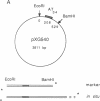
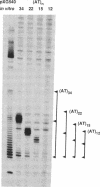
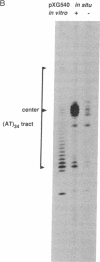
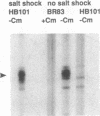
Abstract
Superhelical tension of DNA in living bacteria is believed to be partially constrained by interaction with proteins. Yet DNA topology is a significant factor in a number of genetic functions and is apparently affected by both genetic and environmental influences. We have employed a technique that allows us to estimate the level of unconstrained superhelical tension inside the cell. We study the formation of cruciform structures by alternating adenine-thymine sequences in plasmid DNA by in situ chemical probing. This structural transition is driven by superhelical torsion in the DNA and thus reports directly on the level of such tension in the cellular DNA. We observe that the effect of osmotic shock is an elevation of superhelical tension; quantitative comparison with changes in plasmid linking number indicates that the alteration in DNA topology is all unconstrained. We also show that the synthesis of defective topoisomerase leads to increased superhelical tension in plasmid DNA. These experiments demonstrate that the effect of environmental and genetic influences is felt directly at the level of torsional stress in the cellular DNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bliska J. B., Cozzarelli N. R. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987 Mar 20;194(2):205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Boublíková P., Palecek E. Probing of B-Z junctions in recombinant plasmids in vitro and in the cell with different osmium tetroxide complexes. Gen Physiol Biophys. 1989 Oct;8(5):475–490. [PubMed] [Google Scholar]
- Brill S. J., Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988 Jul 29;54(3):403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K. A., Sternglanz R., Reynolds A. E., Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982 Nov;31(1):43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- Dorman C. J., Lynch A. S., Ni Bhriain N., Higgins C. F. DNA supercoiling in Escherichia coli: topA mutations can be suppressed by DNA amplifications involving the tolC locus. Mol Microbiol. 1989 Apr;3(4):531–540. doi: 10.1111/j.1365-2958.1989.tb00199.x. [DOI] [PubMed] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa N., Bossi L. Transcription induces gyration of the DNA template in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9416–9420. doi: 10.1073/pnas.85.24.9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong J. C., Lilley D. M. Highly selective chemical modification of cruciform loops by diethyl pyrocarbonate. Nucleic Acids Res. 1986 May 27;14(10):3995–4007. doi: 10.1093/nar/14.10.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Ohmori H., Tomizawa J. DNA gyrase and DNA supercoiling. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):35–40. doi: 10.1101/sqb.1979.043.01.007. [DOI] [PubMed] [Google Scholar]
- Gough G. W., Sullivan K. M., Lilley D. M. The structure of cruciforms in supercoiled DNA: probing the single-stranded character of nucleotide bases with bisulphite. EMBO J. 1986 Jan;5(1):191–196. doi: 10.1002/j.1460-2075.1986.tb04195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves D. R., Patient R. K., Lilley D. M. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J Mol Biol. 1985 Oct 5;185(3):461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- Greaves D. R., Patient R. K. RecBC, sbcB independent, (AT)n-mediated deletion of sequences flanking a Xenopus laevis beta globin gene on propagation in E. coli. Nucleic Acids Res. 1986 May 27;14(10):4147–4158. doi: 10.1093/nar/14.10.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. The in-vivo occurrence of Z DNA. J Biomol Struct Dyn. 1983 Dec;1(3):593–609. doi: 10.1080/07391102.1983.10507467. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Transition of a cloned d(AT)n-d(AT)n tract to a cruciform in vivo. Nucleic Acids Res. 1985 Jun 25;13(12):4343–4363. doi: 10.1093/nar/13.12.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W. Diethyl pyrocarbonate: a chemical probe for secondary structure in negatively supercoiled DNA. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8009–8013. doi: 10.1073/pnas.82.23.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., Dorman C. J., Stirling D. A., Waddell L., Booth I. R., May G., Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988 Feb 26;52(4):569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Horowitz D. S., Wang J. C. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984 Feb 15;173(1):75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- Horwitz M. S., Loeb L. A. An E. coli promoter that regulates transcription by DNA superhelix-induced cruciform extrusion. Science. 1988 Aug 5;241(4866):703–705. doi: 10.1126/science.2456617. [DOI] [PubMed] [Google Scholar]
- Jaworski A., Blaho J. A., Larson J. E., Shimizu M., Wells R. D. Tetracycline promoter mutations decrease non-B DNA structural transitions, negative linking differences and deletions in recombinant plasmids in Escherichia coli. J Mol Biol. 1989 Jun 5;207(3):513–526. doi: 10.1016/0022-2836(89)90461-0. [DOI] [PubMed] [Google Scholar]
- Jaworski A., Hsieh W. T., Blaho J. A., Larson J. E., Wells R. D. Left-handed DNA in vivo. Science. 1987 Nov 6;238(4828):773–777. doi: 10.1126/science.3313728. [DOI] [PubMed] [Google Scholar]
- Johnston B. H., Rich A. Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell. 1985 Oct;42(3):713–724. doi: 10.1016/0092-8674(85)90268-5. [DOI] [PubMed] [Google Scholar]
- Johnston B. H. The S1-sensitive form of d(C-T)n.d(A-G)n: chemical evidence for a three-stranded structure in plasmids. Science. 1988 Sep 30;241(4874):1800–1804. doi: 10.1126/science.2845572. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. In vivo consequences of plasmid topology. Nature. 1981 Jul 23;292(5821):380–382. doi: 10.1038/292380a0. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Palecek E. The supercoil-stabilised cruciform of ColE1 is hyper-reactive to osmium tetroxide. EMBO J. 1984 May;3(5):1187–1192. doi: 10.1002/j.1460-2075.1984.tb01949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. Structural perturbation in supercoiled DNA: hypersensitivity to modification by a single-strand-selective chemical reagent conferred by inverted repeat sequences. Nucleic Acids Res. 1983 May 25;11(10):3097–3112. doi: 10.1093/nar/11.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. Bacterial chromatin. A new twist to an old story. Nature. 1986 Mar 6;320(6057):14–15. doi: 10.1038/320014a0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J. A., Lilley D. M. A two-state conformational equilibrium for alternating (A-T)n sequences in negatively supercoiled DNA. J Mol Biol. 1987 Oct 20;197(4):707–721. doi: 10.1016/0022-2836(87)90477-3. [DOI] [PubMed] [Google Scholar]
- McClellan J. A., Palecek E., Lilley D. M. (A-T)n tracts embedded in random sequence DNA--formation of a structure which is chemically reactive and torsionally deformable. Nucleic Acids Res. 1986 Dec 9;14(23):9291–9309. doi: 10.1093/nar/14.23.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejedlý K., Kwinkowski M., Gałazka G., Kłysik J., Palecek E. Recognition of the structural distortions at the junctions between B and Z segments in negatively supercoiled DNA by osmium tetroxide. J Biomol Struct Dyn. 1985 Dec;3(3):467–478. doi: 10.1080/07391102.1985.10508435. [DOI] [PubMed] [Google Scholar]
- Palecek E., Boublíková P., Karlovský P. Osmium tetroxide recognized structural distortions at junctions between right- and left-handed DNA in a bacterial cell. Gen Physiol Biophys. 1987 Dec;6(6):593–608. [PubMed] [Google Scholar]
- Palecek E., Rasovská E., Boublíková P. Probing of DNA polymorphic structure in the cell with osmium tetroxide. Biochem Biophys Res Commun. 1988 Jan 29;150(2):731–738. doi: 10.1016/0006-291x(88)90452-4. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Fontaine A. A native cruciform DNA structure probed in bacteria by recombinant T7 endonuclease. J Biol Chem. 1987 Aug 15;262(23):11364–11368. [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985 Jan;40(1):129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- Pruss G. J., Drlica K. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8952–8956. doi: 10.1073/pnas.83.23.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss G. J., Manes S. H., Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982 Nov;31(1):35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmouni A. R., Wells R. D. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989 Oct 20;246(4928):358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- Richardson S. M., Higgins C. F., Lilley D. M. The genetic control of DNA supercoiling in Salmonella typhimurium. EMBO J. 1984 Aug;3(8):1745–1752. doi: 10.1002/j.1460-2075.1984.tb02041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholten P. M., Nordheim A. Diethyl pyrocarbonate: a chemical probe for DNA cruciforms. Nucleic Acids Res. 1986 May 27;14(10):3981–3993. doi: 10.1093/nar/14.10.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Yamamoto N., Droffner M. L. Mechanisms determining aerobic or anaerobic growth in the facultative anaerobe Salmonella typhimurium. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2077–2081. doi: 10.1073/pnas.82.7.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias W., Jaworski A., Larson J. E., Wells R. D. The B- to Z-DNA equilibrium in vivo is perturbed by biological processes. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7069–7073. doi: 10.1073/pnas.85.19.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]