30 s and 72°C for 30 s, for 30 cycles. The resulting PCR bands were gel purified and confirmed by sequencing using the same primers as were used for the PCR amplification. The follow-up experiment for testing the source of the observed BAC deletions was done as described above.
BACs
Mouse BACs were obtained from Invitrogen. The RP23-341F12 BAC (C57BL/6 DNA) was chosen based on position within the mouse Ebfaz gene using the UCSC genome browser (http://genome.ucsc.edu/). The CITB 50J2 BAC (CJ7/129/SV DNA) was identified by screening a CITB mouse BAC library (Invitrogen) with an Nras genomic probe using standard hybridization methods. The BAC clones were streaked for single colonies and characterized by PCR and restriction analysis before proceeding to the recombineering experiments. Two methods were used for DNA preparation. For BAC minipreps (1–1.5 μg), we used the following protocol: 5 ml overnight LB culture (15 ml Falcon tube) was pelleted for 5 min at 5000 r.p.m., the supernatant removed and the pellet was dissolved in 250 μl buffer P1 (miniprep kit, Qiagen) and transferred to an eppendorf tube. An aliquot of 250 μl P2 buffer was added followed by mixing by inversion and incubation for <5 min at room temperature. An aliquot of 250 μl N3 buffer was added followed by mixing and incubation on ice for 5 min. The supernatant was cleared by two rounds of centrifugation at 13 200 r.p.m. for 5 min in a tabletop centrifuge. Each time the supernatant was transferred to a new tube. DNA was precipitated by adding 750 μl isopropanol, mixing and incubating on ice for 10 min, and centrifugation for 10 min at 13 200 r.p.m. The pellet was washed once in 70% ethanol and the dry pellet was dissolved in 50 μl TE. An aliquot of 40 μl (∼1 μg) was used for restriction analysis, and 1 μl was used as template for PCR analysis or transformation of electrocompetent bacteria. Large-scale preparations of BAC DNA (25–100 μg) were done using the Nucleobond BAC maxi kit from Clontech (BD Biosciences) following the manufacturer's protocol.
RESULTS
Generation of SW102 cells
To see whether we could develop a more efficient selective system for BAC recombineering, we looked for a single selectable marker that could be used for both positive and negative selection. We focused on E.coli galK because both selection steps could be done using galK and its small size makes it easy to amplify by PCR. We previously reported the development of a bacterial strain, DY380 that is readily transformable with BAC DNA due to its DH10B origin (1). DY380 cells also harbor the defective λ prophage required for recombineering. The defective prophage in DY380 was transferred into DH10B with a P1 phage lysate obtained from DY363 cells [for details, see (11)]. DY363 cells carry a 1200 bp IS2 insertion element, gal490, in the mRNA leader sequence of the galactose operon, preventing gal gene transcription. Since the galactose operon is directly proximal to the site of insertion of the defective prophage, the gal490 mutation was also transferred to DY380 during P1 transduction. Thus, DY380 is phenotypically Gal− (galactose minus) and therefore unable to grow on galactose minimal medium.
Recombineering was used to correct this problem. From the wild-type E.coli strain, W3110 (17), we PCR amplified a 441 bp fragment from the wild-type gal promoter, which spans the region containing the IS2 insertion element in DY380. This PCR product was introduced into DY380 and Gal+ recombinants were selected for growth on galactose minimal medium (see Materials and Methods). We named this strain SW101 (Table 1). This strain is identical to DY380 except that it lacks the IS2 insertion element and has been made wild type for the gal operon. We then made a precise deletion of the galK gene, leaving all other genes in the galactose operon intact. This was achieved by PCR amplifying two homology arms of 171 and 335 bp, respectively, flanking the galK ORF and cloning them together into a plasmid using three-way ligation. Since the ORF of galK overlaps that of galM, we left behind the last 33 bp of galK (Figure 1) to allow for proper translation of galM. The targeting cassette, consisting of a 512 bp linear fragment, was electroporated into heat-induced and electrocompetent SW101 cells, and the recombinant clones selected on minimal medium containing glycerol as the carbon source and DOG for selection against galK. The genotype of the resulting strain, SW102 (Table 1), was confirmed by PCR analysis and sequencing of the modified region (Figure 1). This strain now harbors not only the defective λ prophage, but also a functional gal operon, except for the deletion of galK. The λ prophage is located between the galactose operon and the biotin operon, and the bio operon in these cells is nonfunctional, causing a biotin requirement. DH10B, and all derived strains, including SW102, are also deficient in leucine metabolism (18). When using minimal medium, one therefore needs to add both biotin and leucine to the plates to allow SW102 growth (see Materials and Methods).
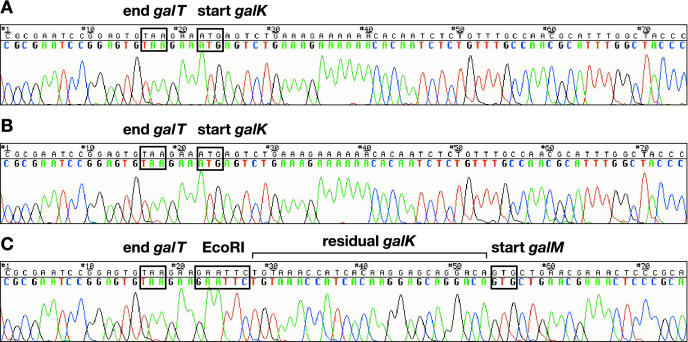
Figure 1.
Sequence analysis of the galactose operon in strains DY380 (A), SW101 (B) and SW102 (C). In SW102, the ORF of galK was deleted, leaving only 33 bp of galK behind to make sure that translation of galM is initiated properly. EcoRI: the restriction site used to clone the 5′ and 3′ homology arms flanking galK.
galK expression cassette
Having produced a bacterial strain, SW102, which is galK defective, the next step was to make a galK expression cassette that could be used to restore the bacteria's ability to grow on galactose by providing galK in trans. This was achieved by PCR amplification of the wild-type galK ORF from W3110 cells. We then added a minimal bacterial promoter, em7, using a two-step PCR approach (see Materials and Methods), and cloned the expression cassette into pBluescript SK−. We call this plasmid pgalK (Table 1). The constitutively active galK expression cassette can easily be amplified by PCR with homology arms added to the primers (see Materials and Methods).
Making a single base pair substitution
The general scheme for making mutations in BACs using galK selection is depicted in Figure 2. To test the galK selection system for BAC recombineering, we decided to introduce a point mutation into a BAC containing the murine Nras gene (CITB clone 50J2). The sequence of the glycine-coding codon 12 is GGT. By changing this codon to GAT we would obtain the desired mutation, G12D. In order to introduce this mutation into the BAC, we first amplified the galK expression cassette by PCR using primers with 50 bp of homology to either side of the second position of the GGT codon. Following homologous recombination, this targeting would introduce a 1 bp deletion into codon 12 in addition to inserting the galK selection cassette. Instead of deleting the basepair, the galK cassette could have been inserted right next to the basepair instead. SW102 cells containing the 50J2 BAC were heat-induced and made electrocompetent, and then electroporated with the galK cassette. Gal+ recombinant colonies were selected for growth on galactose minimal medium with chloramphenicol to maintain the BAC. Bacteria grow more slowly on minimal media than on rich media, and we generally pick colonies after 2–3 days. For this first step, we do not expect any background colonies on the non-induced control plates if the pgalK plasmid is properly eliminated (see Materials and Methods). To purify the Gal+ colonies, we streaked a few colonies on MacConkey galactose indicator plates to obtain single, bright pink/red Gal+ colonies. One of these single-cloned colonies was picked to initiate a culture for the next step, counterselection. We find that there is no need to analyze the Gal+ colonies further, before proceeding to counterselection.
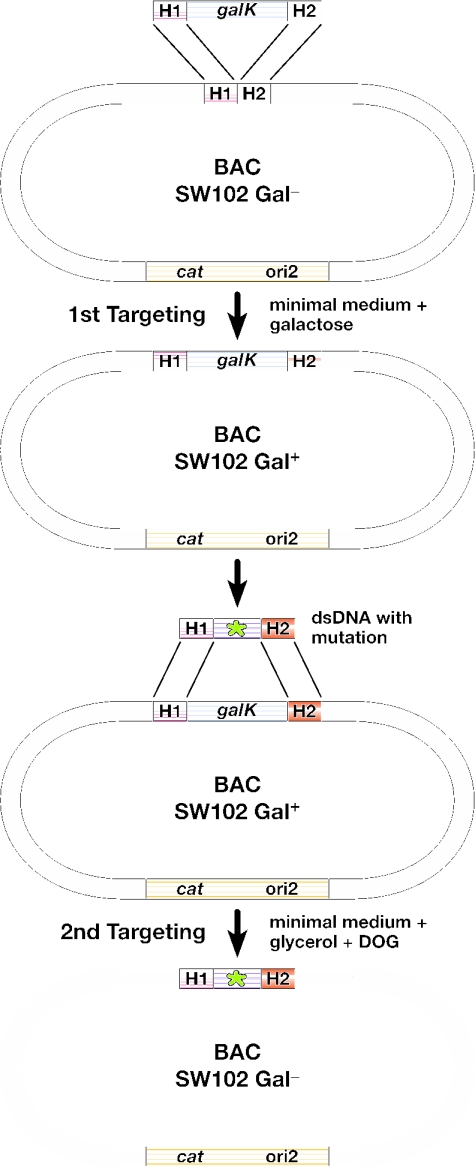
Figure 2.
Overview of the galK selection scheme. The result of the first targeting event is the insertion of constitutively active galK into a defined position on the BAC by selection on minimal medium containing galactose and chloramphenicol to select for the maintenance of the BAC. The bacteria are now phenotypically Gal+. Next, the galK cassette is replaced by a dsDNA oligo, a PCR product, or a cloned dsDNA fragment carrying a desired mutation (indicated by a star) and flanked by the same homology arms used in the first selection step. This is achieved by negative selection using minimal medium containing 2-deoxy-galactose (DOG) with glycerol as the sole carbon source. The bacteria become phenotypically Gal−. H1 and H2, homology arms 1 and 2, respectively; cat, chloramphenicol acetyl transferase gene; ori2, BAC origin of replication; galK, E.coli galactokinase gene driven by a minimal promoter.
A 100 bp dsDNA oligo was then prepared by annealing two complementary oligos having 49 and 50 bp homology, respectively, to either side of the desired mutation, a single A/T bp. An aliquot of 200 ng of this oligo was then electroporated into heat-induced and electrocompetent SW102 Gal+ cells containing the galK modified 50J2 BAC. After electroporation, the bacteria were allowed 4.5 h outgrowth in a 32°C shaking waterbath. The bacteria were then washed in M9 salts to remove any rich medium, and plated on minimal medium with glycerol, DOG and chloramphenicol. The 4.5 h outgrowth is necessary to obtain complete segregation of the recombinant BACs containing the mutation. After 3 days, we obtained colonies with a ratio of 10–100:1 when comparing plates with heat-induced to non-induced bacteria. We picked 12 colonies from the heat-induced plates for BAC minipreps, followed by SpeI restriction analysis (Figure 3A). Ten out of the twelve clones had the same restriction pattern as the unmodified 50J2 BAC DNA, suggesting that the desired replacement of the galK gene by the point mutation had occurred. This was confirmed by PCR amplification followed by sequencing of the modified region of the 10 BAC clones (Figure 3B). All 10 sequenced clones had the desired point mutation. The effective recombination efficiency in this experiment, however, was 8/12 (67%) since 2 of the 10 clones also had an additional single base pair deletion (clones 9 and 11). These deletions probably occurred during oligo synthesis, since we did not purify the oligos beyond desalting (19). Because of the high efficiency in this system, there was no need to pre-screen the selected colonies prior to picking for minipreps.
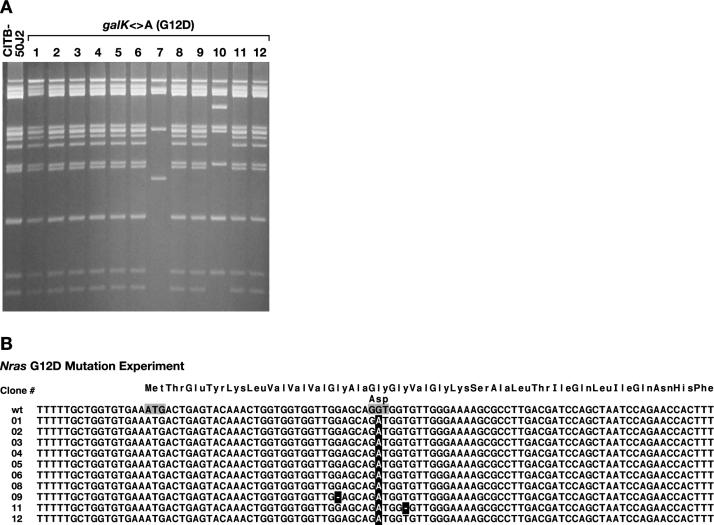
Figure 3.
Introduction of a G12D mutation in the Nras gene. (A) SpeI restriction analysis of BAC miniprep DNA. First lane is the unmodified CITB-50J2 Nras BAC. Lanes 1–12 show digestion patterns of 12 clones counterselected for the substitution of galK with an oligo containing the G→A substitution for the second position of codon 12 of Nras. Clones 7 and 10 had internal deletions, indicating that DOG resistance was achieved by spontaneous deletion and not homologous recombination. These two clones were not analyzed further. (B) Sequence analysis of a PCR product spanning the modified region from clones 1–6, 8–9 and 11–12. All clones had the intended substitution (highlighted). However, clones 9 and 11 also had an internal basepair deletion indicated by a minus (highlighted). The Nras ATG and codon 12 are indicated (shadow).
Large BAC deletions
A drawback of using BACs for the production of transgenic mice is the frequent presence of other genes on the BAC in addition to the gene of interest. This is especially a problem for BAC complementation used in positional cloning, where a BAC is tested for its ability to rescue a loss-of-function mutant phenotype by making BAC transgenic mice. If complementation is achieved, it is impossible to know which gene on the BAC is responsible for the rescue. Therefore, we decided to see whether galK selection could be used to make large, specific and clean deletions in BACs so as to remove unwanted genes (a process called BAC trimming). As a model system, we chose a mouse BAC from the C57BL/6-derived library RP23, RP23-341F12, since we could obtain the BAC sequence directly from the UCSC genomic web browser (see Materials and Methods). We then used PCR to amplify the galK selection cassette with primers containing homology to the BAC. The forward primer contained 50 bp of homology to the very 5′ end of the BAC insert, and this primer was combined with three different reverse primers, having 50 bp of homology to positions 50, 75 and 100 kb away from the forward primer's homology (Figure 4A). SW102 bacteria containing the 341F12 BAC were heat-induced and made electrocompetent, and then transformed with the three different selection cassettes, followed by the selection on minimal medium with galactose. From the many resulting Gal+ colonies, we picked four from each experiment and then did a SpeI restriction analysis of the BAC miniprep DNAs from them (Figure 4B). All four colonies from each experiment had the desired deletion (50, 75 and 100 kb, respectively). The galK selection cassette can then be removed with an oligo using galK negative selection to cleanly delete the galK cassette from the BAC as described in the previous experiment (data not shown).
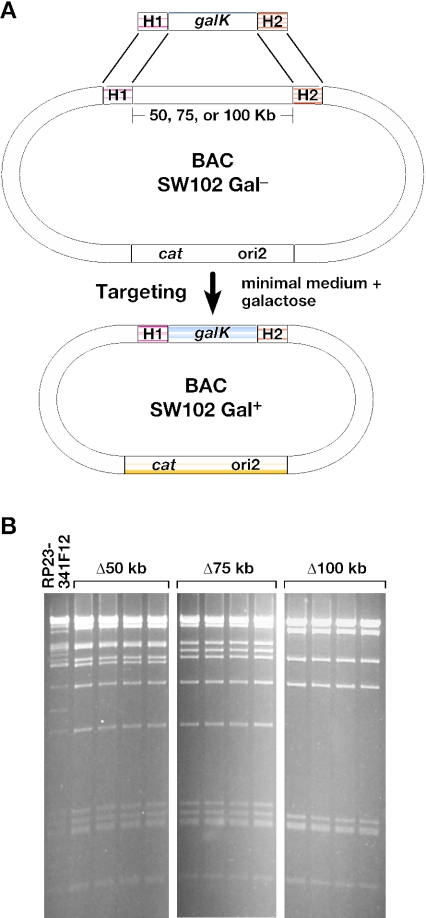
Figure 4.
BAC trimming using galK selection. (A) Illustration of the design of the deletion experiment. Homology arm 1 (H1) was held constant, and H2 was separated from H1 by either 50, 75 or 100 kb. (B) SpeI restriction analysis of BAC miniprep DNA from 12 clones showing deletions of 50, 75 and 100 kb, respectively, after the insertion of the galK selection cassette. The first lane is unmodified RP23-341F12 BAC DNA, which was included as a control. All tested clones had the intended deletion.
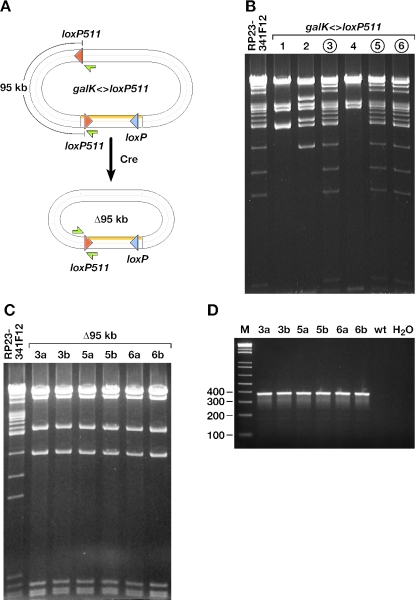
Insertion of a mutant loxP511 site into a BAC
Encouraged by the efficiency of this selection system, we decided to see whether we could introduce a single 34 bp loxP511 site cleanly into BAC DNA using galK positive/negative selection. It has been previously shown that loxP and loxP511 sites cannot recombine with each other; therefore, the introduced loxP511 site can only recombine with the loxP511 site present in the BAC vector backbone and not the wild-type site (Figure 5A) (20). We used galK positive selection to insert a PCR-amplified galK cassette flanked by 50 bp homology arms to a position 95 kb away from the mutant loxP511 site in the BAC RP23-341F12 vector backbone (Figure 5A). We then replaced the galK cassette using DOG counterselection with a double-stranded 100 bp oligo containing a 34 bp loxP511 site flanked by two 33 bp homology arms. In this experiment, we observed less than a 10-fold difference in the number of colonies on the plates from heat-induced and non-induced bacteria, suggesting fewer recombinants. This lower frequency of recombinants, when compared with the Nras G12D substitution experiment, is likely explained by the shorter homology arms used in this latter experiment, as it has been shown that the efficiency of recombination increase four orders of magnitude when homology length is increased from 20 to 40 bp (10). We analyzed six BAC minipreps from potential recombinants by digesting with SpeI and comparing the restriction patterns with that of wild-type RP23-341F12 BAC DNA (Figure 5B). Three of six (50%) colonies had exactly the same restriction pattern as the unmodified RP23-341F12 BAC, suggesting that DOG resistance had selected for the desired homologous recombination products in these three cases, whereas the other three clones apparently became DOG resistant due to the selection of BACs that carry large deletions spanning galK.
Figure 5.
Insertion of a loxP511 site. (A) The location of the wild-type and mutant loxP sites in the BAC backbone are indicated along with the extra mutant loxP511 site that was introduced into the BAC genomic insert via galK counterselection. The 95 kb region deleted by Cre-mediated recombination between the two loxP511 sites is indicated, and PCR primers used to confirm the deletion are shown as small arrows. (B) SpeI restriction analysis of six miniprep clones selected for the replacement of galK with a dsDNA oligo containing the mutant loxP511 site. Clones 3, 5 and 6 (circles) had the same restriction pattern as the unmodified BAC, indicating that DOG resistance occurred due to the intended homologous recombination event. Clones 1, 2 and 4 had large deletions and were not analyzed further. (C) SpeI restriction analysis of BAC miniprep DNA from clones 3, 5 and 6 after transformation into Cre-induced EL350 cells. Two clones from each parental clone were tested. The restriction pattern shows that the 95 kb region flanked by two loxP511 sites is deleted from all clones analyzed, confirming the correct insertion of loxP511 in clones 3, 5 and 6. (D) PCR analysis of the six clones from (C) with one primer mapping to the BAC backbone and the other to a position distal to the inserted loxP511 site.
The 100 bp oligo was designed so that the loxP511 site would be inserted in the same orientation as the loxP511 site in the BAC vector backbone. To confirm that the BACs with the wild-type restriction pattern (3, 5 and 6) had the loxP511 site correctly inserted, we electroporated these three BACs into electrocompetent and l-arabinose induced EL350 cells, which carry an l-arabinose-inducible Cre gene, and then plated the cells on LB plates containing chloramphenicol to select for the BACs. Two colonies generated from each recombinant loxP511 clone were then tested by SpeI digestion of BAC miniprep DNA, and all had the expected, Cre-mediated, 95 kb deletion (Figure 5C). Recombination was confirmed by PCR using a forward primer from the BAC vector backbone and a reverse primer mapping to a position distal to the reverse homology arm used to insert the galK cassette. These primers are 95 kb apart on wild-type RP23-341F12 DNA and only 378 bp apart on Cre/loxP511 recombined DNA. PCR analysis of all six clones produced a band of the expected length, whereas no product could be amplified from unmodified RP23-341F12 BAC DNA (Figure 5D).
Source of background deletions following DOG selection
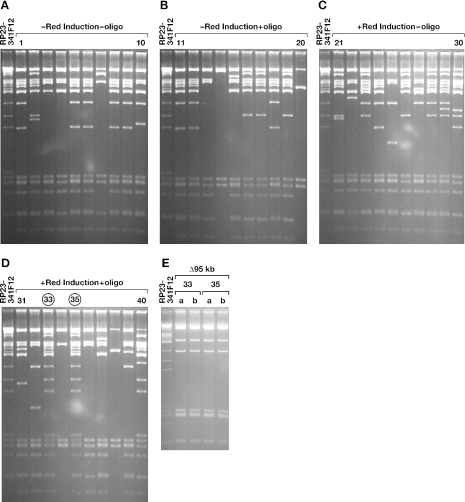
In the Nras G12D substitution and loxP511 insertion experiments, a background of DOG-resistant bacteria, which lacked the desired mutation and instead carried unwanted deletions that spanned the galK selection cassette, were observed. The relative ratio of colonies containing BACs with deletions was also higher when short homology arms were used. This was expected since homologous recombination is more efficient with longer homology arms (10) that would increase the relative frequency of correctly targeted BACs. The source of the deletions could be λ Red-mediated, since proteins capable of mediating recombination are expressed when the Red genes are activated. Alternatively, the deletions could be induced by the large amount of oligos (200 ng) used in these experiments or they could simply represent spontaneous deletions that occur at low level during normal BAC replication and that are found by DOG selection. To distinguish between these possibilities, we repeated the DOG selection step used to insert the loxP511 oligo into BAC RP23-341F12 DNA using four differently treated bacteria samples: non-induced bacteria without the 100 bp loxP511 oligo (Figure 6A), non-induced bacteria with the oligo (Figure 6B), induced bacteria without the oligo (Figure 6C) and induced bacteria with the oligo (Figure 6D). The number of resistant colonies obtained from the four experiments varied by <10-fold (data not shown). Ten colonies from each electroporation were then analyzed by SpeI digestion of BAC miniprep DNA and compared with unmodified RP23-341F12 BAC DNA (Figure 6A–D). Unwanted galK region deletions were observed in all 10 colonies for both of the non-induced samples (Figure 6A and B). These deletions are therefore unlikely to be Red-mediated since these samples were not heat-induced. We also observed deletions in both the absence and presence of oligo, indicating that the deletions are not oligo induced. We therefore conclude that these are spontaneous deletions that occur at low level during normal BAC replication in the DH10B background.
Figure 6.
Same experiment as in Figure 5 with modifications as indicated at the top of each panel. (A) SpeI digest of 10 minipreps from a control experiment without heat-induction and without the loxP511 dsDNA oligo. (B) SpeI digest of 10 minipreps from a control experiment without heat-induction but with the loxP511 dsDNA oligo. (C) SpeI digest of 10 minipreps from a control experiment with heat-induction but without the loxP511 dsDNA oligo. (D) SpeI digest of 10 minipreps from an experiment with heat-induction and with the loxP511 dsDNA oligo (comparable with Figure 5B). Clones with the parental digestion pattern indicating DOG resistance due to homologous recombination (clones 33 and 35, circles) are only seen in (D). DOG resistance in all other clones likely occurred due to internal deletions of the BACs. (E) SpeI restriction analysis of BAC miniprep DNA from clones 33 and 35 after transformation into Cre-induced EL350 cells. Two clones from each parental clone were tested. The restriction pattern shows that the 95 kb region flanked by two loxP511 sites is deleted from all the clones analyzed, confirming the correct insertion of loxP511 in clones 33 and 35 (compare with Figure 5C).
Non-deleted colonies with the parental SpeI fingerprint were only observed in the Red-induced sample that received oligo (Figure 6D, clones 33 and 35). These clones were subsequently electroporated into EL350 Cre-expressing cells to confirm they contained the introduced loxP511 site. Two colonies from each original clone were tested by SpeI digestion of the BAC miniprep DNA. As shown in Figure 6E, each colony has undergone Cre-mediated deletion, confirming that these colonies were correctly targeted and contain the introduced loxP511 site (2 out of 10 clones, a 20% efficiency in this experiment).
Generation of SW105 and SW106 cells
The two DY380-derived bacterial strains EL250 and EL350 contain the defective λ prophage needed for recombineering in addition to l-arabinose inducible Flp or Cre genes, respectively (11). Both strains have proven to be very useful for BAC modification (11), and EL350 is now used routinely for making conditional targeting vectors for ES cell knock-out experiments (4). To further enhance the usefulness of these strains we decided to transfer the galK selection system into them. This was done as previously described for SW102. The galK IS2 element present in both strains was replaced with the wild-type gal promoter and the galK gene in the gal operon deleted using DOG selection. The resulting strains, SW105 (Flp) and SW106 (Cre) (Table 1), were then tested to confirm that they still contained inducible Flp and Cre genes by transforming arabinose-induced and electrocompetent cells with plasmids containing a neo gene flanked by Frt or loxP sites, PL451 and PL452 (4), respectively. Using SW105 or SW106 it is now possible to introduce a point mutation or an informative restriction site into a BAC, retrieve a fragment from this BAC containing the introduced mutation(s) into a plasmid backbone using gap repair, and turn the retrieved fragment into a conditional targeting vector—all using only one bacterial strain, and only a single initial BAC transformation.
DISCUSSION
Here, we describe a new recombineering-based E.coli BAC modification system that makes use of galK positive selection for growth on galactose minimal medium and galK negative selection (counterselection) for growth on DOG. This modification system has several advantages compared with the other related BAC modification systems. First, the galK selection cassette is small (1231 bp + homology arms) compared with, for example, the sacB–neo cassette (3 kb), making PCR amplification and transformation into bacteria easier. Two homology arms are easily added to the galK cassette by including these sequences in the 5′ ends of the primers used for amplification (see Materials and Methods). Second, since galK is used for both selection steps, mutations occurring in galK during PCR amplification will be selected against during positive selection, significantly reducing the risk of DOG resistance from PCR mutations in galK during negative selection. Third, even when very short (33 bp) homology arms were used, the frequency of recombinants among the analyzed DOG-resistant clones was still 20–50%. To our knowledge, successful BAC modification with such short homology arms has never been reported. Finally, since galK recombineering is so efficient, little screening is required following selection, reducing the overall hands-on-time to a minimum. galK selection requires growth on minimal medium in both selection steps; although perhaps not used routinely in most molecular biology laboratories where BAC modification is needed, these plates are fairly simple to make and should not prevent anyone from using this method.
For all such counterselection schemes there will be background since during negative selection any event leading to the loss of the counterselectable marker will result in survival. In our system, virtually all background appears to result from deletions that span the inserted galK gene. These deletions likely occur during BAC replication in the 4.5 h outgrowth phase, since we have shown that these deletions occur independently of Red induction and in the absence or presence of oligo. It is well established that in recA defective bacteria, BACs are very stable compared with other large insert vectors, such as yeast artificial chromosomes (YACs). However, using counterselection, rare spontaneous deletions are seen because of the strong selection force. This spontaneous deletion background is not a problem, however, due to the high frequency of homologous recombination obtained with the defective λ prophage system.
Increasing the length of homology arms used for recombineering will reduce the relative number of background deletions observed following negative selection since the percentage of colonies containing BACs that eliminated galK by homologous recombination will be increased. Oligos with longer homology arms can be produced by annealing two oligos with overlapping 3′ ends (21). These overlapping oligos (5′ single strand overhangs) are then filled in by DNA polymerase in vivo (or in vitro) and subsequently serve as the substrate for the λ Red proteins, exo and bet. Longer homology arms can also be added by traditional cloning using restriction enzymes and DNA ligase (4).
A number of uses can be imagined for the strains described here. In principle, our strains can be used to make virtually any kind of BAC modification one can imagine, including point mutations or clean deletions or insertions of everything from cDNA to loxP sites or small epitope tags. BAC trimming, the specific deletion of BAC DNA flanking a gene of interest could also be used to remove genes from BACs prior to making BAC transgenic mice so that only the gene of interest on the BAC is analyzed. This should be of particular interest for BAC rescue experiments in positional cloning. Of course, trimming and modification can be combined, since the galK selection cassette can be recycled so that the same BAC can be modified several times. BAC modification could also be the starting point for constructing a gene-targeting vector to allow for more sophisticated gene targeting in mouse ES cells. The desired mutation(s) could first be created in the BAC followed by retrieval into a plasmid vector containing a negative-selection marker like thymidine kinase using gap repair (4). Finally, a neo marker could be introduced to allow for positive selection in ES cells. Alternatively, the retrieved fragment could be modified so that the end result is a conditional targeting vector. Strain SW106, which in addition to galK selection, can be induced to express Cre recombinase, makes it possible to perform all of the steps needed to construct a conditional targeting vector starting with only a single BAC transformation step.
Furthermore, in experiments where BAC transgenic mice are used to analyze the effect of deleting long-range regulators of gene expression, galK DOG counterselection can even be used, in the absence of an oligo, to generate a series of deletions around the galK insertion site, and the effect on gene regulation studied.
The Flp or Cre ORFs contained in strains SW105 and SW106, respectively, can also be replaced for any desired gene, thus creating tight arabinose-inducible expression for any gene of interest. This is done by first replacing the Cre/Flpe ORF with galK and select for Gal+ recombinants. The galK cassette is then replaced by the ORF of the gene of interest by DOG selection for Gal− recombinants.
Finally, the strains described here should be very useful for genetic manipulation not only of BACs, but also the E.coli genome itself.
All recombineering reagents discussed in this work are freely available upon request. To obtain these materials, please follow the directions listed on our website (http://recombineering.ncifcrf.gov). Detailed protocols for recombineering, including galK selection, can also be downloaded directly from our website.
Acknowledgments
We thank Allen Kane and Carolyn Whistler from Scientific Publications, Graphics & Media, NCI-Frederick, for help with illustrations and figure preparation. Funding for this research as well as the Open Access publication charges was provided by DHHS, NIH, NCI.
REFERENCES
- 1.Shizuya H., Birren B., Kim U.J., Mancino V., Slepak T., Tachiiri Y., Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl Acad. Sci. USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X.W., Model P., Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat. Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 3.Antoch M.P., Song E.J., Chang A.M., Vitaterna M.H., Zhao Y., Wilsbacher L.D., Sangoram A.M., King D.P., Pinto L.H., Takahashi J.S. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu P., Jenkins N.A., Copeland N.G. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotta-de-Almeida V., Schonhoff S., Shibata T., Leiter A., Snapper S.B. A new method for rapidly generating gene-targeting vectors by engineering BACs through homologous recombination in bacteria. Genome Res. 2003;13:2190–2194. doi: 10.1101/gr.1356503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copeland N.G., Jenkins N.A., Court D.L. Recombineering: a powerful new tool for mouse functional genomics. Nature Rev. Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 7.Court D.L., Sawitzke J.A., Thomason L.C. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Buchholz F., Muyrers J.P., Stewart A.F. A new logic for DNA engineering using recombination in Escherichia coli. Nature Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 9.Muyrers J.P., Zhang Y., Testa G., Stewart A.F. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu D., Ellis H.M., Lee E.C., Jenkins N.A., Copeland N.G., Court D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee E.C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D.A., Court D.L., Jenkins N.A., Copeland N.G. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 12.Gong S., Yang X.W., Li C., Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamsai D., Orford M., Nefedov M., Fucharoen S., Williamson R., Ioannou P.A. Targeted modification of a human beta-globin locus BAC clone using GET Recombination and an I-SceI counterselection cassette. Genomics. 2003;82:68–77. doi: 10.1016/s0888-7543(03)00100-9. [DOI] [PubMed] [Google Scholar]
- 14.Swaminathan S., Ellis H.M., Waters L.S., Yu D., Lee E.C., Court D.L., Sharan S.K. Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis. 2001;29:14–21. doi: 10.1002/1526-968x(200101)29:1<14::aid-gene1001>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 15.Alper M.D., Ames B.N. Positive selection of mutants with deletions of the gal-chl region of the Salmonella chromosome as a screening procedure for mutagens that cause deletions. J. Bacteriol. 1975;121:259–266. doi: 10.1128/jb.121.1.259-266.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons, Hoboken, NJ: Greene Publishing Associates and Wiley-Interscience; 1987. [Google Scholar]
- 17.Bachmann B.J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F.C., Curtiss R. III, Ingraham J.L., Lin E.C.C., Low K.B., Magasanik B., Reznikoff W.S., Riley M., Schaechter M., Umbarger H.E., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Vol. 2. Washington, DC: ASM press; 1996. pp. 2460–2488. [Google Scholar]
- 18.Grant S.G., Jessee J., Bloom F.R., Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl Acad. Sci. USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oppenheim A.B., Rattray A.J., Bubunenko M., Thomason L.C., Court D.L. In vivo recombineering of bacteriophage lambda by PCR fragments and single-strand oligonucleotides. Virology. 2004;319:185–189. doi: 10.1016/j.virol.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Hoess R.H., Wierzbicki A., Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu D., Sawitzke J.A., Ellis H., Court D.L. Recombineering with overlapping single-stranded DNA oligonucleotides: testing a recombination intermediate. Proc. Natl Acad. Sci. USA. 2003;100:7207–7212. doi: 10.1073/pnas.1232375100. [DOI] [PMC free article] [PubMed] [Google Scholar]