Abstract
The completion of the Human Genome Project provides researchers with a reference sequence that covers about 99% of the gene-containing regions and is more than 99.9% accurate. Sequence drafts and completed sequences for several other species are also available to researchers worldwide. The ongoing effort to provide more and more genomic reference information now enables the detection of deviations from this ‘genetic blueprint’. Comparative sequencing projects will play a major role in elucidating the meaning of the genetic code and in establishing a correlation between genotype and phenotype. As part of this effort, a number of projects will focus on distinct functional aspects, like resequencing of exons or HLA determining regions. Typically these target regions are short in length and their analysis does not require long read length. To find an efficient solution for these applications, we developed a novel method that allows simultaneous analysis of multiple independent target regions (Multiplexed Comparative Sequence Analysis) by employing base-specific cleavage biochemistry and MALDI TOF-MS analysis.
INTRODUCTION
We recently introduced a novel technique for comparative sequence analysis that has shown a high specificity and sensitivity for the discovery of SNPs. It utilizes base-specific cleavage of single-stranded nucleic acids and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis (1). The assay starts with PCR amplification of the target region using PCR primers that carry a T7-specific promoter sequence at the 5′ end of either the forward or reverse gene-specific primer. Subsequently, a single-stranded RNA molecule is generated by in vitro transcription of the PCR product. The derived single-stranded RNA is then cleaved to completion at base-specific positions in the sequence. A combination of PCR primer tags and cleavage schemes allows for cleavage after each of the four bases. The developed process is homogeneous and does not require purification of the PCR product or the cleavage product and thus is very amenable to high-throughput (2).
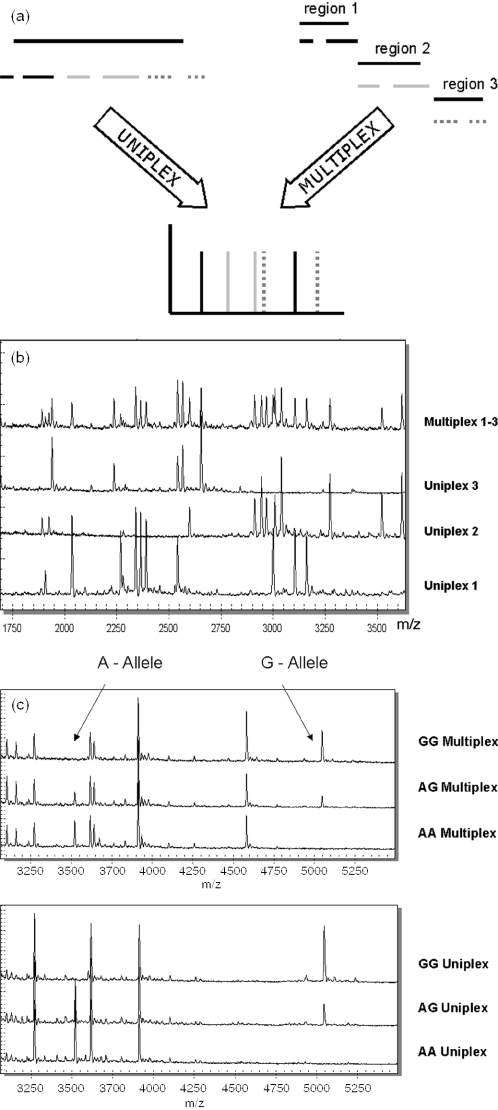
Base-specific cleavage generates a defined experimental mass signal pattern where each mass signal represents at least one fragment evolved from the target sequence. For analysis, this experimental pattern is subsequently compared to an in silico reference mass signal pattern derived from a reference sequence. Differences between the expected and the observed mass signal pattern are interpreted and enable identification of sequence variations. In brief, all unexpected additional signals are collected and their nucleotide compositions are calculated from the detected mass. Then it is calculated from the aggregate of all four-cleavage reactions, which sequence change can account for the observed mass signal changes (3). This procedure involves only part of the reference sequence, namely the affected cleavage product, and not the complete sequence. Hence, one can perform this process at multiple sequences at the same time, provided that the rendered fragments can be mapped to their original sequence. Together with a biochemistry that supports parallel processing of multiple sequences and an analyzer that allows for simultaneous data acquisition, this led us to explore base-specific cleavage/MALDI-TOF MS as a method for multiplexed discovery of sequence polymorphisms (see Figure 1a for visualization of the principle).
Figure 1.
(a) Schematic representation of multiplexed sequence polymorphism discovery enabled through base-specific cleavage and MALDI-TOF MS. Single-stranded RNA molecules are generated from the PCR product and are then cleaved to completion base-specifically. Cleavage products are analyzed in parallel with MALDI-TOF MS. The cleavage products are independent of each other and hence can be generated from one or multiple templates. (b) Mass spectra of the base-specific cleavage reaction for the three uniplex reactions tested compared with the mass spectrum derived from triplexing the same amplicons. All mass signals present in the uniplex reaction are also present in the multiplexed reaction with a homogeneous distribution of signal intensities. (c) Verification of the discovery of an A/G polymorphism in multiplexed base-specific cleavage. The lower set of three mass spectra corresponds to the three possible genotypes for the polymorphism. The upper panel shows the mass spectra of the triplexed reaction for the same polymorphism. The mass signals leading to the discovery of the A/G polymorphism are clearly identified in the multiplexed reaction.
MATERIALS AND METHODS
PCR and in vitro transcription
The target regions were PCR-amplified from human genomic DNA using primers that incorporate the T7 [5′-CAG TAA TAC GAC TCA CTA TAG GGA GA] promoter sequence. For each target region, two sets of primers were designed to incorporate the T7 promoter sequence either to the forward or to the reverse strand. The following PCR primers were used for uniplex and multiplex reactions. Primer sequences are provided with the T7 promoter tag:
MP1_T7_FOR CAGTAATACGACTCACTATAGGGAGAAGGCTGAGCTATTGCGAGAATAAGGAGATG
MP1_10_REV AGGAAGAGAGCGTGTTTGCTGTGCTTGATTG
MP1_T7_REV CAGTAATACGACTCACTATAGGGAGAAGGCTCGTGTTTGCTGTGCTTGATTG
MP1_10_FOR AGGAAGAGAGGAGCTATTGCGAGAATAAGGAGATG
MP2_T7_FOR CAGTAATACGACTCACTATAGGGAGAAGGCTCAAAATAACCAACAACCTCTTCCAG
MP2_10_REV AGGAAGAGAGGCAGAGCTCACAAGGATGGTTAC
MP2_T7_REV CAGTAATACGACTCACTATAGGGAGAAGGCTGCAGAGCTCACAAGGATGGTTAC
MP2_10_FOR AGGAAGAGAGCAAAATAACCAACAACCTCTTCCAG
MP3_T7_FOR CAGTAATACGACTCACTATAGGGAGAAGGCTGAAGCTCAAGTTTAAAGAAGCGTTG
MP3_10_REV AGGAAGAGAGAGCTGATTCCCCTTCAAGACTATTT
MP3_T7_REV CAGTAATACGACTCACTATAGGGAGAAGGCTAGCTGATTCCCCTTCAAGACTATTT
MP3_10_FOR AGGAAGAGAGGAAGCTCAAGTTTAAAGAAGCGTTG
The following PCR primer pairs were used for the CFTR multiplex of exon 10, 21 and 24:
CFTR_ex10_T7_FOR CAGTAATACGACTCACTATAGGGAGAAGGCTTCAGTTTTCCTGGATTATGC
CFTR_ex10_10MER_REV AGGAAGAGAGTTGGCATGCTTTGATGACGC
CFTR_ex10_T7_REV CAGTAATACGACTCACTATAGGGAGAAGGCTTTGGCATGCTTTGATGACGC
CFTR_ex10_10MER_FOR AGGAAGAGAGTCAGTTTTCCTGGATTATGC
CFTR_EX21_T7_FOR CAGTAATACGACTCACTATAGGGAGAAGGCTGAGGTTCATTTACGTCTTTTGTG
CFTR_EX21_10MER_REV AGGAAGAGAGCATAAAAGTTAAAAAGATGATAAGACTTAC
CFTR_EX21_T7_REV CAGTAATACGACTCACTATAGGGAGAAGGCTCATAAAAGTTAAAAAGATGATAAGACTTAC
CFTR_EX21_10MER_FOR AGGAAGAGAGGAGGTTCATTTACGTCTTTTGTG
CFTR_ex24_T7_FOR CAGTAATACGACTCACTATAGGGAGAAGGCTTTTCTTCTTCTTTTCTTTTTTGCTATAG
CFTR_ex24_10MER_REV AGGAAGAGAGCCCTTTCAAAATCATTTCAGTTA
CFTR_ex24_T7_REV CAGTAATACGACTCACTATAGGGAGAAGGCTCCCTTTCAAAATCATTTCAGTTA
CFTR_ex24_10MER_FOR AGGAAGAGAGTTTCTTCTTCTTTTCTTTTTTGCTATAG
The PCR reactions were carried out in a total volume of 5 μl using 1 pmol of each primer, 40 μM dNTP, 0.1 U Hot Star Taq DNA polymerase (Qiagen), 1.5 mM MgCl2 and buffer supplied with the enzyme (final concentration 1×). The reaction mix was pre-activated for 15 min at 95°C. The reactions were amplified in 45 cycles of 95°C for 20 s, 62°C for 30 s and 72°C for 30 s followed by 72°C for 3 min. Unincorporated dNTPs were dephosphorylated by adding 1.7 μl H2O and 0.3 U Shrimp Alkaline Phosphatase. The reaction was incubated at 37°C for 20 min.
Typically, 2 μl of the PCR reaction was directly used as template in a 4-μl transcription reaction. Twenty units of T7 R&DNA polymerase (Epicentre, Madison, WI) were used to incorporate either dCTP or dTTP in the transcripts. Ribonucleotides were used at 1 mM and the dNTP substrate at 2.5 mM; other components in the reaction were as recommended by the supplier. Following the in vitro transcription, RNase was added to cleave the in vitro transcript. The mixture was then further diluted with H2O to a final volume of 27 μl. Conditioning of the phosphate backbone prior to MALDI-TOF MS was achieved by the addition of 6 mg CLEAN Resin (Sequenom Inc., San Diego, CA). Further experimental details have been described elsewhere (1,2).
Mass spectrometry measurements
Fifteen nanoliters of the cleavage reaction was robotically dispensed onto silicon chips preloaded with matrix (SpectroCHIP® bioarrays; Sequenom Inc., San Diego, CA). Mass spectra were collected using a MassARRAY™ system mass spectrometer (SEQUENOM).
SNP database entries
| Amplicon MP1 | rs18120224 | Substitution A/G at position 27 |
| rs5995250 | Substitution A/G at position 116 | |
| rs5995251 | Substitution A/T at position 136 | |
| rs6000189 | Substitution A/G at position 194 | |
| rs6000190 | Substitution A/G at position 221 | |
| Amplicon MP2 | rs2073989 | Substitution C/T at position 115 |
| rs178290 | Substitution A/G at position 136 | |
| Amplicon MP3 | rs710192 | Substitution C/T at position 36 |
RESULTS
We approached the realization of multiplexing two or more target regions by considering three main aspects. First, the combinatorial foundation of base-specific cleavage/MALDI-TOF MS; second, aspects of molecular biology/biochemistry for multiplexing PCR and base-specific cleavage; third, mass spectrometry related challenges.
To define the combinatorial foundation of multiplexed sequence analysis by base-specific cleavage/MS, we looked at the relationship between mass signals, nucleotide composition, signal density and detection sensitivity. The mass of each cleavage product is defined by its nucleotide composition (or compomer), but not the order of nucleotides. All fragments with identical nucleotide composition have identical mass and, hence, will result in the same mass signal. For example, a cleavage product consisting of two adenines and one guanine corresponds to three fragment sequences (AAG, AGA and GAA) but results in the same mass signal. A longer target sequence generates more cleavage products and thus the ‘density’ of signals within a spectrum increases. This results in a higher probability that two or more fragments show identical or indistinguishable masses. Consequently, the density of mass signals has a great impact on the ability to detect a sequence change. The higher the signal density in a spectrum, the higher the chance that a new signal overlaps with an existing signal and its detection is compromised. The amplicon lengths used in our standard uniplex resequencing reactions vary between 50 and 1000 nt. For this amplicon, length detection rates between 95% and 100% are achieved when all ‘theoretically possible’ SNPs (see below) are considered. These detection rates should remain unchanged when multiplexing amplicons, as long as the overall nucleotide count is similar and as long as the amplicons do not contain repeat sequences.
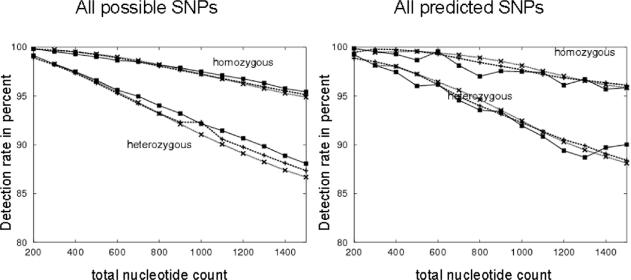
To further clarify the influence of multiplexing on SNP detection rates, we performed in silico simulations. We obtained all exonic sequences from the human Ensembl database (34b) including UTR regions, plus 20 nt on either side to account for primer design. In total, we used 241 297 sequences with 62 million overall nucleotide count. For a uniplex sequence, we simulated all aspects of the four utilized cleavage reactions described earlier (1). We used the resulting simulated mass spectra as a reference, and for every SNP under consideration, we compared the resulting mass spectra to these references. This comparison took into account the resolution of the mass spectrometer, peak separation and several other aspects. For heterozygous analysis, we defined that a SNP can be detected if we find at least one new mass signal in one of the corresponding mass spectra, which is not ‘silenced’ by a close peak in the reference mass spectrum. For homozygous analysis, we also used those mass signals that are present in a reference mass spectrum but missing in the SNPs mass spectrum. The complete details of the simulation will be reported elsewhere (S. Böcker, manuscript in preparation).
Next, we simulated SNP detection rates for 3-plex and 5-plex reactions. We randomly chose 3 (or 5) amplicons such that the sum of lengths equals the desired total length of 100–1500 nt. For every parameter set, 100 000 multiplexes were drawn uniformly from the set of all possible multiplexes of the desired length. Reference mass spectra for all 3 or 5 multiplexed sequences were generated and combined, and SNP mass spectra were compared to these references. Simulations were performed for all SNPs, and for all known SNPs. With respect to this simulation, known SNPs are defined as those SNPs (validated as well as predicted) found in the Ensembl database. All SNPs, on the other hand, means that at every position of the reference sequence (excluding the primer region of 20 nt at the ends) we substituted, inserted or deleted a base if possible. Those insertions and deletions that lead to the same sequence (e.g. inserting an A into the sequence TAAC after positions 1, 2 or 3 all lead to the same sequence TAAAC) were only counted once. Hence, we find three substitutions, and at most three insertions and one deletion per position.
The simulation results demonstrate that multiplexing does not significantly decrease detection rates (Figure 2). Furthermore, we have seen that by optimizing the choice of sequences that are multiplexed, we can significantly increase SNP detection rates (data not shown). The uniplexing assays show a noticeable fluctuation of the discovery rates for known SNPs. To calculate the uniplex discovery rates for length 1 bp, we filter all exons and extract those with length l ± 20 bp. The number of exons that pass this test and contain one or more predicted SNPs is usually very small. The fluctuation thus represents solely a statistical artifact that would be eliminated if we choose a larger set of analyzed amplicons. For triplexes and pentaplexes, we draw three or five sequences from the pool of potential sequences. These numbers are much higher compared to the number of potential sequences itself and the corresponding discovery rates of multiplexes exhibit a smoother curve.
Figure 2.
Simulation results for predicted SNPs (‘known’ SNPs from ensembl database) and all theoretically possible SNPs using the reference sequence for all exons of Ensembl (build 34b). The y-axis provides the percentage of sequence change events that can be detected with base-specific cleavage (using four cleavage reactions). The x-axis provides the simulation results for overall amplicon length 200 up to 1.5 kb. Solid lines represent uniplexing results, dashed lines represent triplexed re-sequencing, dotted lines pentaplexed re-sequencing.
The experimental implementation of multiplexed sequence analysis requires that two or more target regions are generated and analyzed concurrently. Simultaneous amplification of multiple genetic loci in a multiplexed PCR reaction has been used broadly for high-throughput genotyping, especially at the plexing level considered here (3- to 5-plex). We thus did not investigate sophisticated PCR amplification strategies in this framework. The post-PCR process consists of RNA transcription and RNase cleavage. Both processes are ideally suited for multiplexing. Each amplicon is tagged with the same promoter sequence during PCR. This should lead to uniform generation of transcription-initiation complexes with the T7 RNA polymerase, and thus avoids complications related to primer extension methods, such as different primer-template kinetics, formation of primer hairpins and primer cross-interaction. Lastly, the use of RNAse A guarantees specificity to cleavage bases C and/or U. Performing the cleavage reaction to completion assures that each cleavage product is untied from the surrounding sequence and can be studied independent of its origin. Finally, the cleavage products are detected by MALDI-TOF MS, which allows simultaneous data acquisition for all generated cleavage products (see Figure 1a and b).
The practical implementation of multiplex sequence analysis is also determined by the mass spectra quality. Because the discovery of SNPs requires a detectable difference between predicted and observed mass signal patterns, any arbitrarily created mass signal change could compromise the detection accuracy. Non-predicted additional signals can occur from unspecific amplification during multiplex PCR amplification, primer dimers, unpredictable transcription termination or unspecific cleavage. Issues related to misamplification, primer dimers and primer cross-talk can nowadays be minimized using a variety of bioinformatic tools. The availability of genome sequences can assist greatly in assuring that primers hit the genome at a specific site and only once. These in silico tools provided, we evaluated the robustness of multiplex hMC reactions with a focus on the end point, the mass spectra quality. The spectra quality is characterized by how well the observed mass signal pattern fits the expected pattern. The number of additional signals, missing signals as well as several recalibration parameters (e.g. average observed mass delta or average observed intensity delta) can be summarized in a single confidence score. This confidence score represents how confident an algorithm can fit the observed mass spectrum to the expected. It ranges from 0 (no confidence) to 5 (high confidence) and is a basic element of the real time quality judgment used in our current analysis software (Discovery RT). Under the assumption of similar mass spectra quality, the accuracy/sensitivity of detecting a sequence change is mainly a function of the overall nucleotide count. This is demonstrated in our simulations.
To experimentally evaluate the performance of simultaneous discovery of sequence polymorphisms in multiple target regions, we selected a set of three amplicons. All amplicons are located on the long arm of chromosome 22 and ranged from 140 to 250 bp in length. We have previously analyzed these target regions using the MassARRAY™ system for SNP Discovery as well as Sanger sequencing. This process led to the discovery of 8 SNPs (the corresponding rs numbers are provided in the Methods). One amplicon carried five, one amplicon carried two and one amplicon carried one polymorphic site. All discovered sequence variations were validated using a primer extension genotyping assay and MALDI-TOF analysis (MassEXTEND® method) (4). Optimal multiplex PCR conditions were assured by adjusting all amplification primers to uniform PCR conditions using Primer3. Each individual DNA was analyzed in one multiplex and three uniplex reactions.
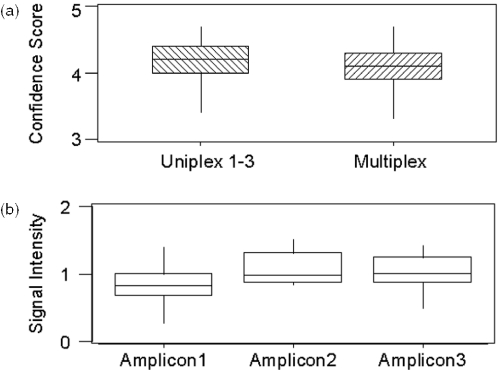
We mentioned earlier that the theoretical aspects of multiplexing several target regions can be assessed upfront in silico. The mass spectra quality is one remaining key component for the discovery of sequence polymorphisms. Correspondingly, we first evaluated whether multiplex base-specific cleavage reactions yield equivalent quality when the spectra quality of uniplex and multiplex reactions are compared. To generate sufficient data, we processed 12 DNA samples in triplicate with 4 cleavage reactions for each uniplexed amplicon and the triplexed design. We based the spectra quality comparison on the confidence scores explained above. Given this experimental setup, our statistical analysis of the spectra quality was based on 144 mass spectra for each of the uniplex reactions (yielding a combined total of 432 mass spectra) and 144 spectra for the multiplex reactions. The comparison revealed no significant difference in the confidence scores for the set of uniplex reaction and the multiplexed reaction employed (ANOVA, p = 0.71; see Figure 3a).
Figure 3.
Comparison of spectra quality based on (a) confidence scores in uniplex and triplex reactions and (b) signal intensities for each individual amplicon derived from the triplexed reactions. The distribution of confidence scores and signal intensities are presented as box and whisker plots. The box extends from the first to the third quartile (interquartile range). The line inside the box marks the median and the lines extending out on either side indicate the minimum and maximum values.
We were further interested in the distribution of signal intensities between cleavage products originating from different amplicons. This aspect is of importance as the dynamic range of a MALDI-TOF MS spectrum is limited to about two logs. Although this gives considerably more flexibility than a standard capillary sequencer, one still has to assure that target regions amplify with similar efficiency. To assess this aspect, we collected and grouped multiple signals by their original target region. We then compared the signal intensities to explore if one group is significantly different from the rest. For this analysis, we collected five representative mass signals from each target region in the triplex reaction. Based on the experimental setup described above, this led to 720 observations (144 mass spectra × 5). A statistical analysis of the results revealed that there is no significant difference between the three groups of signal intensities (ANOVA, p = 0.11; Figure 3b). An overlay of three spectra representing uniplex reactions with one spectrum derived from the multiplexed reaction illustrates this (Figure 1b).
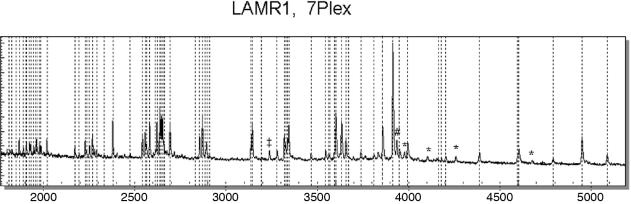
Figure 1c shows that all polymorphisms initially discovered in the uniplexed base-specific cleavage reactions have also been detected automatically in the multiplexed reaction. We cannot derive the optimal multiplexing level from these initial experiments. It is, however, worth mentioning that we routinely perform 3- to 5-plexes with an overall nucleotide count from 300 to 1000 nt. We have also performed 7-plex reactions covering the complete coding region of the laminin receptor gene (LAMR, overall nucleotide count 1800 nt) and, on a more experimental basis, we analyzed the complete coding region for the SURFB gene (10-plex) in a single reaction (overall nucleotide count 1850 nt). Figure 4 depicts representative results obtained with the LAMR 7-plex SNP Discovery assay. All expected cleavage products are indicated by dashed lines. Mass signals marked with footnotes represent artifacts not related to sequence changes and can be explained either by transcription from primer dimer formation (*), by abortive cycling products (‡) (2) or by salt adducts (#), in particular sodium adducts.
Figure 4.
Representative mass spectrum of a heptaplexed base-specific cleavage reaction of discovery of sequence polymorphism in the LAMR1 gene. The overall nucleotide count in this multiplex totaled to 1850 bases. All expected mass signals are marked with a dashed line. The mass signal labeled ‘*’ represents an additional signal derived from primer dimer formation, the signal labeled ‘‡’ represents an abortive cycling product and the signal labeled ‘#’ represents a sodium adduct of the cleavage product GGGAGAAGGCT at 3913 Da (+22 Da).
To evaluate situations where only a limited number of target regions are available for multiplexing, we used the CFTR gene as a model system. A restriction of primer design to defined number and length of target regions does not always allow optimal combination of primer pairs. We created a triplex assay, which allows analysis of the most relevant CFTR mutation, ΔF508, in combination with other mutations occurring in exon 10, exon 21 and exon 24.
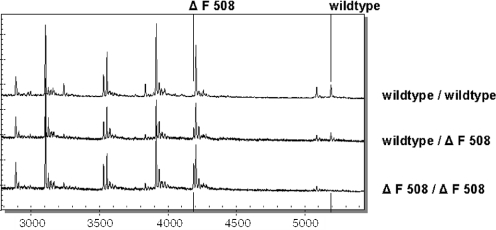
Figure 5 shows a mass spectrum of the T-cleavage reaction of the reverse strand for the described triplex. The mass spectra of three different individuals with varying ΔF508 genotype are overlaid. Mass signals indicating the presence of ΔF508 are labeled above and below the spectrum.
Figure 5.
Overlay of three mass spectra showing the ΔF508-specific mass signal changes generated in the T-specific cleavage reaction of the reverse strands. The mass signal pattern is derived from a triplexed reaction encompassing exon 10, 21 and 24 of the CFTR gene. Three different individuals were analyzed, the upper mass spectrum representing a homozygous wild-type for ΔF508, a carrier of the ΔF508 mutation (middle) and an individual homoyzgous mutant ΔF508 (lower mass spectrum). Mass signals specific for the mutation are indicated with a line.
DISCUSSION
Our experiments have demonstrated that multiplexed discovery of sequence polymorphisms and mutations can be achieved when base-specific cleavage and MALDI-TOF MS analysis are employed. The method provides very fast and accurate detection of sequence variations. In the multiplex reactions tested here, we did not observe performance limitations when compared to uniplex reactions. However, the balanced amplification of individual PCRs in the multiplex is potentially a critical factor. Potential solutions for multiplexes showing unbalanced amplification can be either re-design of PCR primers, re-plexing of amplicons or, in case of limited flexibility, PCR primer amounts can be adjusted.
To date chain terminator sequencing is without doubt the most common method for de novo and comparative sequencing. The method relies on the subsequent readout of nucleic bases in any form of space separated extension terminated products. Given the current restrictions in fluorescent labels suitable for laser readout, it is almost impossible to achieve simultaneous analysis of generated sequencing fragments from different origins, unless more complicated schemes like nylon blotting are performed (5–7). Resequencing of one target region will always require one complete Sanger sequencing reaction, independent of the target length. For very long read length, this is still the gold standard, but for the analysis of shorter target regions the possibilities to reduce cost and save time are very limited. We feel that a method that allows multiplexed PCR sequencing in a high-throughput fashion can be a highly valuable asset in several research activities. The human genome, for example, still contains multiple SNP deserts. The current effort to evaluate the degree of linkage disequilibrium in the human genome and to establish a map of haplotype tag SNPs will most likely require to fill these deserts with SNPs at a reasonable density. We see further relevance of our method in resequencing strategies that focus on the analysis of multiple short target regions like the coding sections of the mammalian genome. Exons are typically below 200 bp in length. Using base-specific cleavage, target regions of this length can easily be analyzed in a 3- to 5-plex with a detection accuracy of 95% or greater. Hence, base-specific cleavage allows reducing the cost 3- to 5-fold by using multiplexed reactions.
In the post-genomic era, where diagnostic sequencing and variant sequencing will increasingly replace de novo sequencing in most laboratories, many more applications will benefit from the opportunity to perform multiplex comparative sequencing assays. The initial adaptation of this method to multiplexed mutation analysis of CFTR exons is a first step towards this goal. Further assay optimization and improved data analysis algorithms have to be developed alongside for this method to become routine use. We currently also evaluate the applicability of multiplexed base-specific cleavage for the combined analysis of marker regions in pathogen identification and resistance typing and see also great potential in multiplexed analysis of CpG islands using base-specific cleavage.
Acknowledgments
We thank Matthew R. Nelson for critical reading of the manuscript and helpful suggestions. Funding to pay the Open Access publication charges for this article was provided by SEQUENOM Inc., San Diego.
REFERENCES
- 1.Stanssens P., Zabeau M., Meersseman G., Remes G., Gansemans Y., Storm N., Hartmer R., Honisch C., Rodi C.P., Böcker S., van den Boom D. High-throughput MALDI-TOF discovery of genomic sequence polymorphisms. Genome Res. 2004;14:126–133. doi: 10.1101/gr.1692304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartmer R., Storm N., Böcker S., Rodi C.P., Hillenkamp F., Jurinke C., van den Boom D. RNase T1 mediated base-specific cleavage and MALDI-TOF MS for high-throughput comparative sequence analysis. Nucleic Acids Res. 2003;31:e47. doi: 10.1093/nar/gng047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böcker S. SNP and mutation discovery using base-specific cleavage and MALDI-TOF mass spectrometry. Bioinformatics. 2003;19(Suppl. 1):I44–I53. doi: 10.1093/bioinformatics/btg1004. [DOI] [PubMed] [Google Scholar]
- 4.Storm N., Darnhofer-Patel B., van den Boom D., Rodi C.P. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol. Biol. 2003;212:241–262. doi: 10.1385/1-59259-327-5:241. [DOI] [PubMed] [Google Scholar]
- 5.Chee M. Enzymatic multiplex DNA sequencing. Nucleic Acids Res. 1991;19:3301–3305. doi: 10.1093/nar/19.12.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olesen C.E., Martin C.S., Bronstein I. Chemiluminescent DNA sequencing with multiplex labeling. Biotechniques. 1993;15:480–485. [PubMed] [Google Scholar]
- 7.Cherry J.L., Young H., Di Sera L.J., Ferguson F.M., Kimball A.W., Dunn D.M., Gesteland R.F., Weiss R.B. Enzyme-linked fluorescent detection for automated multiplex DNA sequencing. Genomics. 1994;20:68–74. doi: 10.1006/geno.1994.1128. [DOI] [PubMed] [Google Scholar]