Abstract
The existence of two sophisticated parallel splicing machineries in multicellular organisms has raised intriguing questions—ranging from their impact on proteome expansion to the evolution of splicing and of metazoan genomes. Exploring roles for the distinct splicing systems in vivo has, however, been restricted by the lack of techniques to selectively inhibit their function in cells. In this study, we show that morpholino oligomers complementary to the branch-site recognition elements of U2 or U12 small nuclear RNA specifically suppress the function of the two splicing systems in mammalian cells. The data provide the first evidence for a role of distinct spliceosomes in pre-mRNA splicing from endogenous mammalian genes and establish a tool to define roles for the different splicing machineries in vivo.
INTRODUCTION
Eukaryotic gene expression requires the removal of non-coding gene sequences (introns) in the course of mRNA processing. This splicing reaction is catalyzed by a complex machinery made up of small nuclear ribonucleoprotein particles (snRNPs) and a large number of non-snRNP proteins, the spliceosome [for a recent review see (1)]. snRNPs form the catalytic core of the spliceosome and recognize introns by short consensus sequences (splice sites) located at the intron ends, a process that involves base-pairing with complementary sequences in uridine-rich (U) snRNA molecules, the RNA component of the snRNPs. Among the five snRNPs (U1, U2, U4, U5 and U6), the U1 snRNP recognizes the 5′ splice site while the U2 snRNP binds to the branch point sequence, which is close to the 3′ boundary of the intron.
In the mid-1990s, non-canonical splice sites led to the discovery of a second spliceosome in multicellular organisms. In addition to the common U5 snRNP, this second spliceosome contains distinct, low-abundance snRNPs (U11, U12, U4atac and U6atac), which functionally correspond to the U1, U2, U4 and U6 snRNPs of the classical (major class or U2-type) splicing machinery [reviewed in (2)]. Although evolutionarily highly conserved, only a minute proportion of introns (e.g. 0.15–0.34% in primate and human genomes) have been predicted to be targets for this second (minor class or U12-type) spliceosome, based on the presence of the distinct and highly conserved 5′ splice site and branch-point sequences (3,4). The extremely small proportion of putative substrates and the conversion from U12-type to U2-type introns (3) have raised the question why a sophisticated parallel splicing apparatus has been conserved during evolution. One answer might be that the remaining U12-type introns have indispensable functions in metazoan cells, e.g. in the regulation of gene expression or in generating unique splice variants. Alternatively, or in addition, there could be a wider spectrum of action for minor spliceosomes or snRNPs than predicted. In fact, hints in favor of the latter possibility have been emerging (5,6).
An understanding of why and how metazoan genomes are interpreted by distinct splicing systems during transcription will require their selective inhibition in vivo. In this paper, we report on a technique to selectively ‘knockdown’ the function of the different splicing machineries in living cells, thereby permitting to analyze their role in mRNA processing from endogenous transcripts.
MATERIALS AND METHODS
Cell culture and transfections
EL4 cells were grown in DMEM and Jurkat cells in RPMI medium; both supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin.
On the day before transfection, cells were plated at a density of 8 × 105 cells ml−1 and 2–3 × 107 cells were used for electroporation. EL4 cells were washed once with PBS (with Ca2+/Mg2+) and resuspended in 350 μl PBS (with Ca2+/Mg2+)/2 mM 2-mercaptoethanol. Jurkat cells were directly resuspended in 350 μl of plain RPMI medium. After mixing the cells with the morpholino (and 20 μg of P120 minigene plasmid for co-transfections) in a 1.5 ml polypropylene reaction tube (Eppendorf) and incubation at room temperature for 5–10 min, they were transferred to a 4-mm-gap electroporation cuvette (Peqlab). Electroporation was performed using a Bio-Rad Gene Pulser at 250 μF and 320 V for EL4 cells or at 960 μF and 250 V for Jurkat cells. After electroporation, cells were plated in 10 ml growth medium. Electroporation conditions were determined empirically, based on the established protocols for the transfection of plasmid DNA into the two cell lines. From our experience with different lymphoma and leukemia cell lines (EL4, Jurkat and LB17), it appears that morpholino electroporation requires no special solutions or conditions as compared with plasmid electroporation; still the transfection efficiency of morpholinos is very high (80–95%, as determined by fluorescein-labeled morpholinos), even if the transfection of plasmid DNA is markedly less efficient under those conditions.
Morpholino sequences
u12MO, TCGTTATTTTCCTTACTCATAAGTT (complementary to U12 nucleotides 9–33); u2MO, TGATAAGAACAGATACTACACTTGA (complementary to U2 nucleotides 27–51); and cMO, CCTCTTACCTCAGTTACAATTTATA. Morpholinos were obtained from GeneTools, Inc. (Philomath, OR) and were dissolved in water at a concentration of 0.5 nmol μl−1.
RT–PCR analysis
Total RNA was prepared using guanidinium isothiocyanate/phenol (RNAPure; Peqlab). An aliquot of 0.8 μg of DNase-treated RNA was reverse transcribed (20 μl reactions) in MMLV-RT buffer (Promega) with 100 ng oligo-d(T)15 and 200 U MMLV-RNase-H minus reverse transcriptase (Promega) at 42°C for 45 min.
PCRs were performed in 50 μl GoTaq-reaction buffer (Promega) containing 5 μl of the diluted (1:10) reverse transcriptase reactions, 250 μM dNTP each, 10 pmol primers and 2.5 U GoTaq DNA polymerase (Promega). A total of 28 cycles were performed for the amplification of P120 minigene transcripts and 31 cycles for endogenous P120 and ADPRT transcripts. Each cycle consisted of 95°C for 1 min, 59°C (P120) or 58°C (ADPRT) for 1 min and 72°C for 1.5 min.
Primer sequences were as follows: P120, GAAAGAAGTGACCCCTGAGTCAGG and CAGATCCTTCTTGAGCCGGTTCAG (splicing of intron F), CTGAACCGGCTCAAGAAGGATCTG and GTCTCGGCGTCGGGTTTTCAAGG (splicing of intron G); ADPRT, GATCCTTCAGCTAACATTAGTCTG and GCTACCTCTCCCAATTACCACAG.
RESULTS
Antisense RNA oligonucleotides to spliceosomal snRNAs, e.g. masking those regions of U2 or U12 snRNAs complementary to the intron branch site, can selectively interfere with assembly of the two splicing machineries in in vitro splicing reactions using nuclear extracts and synthetic RNA substrates (7,8). For a strategy to selectively target spliceosomal snRNAs in living cells, we decided to use non-ionic antisense morpholino oligomers (9). Morpholinos bind with high specificity to transcripts and have been used to block splice sites in pre-mRNA (10,11) and mRNA translation (12,13) in vivo. To selectively interfere with U12- and U2-type splicing, we chose morpholinos complementary to sequences in U12 and U2 snRNA base-pairing with the distinct branch sites of the two splicing systems (Figure 1).
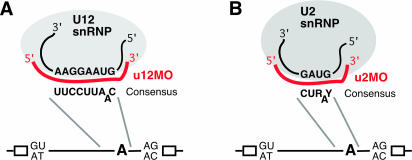
Figure 1.
Schematic representation of morpholino-mediated targeting of U12 (A) and U2 snRNA (B). U12 and U2 snRNPs with snRNA sequences base-pairing with the corresponding branch point consensus sequence (19) in pre-mRNA are shown (R, purine; Y, pyrimidine; and bulged A, branch site adenosine). Antisense-morpholino (MO) oligomers to these snRNA sequences are depicted in red (u12MO, U12 nucleotides 9–33; and u2MO, U2 nucleotides 27–51). Corresponding antisense RNA oligonucleotides block U12 or U2 snRNP binding to pre-mRNA in vitro (7,8). An overview on the localization in pre-mRNA of the sequences is shown in the lower part, with exons (open boxes) and introns [lines, with possible terminal nucleotides (19) and branch-point adenosine, A]. For morpholino sequences, see Materials and Methods.
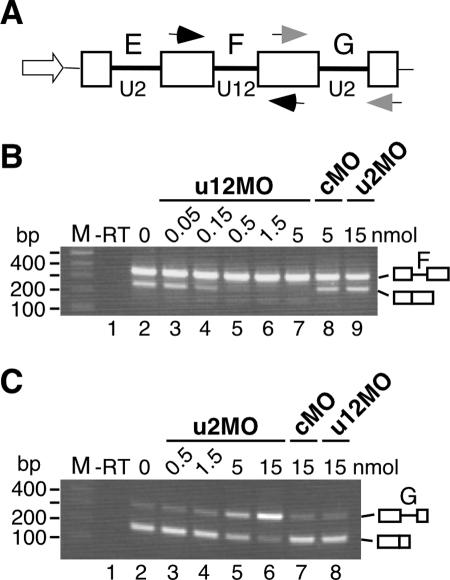
In studies using lymphoma cells, we recently found that plain morpholinos, although being non-ionic, can be efficiently and cheaply transfected by electroporation, even into cells for which electroporation of plasmid DNA is much less efficient [(14); and our unpublished data)]. To test the potential of our morpholino oligomers to interfere with either splicing apparatus in vivo, they were electroporated into mouse EL4 T-lymphoma cells together with a four-exon minigene construct derived from the human P120 gene (15) (Figure 2A), containing U2-dependent introns (E and G) and a U12-type intron (F). Splicing of introns F (U12-type) and G (U2-type) from the minigene RNA was then monitored after RT of total cellular RNA and PCR using primers (see Figure 2A) hybridizing in the upstream and downstream exons, respectively. Intron F removal, reflected by the amplification product lacking the intron, was inhibited with increasing amounts of the antisense-U12 oligomer (u12MO) co-transfected (Figure 2B, lanes 2–7). A non-specific control morpholino (cMO, lane 8) and the antisense-U2 sequence (u2MO, lane 9), in a concentration inhibiting U2-type splicing (cf. Figure 2C, lane 6), had no effect. Conversely, increasing doses of the u2MO sequence interfered with splicing of U2-dependent intron G (Figure 2C, lanes 2–6), while the control morpholino and the antisense-U12 morpholino in the highest amount did not affect the removal of the intron (lanes 7 and 8). Inhibition of U2-type splicing, however, required several-fold higher morpholino doses (Figure 2B and C), which could be caused by differences in the efficiency of morpholino hybridization or reflect the high abundance of U2 snRNP relative to U12 snRNP (16). The low abundance of the U12-type spliceosome, together with the high levels of minigene expression, may also explain the incomplete splicing of intron F from the minigene transcripts (Figure 2B, lane 2) relative to endogenous P120 pre-mRNA (Figure 3A and B, upper panel, lane 2; the same result was also obtained in the EL4 cells, data not shown). In conclusion, the findings indicate selective inhibition of U2- and U12-type splicing by delivering the corresponding antisense-morpholino sequences to cells.
Figure 2.
Selective inhibition of U2- and U12-type splicing in vivo. (A) Schematic representation of human P120 minigene (15) with promoter (open arrow), exons (open boxes) and U2- and U12-type intron sequences (E–G, bold lines). Arrowheads indicate positions of primers used in RT–PCR. (B and C) RT–PCR analysis of intron F (B) and intron G (C) splicing from the P120 minigene co-transfected into murine EL4 lymphoma cells with different amounts (in nmol) of the morpholino (MO) oligomers indicated. RNA was prepared 6 h after transfection. Lane 1 (-RT) corresponds to RT–PCR shown in lane 2, but lacking reverse transcriptase. Amplification products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. M, 100 bp DNA ladder; numbers on the left indicate the size of marker bands (in bp). Splicing products are shown on the right-hand side as schematic diagrams (exons, open boxes; and introns, lines). Predicted sizes for unspliced and spliced products are 349 and 250 bp (intron F splicing), and 315 and 171 bp (intron G splicing), respectively. Amplification reactions were not in plateau phase as verified by fewer PCR cycles. The same results were obtained in independent transfections followed by RT–PCR assays.
Figure 3.
Inhibition of U12-type splicing from endogenous genes. (A) Antisense-U12 morpholino-mediated inhibition of splicing at different time points after morpholino transfection. Human Jurkat-leukemia cells were transfected with 15 nmol of the u12MO or of a non-specific (cMO) morpholino oligomer. Splicing of intron F from P120 pre-mRNA (upper panel) and of intron 22 from ADPRT pre-mRNA (lower panel) was analyzed by RT–PCR after the indicated times. (B) Dose-dependence of u12MO-mediated splicing suppression. Morpholino oligomers were delivered to Jurkat cells in the amounts indicated (in nmol), and splicing of P120 intron F and of ADPRT intron 22 was analyzed 18 h after transfection. Expected sizes for unspliced and spliced products are 349 and 250 bp (P120) and 628 and 191 bp (ADPRT), respectively. Analysis, DNA marker and symbols are as in Figure 2. The weak band marked by the asterisk could not be obtained sufficiently pure for unequivocal identification by sequencing, but is most probably due to cryptic splicing. Amplification reactions were not in plateau phase as verified by fewer PCR cycles. The same results were obtained in independent transfections.
To test the suitability of the method for analyzing splicing of endogenous pre-mRNAs, we delivered the antisense-U12 oligomer to the commonly used human Jurkat-leukemia cell line by electroporation and monitored intron F removal from the human P120 pre-mRNA at different time points after morpholino transfection (Figure 3A, upper panel). While intron removal was complete immediately after transfection (lane 2), transcripts without intron F became almost undetectable after 6–12 h; instead, the intron-containing RNA accumulated (lanes 2–4), indicating the suppression of intron F splicing. The non-specific control sequence (lane 6) and the antisense-u2MO oligomer (not shown in this panel, but see Figure 3B) had no effect. P120 intron F represents U12-type introns having AT and AC dinucleotides at their 5′ and 3′ boundaries, respectively (15,17). As an example for an U12-type intron with GT and AG terminal nucleotides, we examined splicing of intron 22 of the human poly-ADP-ribosyl transferase (ADPRT) gene (17) (Figure 3A, lower panel). Again, transfection of the antisense-U12 morpholino blocked original complete splicing of the intron (lane 2), and led to the accumulation of the unspliced transcript over a period of 18 h (lanes 2–5). The suppression of U12-type splicing by the u12MO oligomer from both endogenous genes occurred in a dose-dependent manner (Figure 3B, lanes 2–8). Specificity is indicated by the non-specific control morpholino (lane 9) and the antisense-U2 sequence (lane 10), which did not affect intron removal. These results indicate that the approach is effective enough for restraining spliceosome function on endogenous transcripts. At the same time, they establish a role for U12 snRNA in the splicing of endogenous pre-mRNAs.
DISCUSSION
In mammalian cells, the existence of a second spliceosome has been established by forced co-expression of recombinant mutated minigenes and snRNAs carrying compensatory mutations (15,18). Employing snRNA as a target to block spliceosome function in cells, the functional ‘knockdown’ strategy presented here proves the second spliceosome to be required for pre-mRNA splicing from endogenous genes, and provides a new method to explore roles for the different splicing systems in vivo.
In keeping with a block of U12 snRNA interaction with the branch site, as shown for corresponding antisense-U12 RNA oligonucleotides in vitro (8), we found U12 snRNA levels unaltered after transfection of the u12MO oligomer (data not shown). In contrast to the highly conserved U12-type branch-point consensus (19), metazoan U2-type branch sites are only poorly preserved (20). Therefore, dependence of spliceosome assembly on base-pairing with U2 snRNA is most probably variable between U2-type introns. Weak base-pairing may be compensated through the recruitment of U2 snRNP by auxiliary proteins that bind to the adjacent polypyrimidine tract or the downstream 5′ splice site (21,22). Notably, splicing of U2-dependent intron G was inhibited to a lesser degree by the antisense-U2 morpholino in endogenous P120 pre-mRNA compared with P120 minigene RNA (see Supplementary Figure). This could reflect help in U2 snRNP recruitment to the branch point of intron G from the downstream 5′ splice site, which is not present in the minigene construct (15). Thus, using interference with branch site base-pairing, the U12-dependent system with its highly conserved branch points appears to be the better target for functionally depriving cells of one splicing machinery. Alternatively, antisense sequences interfering with U6 or U6atac snRNA interactions may be used, which were shown to inhibit other steps of spliceosome assembly in vitro (7,8).
Since several methods are now available to deliver morpholino oligomers to cultured cells (for current update, see http://www.gene-tools.com), morpholino-mediated interference with spliceosomal snRNA function should be applicable to many cell lines. Also, model organisms in which morpholinos have become popular for gene ‘knockdowns’ to study embryonal development (13), including frog (Xenopus species), chicken and zebrafish, should become accessible for the technique. Disruption of the U12 snRNA gene by insertional mutagenesis in Drosophila was lethal very early in embryogenesis, while an insertion generating partially active U6atac snRNA developed till larval stages (23). In contrast to most gene disruptions, morpholinos can phenocopy graded series of alleles (13) that may facilitate functional analyses. Together with techniques to introduce morpholinos into specific populations of embryo cells (24,25), the approach could thus provide a tool to study the role of different splicing systems in developmental processes. Finally, cells functionally deprived of one spliceosomal system may provide the basis for defining genome-wide spectra of splicing events (26,27) dependent on the distinct spliceosomes, casting light on their role in gene expression and in the evolution of proteome complexity in multicellular organisms.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank R. Padgett for providing the P120 minigene construct, H. Olinger for technical assistance and N. Johnsson for comments on the manuscript. This work was supported by Deutsche Forschungsgemeinschaft. Funding to pay the Open Access publication charges for this article was provided by Forschungszentrum Karlsruhe GmbH.
REFERENCES
- 1.Jurica M.S., Moore M.J. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 2.Patel A.A., Steitz J.A. Splicing double: insights from the second spliceosome. Nature Rev. Mol. Cell. Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 3.Burge C.B., Padgett R.A., Sharp P.A. Evolutionary fates and origins of U12-type introns. Mol. Cell. 1998;2:773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- 4.Levine A., Durbin R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 2001;29:4006–4013. doi: 10.1093/nar/29.19.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gontarek R.R., McNally M.T., Beemon K. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes Dev. 1993;7:1926–1936. doi: 10.1101/gad.7.10.1926. [DOI] [PubMed] [Google Scholar]
- 6.Roesser J.R. Both U2 snRNA and U12 snRNA are required for accurate splicing of exon 5 of the rat calcitonin/CGRP gene. RNA. 2004;10:1243–1250. doi: 10.1261/rna.5210404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamond A.I., Sproat B., Ryder U., Hamm J. Probing the structure and function of U2 snRNP with antisense oligonucleotides made of 2′-OMe RNA. Cell. 1989;58:383–390. doi: 10.1016/0092-8674(89)90852-0. [DOI] [PubMed] [Google Scholar]
- 8.Frilander M.J., Steitz J.A. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev. 1999;13:851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- 10.Lacerra G., Sierakowska H., Carestia C., Fucharoen S., Summerton J., Weller D., Kole R. Restoration of hemoglobin A synthesis in erythroid cells from peripheral blood of thalassemic patients. Proc. Natl Acad. Sci. USA. 2000;97:9591–9596. doi: 10.1073/pnas.97.17.9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sazani P., Gemignani F., Kang S.H., Maier M.A., Manoharan M., Persmark M., Bortner D., Kole R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 12.Nasevicius A., Ekker S.C. Effective targeted gene ‘knockdown’ in zebrafish. Nature Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 13.Heasman J. Morpholino oligos: making sense of antisense? Dev. Biol. 2002;243:209–2014. doi: 10.1006/dbio.2001.0565. [DOI] [PubMed] [Google Scholar]
- 14.Matter N., Herrlich P., König H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 15.Hall S.L., Padgett R.A. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 16.Montzka K.A., Steitz J.A. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc. Natl Acad. Sci. USA. 1988;85:8885–8889. doi: 10.1073/pnas.85.23.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietrich R., Incorvaia R., Padgett R.A. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell. 1997;1:151–160. doi: 10.1016/s1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- 18.Kolossova I., Padgett R.A. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) pre-mRNA introns. RNA. 1997;3:227–223. [PMC free article] [PubMed] [Google Scholar]
- 19.Sharp P.A., Burge C.B. Classification of introns: U2-type or U12-type. Cell. 1997;91:875–879. doi: 10.1016/s0092-8674(00)80479-1. [DOI] [PubMed] [Google Scholar]
- 20.Harris N.L., Senapathy P. Distribution and consensus of branch point signals in eukaryotic genes: a computerized statistical analysis. Nucleic Acids Res. 1990;18:3015–3019. doi: 10.1093/nar/18.10.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valcarcel J., Gaur R.K., Singh R., Green M.R. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 22.Robberson B.L., Cote G.J., Berget S.M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otake L.R., Scamborova P., Hashimoto C., Steitz J.A. The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila. Mol. Cell. 2002;9:439–446. doi: 10.1016/s1097-2765(02)00441-0. [DOI] [PubMed] [Google Scholar]
- 24.Kos R., Tucker R.P., Hall R., Duong T.D., Erickson C.A. Methods for introducing morpholinos into the chicken embryo. Dev. Dyn. 2003;226:470–477. doi: 10.1002/dvdy.10254. [DOI] [PubMed] [Google Scholar]
- 25.Mellitzer G., Hallonet M., Chen L., Ang S.L. Spatial and temporal ‘knockdown’ of gene expression by electroporation of double-stranded RNA and morpholinos into early postimplantation mouse embryos. Mech. Dev. 2002;118:57–63. doi: 10.1016/s0925-4773(02)00191-0. [DOI] [PubMed] [Google Scholar]
- 26.Yeakley J.M., Fan J.B., Doucet D., Luo L., Wickham E., Ye Z., Chee M.S., Fu X.D. Profiling alternative splicing on fiber-optic arrays. Nat. Biotechnol. 2002;20:353–358. doi: 10.1038/nbt0402-353. [DOI] [PubMed] [Google Scholar]
- 27.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.