Abstract
Zika virus (ZIKV) and the 4 dengue virus (DENV) serotypes are mosquito-borne Flaviviruses that are associated with severe neuronal and hemorrhagic syndromes. The mature flavivirus infectious virion has 90 envelope (E) protein homo-dimers that pack tightly to form a smooth protein coat with icosahedral symmetry. Human antibodies that strongly neutralize ZIKV and DENVs recognize complex quaternary structure epitopes displayed on E-homo-dimers and higher order structures. The ZIKV and DENV E protein expressed as a soluble protein is mainly a monomer that does not display quaternary epitopes, which may explain the modest success with soluble recombinant E (sRecE) as a vaccine and diagnostic antigen. New strategies are needed to design recombinant immunogens that display these critical immune targets. Here we present two novel methods for building or stabilizing in vitro E-protein homo-dimers that display quaternary epitopes. In the first approach we immobilize sRecE to enable subsequent dimer generation. As an alternate method, we describe the use of human mAbs to stabilize homo-dimers in solution. The ability to produce recombinant E protein dimers displaying quaternary structure epitopes is an important advance with applications in flavivirus diagnostics and vaccine development.
Introduction
Zika virus (ZIKV) and the dengue viruses (DENVs) are mosquito-borne members of the flaviviridea family, which can cause severe neurological and hemorrhagic syndromes in humans1–3. It is estimated that 400 million DENV infections occur each year and that over half of the world’s population live in countries with active DENV or ZIKV transmission3.
The continuing threat of DENVs and, more recently, ZIKV has stimulated much work on different vaccine platforms including live attenuated virus, inactivated whole virus, protein subunit and DNA vaccines4–12. The congenital malformations caused by ZIKV infection during pregnancy has stimulated work on subunit vaccines, as live attenuated and other replicating virus vaccines are contraindicated during pregnancy. Advances in molecular biology and bio/nanotechnology have led to the production of recombinant viral proteins with applications in diagnostics and vaccinology. The flavivirus envelope (E) protein is a major target of neutralizing and protective human antibodies, but recombinantly expressed soluble E protein without the C-terminal transmembrane domains has not proven to be particularly effective as a vaccine antigen13–16. For DENV and, more recently ZIKV, we see growing evidence that E protein domains or E proteins expressed as a soluble recombinant antigen (sRecE) fails to induce robust protective responses unlike whole virus or virus-like particle (VLP) vaccine antigens5, 6, 17–19. Recent studies have established that complex quaternary structure epitopes displayed by E oligomers on the viral surface but not E monomers are targets of strongly neutralizing and protective human antibodies6, 7. In solution, sRecE from flaviviruses are in a dynamic equilibrium that favors the monomer over the dimer, which likely explains the poor binding of strongly neutralizing quaternary epitope directed human antibodies and the overall poor immunogenicity in pre-clinical studies6, 20, 21.
From a patient who had recovered from a primary DENV serotype 2 infection, we previously isolated and characterized a DENV2 serotype-specific strongly neutralizing mAb, 2D22, that binds to a E protein dimer-dependent quaternary epitope (Fig. 1A)22–24. Human mAb 2D22 does not bind to the DENV2 sRecE monomer24. Additionally, analysis of B-cells from people exposed to repeated DENV infections has led to the discovery of mAbs that target envelope dimer epitopes (EDE) that are conserved between the 4 DENV serotypes and ZIKV25, 26. EDE mAbs (Fig. 1A), which cross-neutralize different DENV serotypes and ZIKV to varying degrees, have been divided into two groups (EDE1 and EDE2), based on their binding footprint and sensitivity to the presence or absence of an N-linked glycan in domain I (EDI) of E protein25.
Figure 1.
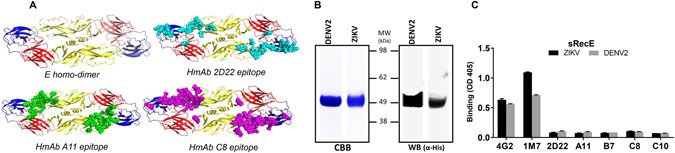
DENV2 and ZIKV sRecE expression and characterization. (A) The flavivirus E protein consists of three beta-barrel domains designated domains I (red), II (yellow) and III (blue), with the native protein forming a head-to-tail homo-dimer. The quaternary epitopes recognized by human dimer-dependent Mabs 2D22 (cyan), A11 (green) and C8 (magenta) are indicated22, 35. (B) DENV and ZIKV sRecE expression was analyzed by Western Blot (anti-His mAbs), CBB (C) and by ELISA using 4G2, 1M7, 2D22, A11, B7 (EDE2), C8 and C10 (EDE1) mAbs.
In this study we describe novel methods based on the immobilization of sRecE on a matrix or the use dimer-specific human mAbs to promote the temperature dependent assembly and stabilization of sRecE dimers displaying quaternary structure antibody epitopes targeted by human antibodies. The ability to assemble E homo dimers displaying quaternary structure antibody epitopes is an important advance with applications in flavivirus vaccine development and diagnostics.
Results
Expression of DENV2 and ZIKV recombinant E-proteins
The ectodomains of the DENV2 and ZIKV E-proteins were expressed using the EXPI293 mammalian transient expression system, containing their native prM sequences, an N-terminal IL2 secretion leader peptide and a C-terminal 6xHis tail for Ni-affinity purification of the recombinant protein. The expression and purity of sRecE monomers from both DENV2 and ZIKV was analyzed by SDS-PAGE and western blot using anti-His mabs (Fig. 1B). Both Coomassie Brilliant Blue staining and western blotting show the presence of pure proteins with the predicted molecular mass for DENV2 (~48 kDa) and ZIKV sRecE (~47 kDa). The purified proteins were subsequently analyzed by ELISA using a panel of mAbs (Table 1) that recognize epitopes on the monomer (4G2 and 1M7) or homo-dimer (2D22, A11, B7, C8 and C10) (Fig. 1A,C). Binding was observed for mAbs 4G2 and 1M7 but not for mAb 2D22, and the EDE mAbs confirming that the monomer was more abundant than the dimer at equilibrium.
Table 1.
Binding characteristics of used mAbs.
| Mab | M/H | Binding | Neutralization (W/M/S) | Binding region | Binding DENV serotypes | Ref. | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| DV1 | DV2 | DV3 | DV4 | ZIKV | ||||||
| 4G2 | M | F-CR | W | DII FL | ++ | ++ | +++ | +++ | +++ | 41 |
| 2D22 | H | DV2 | DV2:S ZIKV:W | DI/DII Q | − | ++ | − | − | − | 22, 36 |
| 1M7 | H | F-CR | M | DII FL | +++ | ++ | +++ | +++ | +++ | 27 |
| A11 | H | F-CR | DV:S ZIKV:W | DI/DII/DII Q | +++ | +++ | +++ | +++ | + | 38 |
| B7 | H | F-CR | DV:S ZIKV:W | DI/DII/DII Q | +++ | +++ | +++ | +++ | + | 38 |
| C8 | H | F-CR | DV:S ZIKV:S | DI/DII/DII Q | +++ | +++ | +++ | +++ | ++ | 38 |
| C10 | H | F-CR | DV:S ZIKV:S | DI/DII/DII Q | +++ | +++ | +++ | +++ | ++ | 38 |
Characteristics of the mAbs used for dimer assembly. Several mouse or human (M/H) derived mAbs were used for sRecE dimer assembly, indicating their DENV and ZIKV binding potential. Flavivirus cross reactive (F-CR), weakly, medium or strong (W/M/S) neutralizing, E-domain I, II, III (DI, DII, DIII), fusion loop (FL), quaternary epitope (Q).
Assembly of sRecE dimers and generation of DENV2 specific quaternary epitopes
As the DENV sRecE antigen does not form stable homo-dimers in solution, we used the C-terminal 6X His tail to immobilize the protein to a Ni2+-coated surface under different conditions to promote dimerization. The Ni2+-coated surfaces were loaded with different amounts of DENV2 sRecE (50, 500 or 1000 ng/well), blocked and then reloaded with 0, 50 or 500 ng/well of sRecE (Fig. 2A). Dimer formation was assessed by calculation the ratio of dimeric E protein (mAb 2D22 signal) to total E protein (mAb 4G2 signal). Neither mAb bound non-specifically to the Ni2+ surface in the absence of antigen (Fig. 2A). Initial loading with 50 or 500 ng of antigen without reloading resulted in a low ratio of 2D22/4G2 binding, indicating the protein was captured and mainly retained as a monomer. When the plates were reloaded with 500 ng of sRecE, we observed a four to seven fold increase in the 2D22/4G2 ratio (Fig. 2A). Augmented 2D22 binding is not a mere antigen concentration effect, since a 1000 ng sRecE load without reload did not show an increased 2D22 signal, whereas a load of 500 ng followed by a reload of 500 ng led to a large increase in 2D22 binding (Fig. 2A). In addition to 2D22, EDE mAbs were used to measure dimer formation of DENV2 sRecE (Fig. 2B). All EDE1 and EDE2 mAbs tested bound well to the 500 ng load/500 ng reload group compared to the other groups confirming the formation of dimers (Fig. 2B ). This dimer-building platform was also used to analyze the binding of DENV2 serotype-specific and strongly neutralizing human mAbs 1L12 and 3F9, which bind to complex epitopes that have not been fully mapped yet (Fig. 2C)27. Under conditions that favor DENV2 sRecE dimer formation, we observed increased binding of both 1L12 and 3F9.
Figure 2.
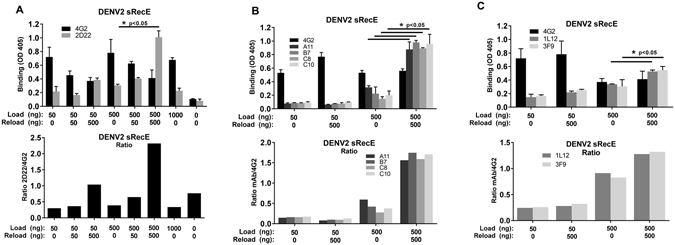
DENV2 sRecE dimer assembly on Ni2+-coated surfaces. (A) Indicated amounts of DENV2 sRecE were loaded and reloaded on Ni2+-coated plates and analyzed for monomeric (4G2) or dimeric protein structures (2D22). Increased dimer formation is displayed as the 2D22/4G2 signal ratio. (B) The same assay was performed with EDE1 (A11, B7), EDE2 (C8, C10) and C) 3F9 and 1M7 mAbs. No antibody signals were detected when no protein was loaded or reloaded and groups 500 + 0 and 500 + 500 were statistically compared by student T-test (p < 0.05).
sRecE-dimer formation is a temperature dependent process
To assess the effect of temperature on E dimer formation on Ni2+ plates, DENV2 sRecE was immobilized at 37 °C or 21 °C. After blocking, the wells were reloaded at 37 °C or 21 °C and analyzed with 4G2 and 2D22. A decrease in dimer formation was observed when sRecE was incubated at 37 °C during the immobilization and reloading step (Fig. 3A). Reloading the protein at 21 °C resulted in a modest increase in dimerization (Fig. 3B). A similar increase in the 4G2/2D22 ratio was observed when sRecE was immobilized at 21 °C and reloaded at 37 °C (Fig. 3C). Dimer formation was most efficient when the immobilization and reloading steps were conducted at 21 °C (Fig. 3D), indicating a strong temperature dependency towards the formation of dimer dependent epitopes in this assay.
Figure 3.
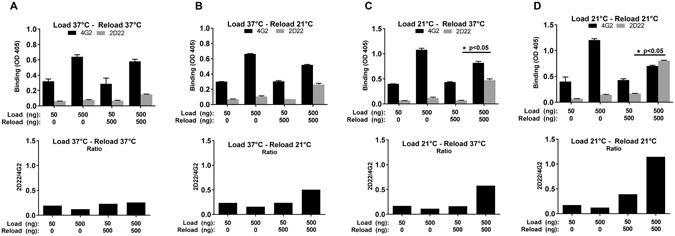
Temperature effects on sRecE dimer formation. Indicated amounts of DENV2 sRecE were loaded and reloaded at different temperatures, resulting in the following temperature regiments: (A) Load at 37 °C and reload at 37 °C, (B) load at 37 °C and reload at 21 °C, (C) load at 21 °C and reload at 37 °C and (D) load at 21 °C and reload at 21 °C. Dimer formation was analyzed by 4G2 and 2D22 binding. Groups 500 + 0 and 500 + 500 were statistically compared by student T-test (p < 0.05).
Antibody mediated dimer stabilization
In solution sRecE is likely to be present as both monomers and dimers with the equilibrium favoring the monomers under our experimental conditions. As an alternative approach to producing stable E protein dimers, we incubated sRecE in solution with dimer-dependent mAbs. sRecE (500 ng) was incubated with 500 ng of mAbs 2D22, A11 and B7 (EDE2), and C8 and C10 (EDE1). sRecE/mAb complexes were subsequently captured through His tag at the C terminal of sRecE to Ni2+-coated ELISA plate. Any mAb bound to the captured sRecE was detected using a secondary goat anti-human IgG-AP conjugated antibody. When 2D22 or 4G2 were incubated without DENV or ZIKV sRecE subunits (0 ng/well), no mAb-protein complex was captured indicating that the mAbs did not directly bind to Ni2+ plates (Fig. 4A). A large increase in the detection of mAb 2D22 was observed after incubation with DENV2 sRecE, and, as expected, 2D22 failed to bind ZIKV sRecE (Fig. 4A ). Similar binding patterns were obtained after EDE mAbs were mixed with DENV2 sRecE proteins in solution (Fig. 4B). These results indicate that dimer dependent human mAbs can stabilize DENV2 E dimers in solution and preserve the dimers during capture to Ni2+-plates. Interestingly for ZIKV sRecE, only EDE1 mAbs C8 and C10 were detected, while EDE2 antibodies did not bind.
Figure 4.
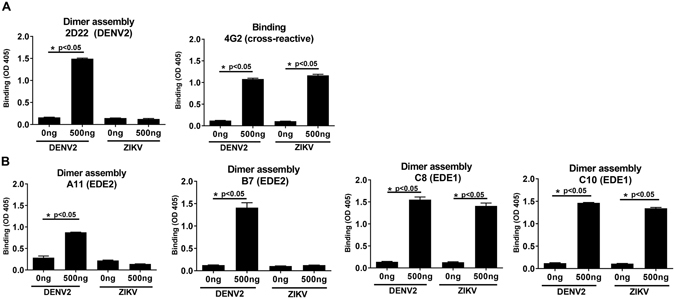
Antibody mediated dimer assembly. Indicated amounts of DENV and ZIKV sRecE were incubated with 500 ng of (A) 2D22, 4G2, (B) A11, B7, C8 and C10 mAbs in solution. Protein/mAb complexes were loaded on Ni2+-coated plates and captured antibodies were detected. Statistical analysis was performed using the student T-test.
The binding assay was repeated by incubating the mAbs and sRecE proteins at 21 °C and 37 °C to determine the impact of temperature on dimer formation and mAb mediated stabilization in solution (Fig. 5). The dimer dependent mAbs stabilized both DENV2 and ZIKV E dimers poorly at 37 °C compared to 21 °C. Similar to the assembly of sRecE dimers on Ni2+-surfaces, these results indicate a strong temperature dependence on the formation and mAb mediated stabilization of E dimers.
Figure 5.
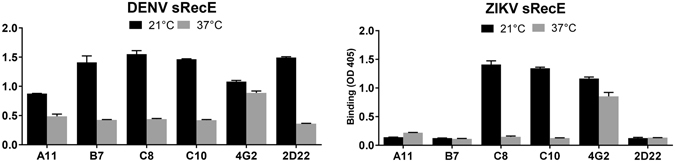
Antibody mediated dimer assembly is a temperature dependent process. The effect of temperature on mAb mediated dimer assembly was analyzed by incubating 500 ng mAb with 500 ng DENV2 or ZIKV sRecE. sRecE-mAb incubations were performed at 21 °C or 37 °C and loaded on Ni2+-coated plates and captured antibodies were detected.
Discussion
Recent studies show that flavivirus virions or virus-like particles (VLPs) are better vaccine antigens than sRecE5, 6. The poor immunogenicity of sRecE may be, in part, due to the failure to efficiently display E protein quaternary structure epitopes displayed on virions and VLPs that are known to be targets of strongly neutralizing human mAbs. In this study, we present novel methods to assemble or stabilize dimer-dependent quaternary epitopes out of monomeric sRecE building blocks.
The binding of 2D22 and the EDE mAbs to reloaded wells demonstrates the formation of E homo-dimers displaying quaternary epitopes, that were absent in the non-reloaded wells. During the initial sRecE load, proteins are chelated to the Ni2+-surface through their C-terminal His-tag. This interaction is anticipated to immobilize the proteins in a confirmation that enables interaction and dimer formation with the reloaded sRecE (Fig. 6). In contrast to the homo-dimers, the dimer-dependent binding mAbs might bind the partial epitope present on monomers with no or very low binding affinity. The C-terminal attachment of the proteins to the Ni2+-surface replicates the native C-terminal anchoring of E-proteins to lipid membranes. To transfer this technology for vaccine purposes, further platform development is required where the Ni2+-His immobilization is replaced by more biological compounds suitable for human administration.
Figure 6.
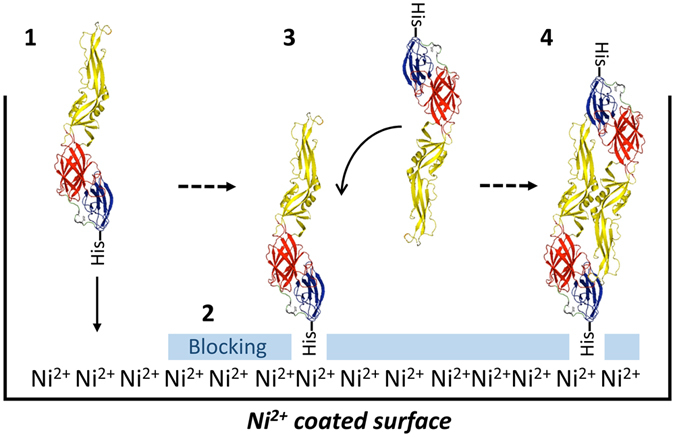
Schematic representation of the postulated model for sRecE-dimer assembly from immobilized monomers. (1) sRecE is chelated to Ni2+-coated plates. The immobilization of sRecE at its C-terminal end presumably locks the protein in a specific conformation. (2) The plates are subsequently blocked and (3) reloaded with sRecE at high protein concentrations. (4) This enables interaction of the immobilized sRecE with the reloaded proteins and generates quaternary epitopes that can be recognized by E-dimer epitope dependent mAbs.
DENV virion morphology is affected by several factors such as the binding of mAbs, pH and the proteolytic processing of envelope proteins28–30. In addition, some DENV strains have been shown to undergo temperature dependent conformational changes in their envelope structure. At lower temperatures, the DENV2 E protein lies flat on the virus surface in a compact manner, resulting in a smooth appearance. At 37 °C however, E protein becomes less compact and undergoes conformational changes leading to a ‘bumpy’ surface31–33. Similar effects on proteins structure and organization have recently been seen in other sRecE expression studies34. The effect of temperature in the assembly of DENV2 and ZIKV sRecE dimers on artificial surfaces is apparent. Temperature might affect the flexibility of sRecE to be able to pair with the reloaded or immobilized counterpart. In this case, the actual number of dimers will be affected. Another explanation would be that at both temperatures dimers are formed with equal efficiency, but at the higher temperature the 2D22 epitope may be disrupted or hidden.
The sRecE proteins are expressed in an equilibrium between dimers and monomers, with an emphasis towards the monomeric state. Using static light scattering after size exclusion chromatography, we confirmed that the majority of sRecE was monomeric in solution and that the addition of dimer dependent mAbs in solution stabilized E dimer/mAb complexes (Fig. S1). For DENV2 sRecE dimers, all tested mAbs stabilized dimers prior to capture on Ni2+-plates. For ZIKV sRecE however, only EDE1 mAbs were able to stabilize dimers, whereas EDE2 mAbs failed to do so. The ability of EDE1 mAbs to stabilize ZIKV sRecE-dimers is consistent with previous work showing better binding and neutralization of ZIKV by EDE1 compared to EDE2 mAbs35–37. For DENV, all EDE mAbs occupy two binding sites within one E-dimer38. In case of ZIKV however, EDEs dock in different angels compared to DENV. Where EDE1 still occupies two binding sites, EDE2 A11 only occupies one site. Docking of a second Fab is most likely inhibited by steric hindrance by proximate complexes35. This might be a reason why the low affinity binding EDE2 mAbs are not able to stablize ZIKV sRecE dimers in solution.
The magnitude, explosive spread and severe clinical manifestations of DENV and ZIKV epidemics demand research towards reliable diagnostic tools and vaccines. Considering that pregnant women could be a potential target group for ZIKV vaccines, safe alternatives to live attenuated and replicating virus vaccines, such as subunit approaches, are in urgent demand. More broadly, investigators are focusing on methods to produce recombinant antigens that display complex quaternary epitopes on the viral surface targeted by the human immune response39, 40. Here we demonstrate the feasibility of assembling flavivirus E protein structures that display complex quaternary epitopes recognized by both virus specific and cross-reactive, broadly neutralizing antibodies. Flavivirus sRecE protein dimers hold much promise as antigens for advancing diagnostics and subunit vaccines.
Methods
Expression and purification of DENV2 and ZIKV sRecE
The soluble recombinant DENV2 (sRecE, aa 1–395) and ZIKV (aa 1–404) envelope proteins were expressed by the EXPI293 transient expression system (ThermoFisher) as previously described24. In short: DENV2 and ZIKV sRecE was equipped with a C-terminal 6x His tag and expression was driven by a CAG promoter. Through tangential flow filtration, supernatants were concentrated and buffer exchanged into a Ni2+ binding buffer (50 mM NaPO4, 500 mM NaCl, 25 mM imidazole, 0.02% Na-Azide) and subsequently Ni2+ affinity purified. After washing, the Ni2+ column was serially eluted with elution buffer (50 mM NaPO4, 500 mM NaCl, 500 mM Imidazole 0.02%, Na-Azide, 10% glycerol). Fractions containing envelope proteins were pooled subjected to size exclusion chromatography using a 16/60 Superdex S200TM column. Fractions with envelope protein were pooled in PBS + 10% glycerol, concentrated and flash frozen in liquid N2, and stored at −80 °C. The oligomeric state of the purified proteins were analyzed size exclusion chromatography using a Superdex S200TM column connected to multi angle light scattering instrument (Wyatt DAWN HELEOS-II) with an OptiLab T-rex refractometer. 200 µg of DENV2 sRecE was analyzed with or without 40 µg of 2D22 in 100 µl PBS.
Protein analysis by Western blot and CBB stain
DENV2 and ZIKV sRecE expression was analyzed by Coomassie Brilliant Blue (CBB) staining and Western blot. Protein samples were added to Laemmli sample buffer containing 20 mM ß-mercaptoethanol. Samples were boiled for 3 mins and loaded on a NuPAGETM, NOVEXTM 4–12% Bis-Tris protein gel (Invitrogen). Proteins were transferred to a polyvinylidene difluoride (PDVF) membrane and blocked for 1 hr with 0.5% skim milk in PBS + 0.1% Tween 20 (PBST). Next, membranes were incubated with anti-His-HRP conjugated antibody (Invitrogen) 1:5000 diluted in PBST for 1 hr. Membranes were washed 3 times with PBST and developed with Amersham ECL Prime Wester Blotting Detection Reagent (GE Healthcare).
Protein analysis by ELISA
100 ng/well (in TBS) of purified DENV2 or ZIKV sRecE was loaded on Ni2+-coated wells (Pierce) for 1 hr at RT. Next, wells were blocked with 3% skim milk in TBS + 0.05% Tween-20 (TBST) for 1 hr at RT and after washing with TBST, wells were incubated with 100 ng/well of 4G2, 1M7, 2D22, EDE1 (C8, C10) or EDE2 (A11, B7) mAbs in TBST for 1 hr at RT. Wells were subsequently washed and 4G2 treated wells were incubated with 1:1000 anti-mouse IgG-AP conjugated (Sigma) and 1M7, 2D22, EDE1 and EDE2 treated wells were incubated with 1:2500 anti-human IgG-AP conjugated (Sigma). After washing, plates were developed using AP-substrate (Sigma). Absorbance was measured at 405 nm.
Quaternary epitope assembly ELISA
To assemble DENV2 sRecE dimers, 50 ng and 500 ng/well of sRecE was loaded on clear Ni2+-coated ELISA plates (pre-blocked with BSA, 9 pmol binding capacity, purchased from Pierce, #15142) in 50 µl/well TBS for 1 hr at room temperature (RT, 21 °C). Wells were subsequently blocked with 3% skim milk in TBS + 0.05% Tween-20 (TBST) for 1 hr at RT. Wells were washed in TBST and reloaded with 0 ng, 50 ng or 500 ng/well sRecE and incubated for 1 hr at RT. Next, wells were washed 3 times with TBST and incubated with 100 ng/well of 4G2, 2D22, EDE1 (C8, C10), EDE2 (A11, B7), 1L12 or 3F9 mAbs in blocking buffer for 1 hr at RT. After washing, 4G2 treated wells were incubated with 1:1000 anti-mouse IgG-AP conjugated (Sigma) and 2D22, EDE1, EDE2, 1L12 and 3F9 treated wells were incubated with 1:2500 anti-human IgG-AP conjugated (Sigma). Plates were subsequently washed and developed using AP-substrate (Sigma). Absorbance was measured at 405 nm.
To analyze the effect of temperature on dimer-assembly, Ni2+-coated ELISA plates were loaded with 50 ng or 500 ng/well of sRecE in TBS for 1 hr at 37 °C or 21 °C. Plates were blocked with 3% skim milk in TBST for 1 hr at 21 °C or 37 °C. After washing in TBST, wells were reloaded with 0 ng or 500 ng/well sRecE in TBST for 1 hr at 21 °C or 37 °C. This way, the following loading-reloading regiments were created: Load at 37 °C and reload at 37 °C, load at 37 °C and reload at 21 °C, load at 21 °C and reload at 37 °C, load at 21 °C and reload at 21 °C. After washing, plates were incubated with 100 ng/well of 4G2 or 2D22 in blocking buffer for 1 hr at RT. Next, plates were washed and 4G2 treated wells were incubated with 1:1000 anti-mouse IgG-AP conjugated (Sigma), EDE2, 1L12 and 3F9 treated wells were incubated with 1:2500 anti-human IgG-AP conjugated (Sigma). Plates were subsequently washed and developed using AP-substrate (Sigma). Absorbance was measured at 405 nm. The binding ratios (mAb/4G2) were determined by dividing the means of the mAb binding signals.
Antibody mediated dimer stabilization
0 ng, or 500 ng of sRecE in TBS + Tween-20 (TBST) was incubated with 500 ng of EDE1 (C8, C10), EDE2 (A11, B7), 2D22 and 4G2 mAbs (in TBST) for 1 hr, shaking at RT (21 °C). The sRecE/mAb mix was plated on Ni2+-coated plates and incubated for 1 hr at RT. Next, wells were blocked in 3% skim milk in TBS + 0.05% Tween-20 for 1 hr at RT. After washing with TBST, 4G2 treated wells were incubated with 1:1000 anti-mouse IgG-AP conjugated (Sigma) and EDE1, EDE2 and 2D22 treated wells were incubated with 1:2500 anti-human IgG-AP conjugated (Sigma). Following washing, wells were developed using AP-substrate (Sigma). Absorbance was measured at 405 nm. To analyze the effect of temperature on mAb mediated dimer formation, 500 ng of sRecE was incubated with 500 ng of mAb. The previously described ELISA was repeated with incubation steps performed at 21 °C or 37 °C.
Data availability statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Electronic supplementary material
Acknowledgements
We would like to thank Prof. Ashutosh Tripathy from the UNC for his help in protein characterization. These studies were supported by NIAID grants 1-R01-AI107731-01 (PI A. de Silva, UNC), U19 AI109784-01 (PI J. Ting, UNC), NCI-NIH P30CA016086 and the European Union Zika Plan Research Consortium. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Contributions
S.M. designed the study, wrote the manuscript and performed the experiments. E.G. and R.B. supplied key reagents to perform the study. A.B. and M.M. produced and purified the recombinant proteins. L.P. helped designing experiments. A. dS helped designing the study, and helped writing the manuscript.
Competing Interests
Aravinda de Silva has consulted on dengue vaccines for Takeda Vaccines, GSK and Merck Pharmaceuticals. He is also an inventor in patents related to dengue vaccines.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04767-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Stefan W. Metz, Email: swmetz@med.unc.edu
Aravinda M. de Silva, Email: aravinda_desilva@med.unc.edu
References
- 1.Fauci AS, Morens DM. Zika Virus in the Americas–Yet Another Arbovirus Threat. N Engl J Med. 2016;374:601–604. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- 2.Peterson, A. T., Osorio, J., Qiao, H. & Escobar, L. E. Zika Virus, Elevation, and Transmission Risk. PLoS currents8 (2016). [DOI] [PMC free article] [PubMed]
- 3.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durbin AP. Vaccine Development for Zika Virus-Timelines and Strategies. Seminars in reproductive medicine. 2016;34:299–304. doi: 10.1055/s-0036-1592070. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Liu J, Cheng G. Vaccines and immunization strategies for dengue prevention. Emerging microbes & infections. 2016;5:77–83. doi: 10.1038/emi.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larocca RA, et al. Vaccine protection against Zika virus from Brazil. Nature. 2016;536:474–478. doi: 10.1038/nature18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbink P, et al. Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science. 2016;353:1129–1132. doi: 10.1126/science.aah6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirkpatrick BD, et al. Robust and Balanced Immune Responses to All 4 Dengue Virus Serotypes Following Administration of a Single Dose of a Live Attenuated Tetravalent Dengue Vaccine to Healthy, Flavivirus-Naive Adults. J Infect Dis. 2015;212:702–710. doi: 10.1093/infdis/jiv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkpatrick, B. D. et al. The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Science translational medicine8, 330ra336 (2016). [DOI] [PubMed]
- 10.Abbink P, et al. Construction and evaluation of novel rhesus monkey adenovirus vaccine vectors. J Virol. 2015;89:1512–1522. doi: 10.1128/JVI.02950-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chahal JS, et al. An RNA nanoparticle vaccine against Zika virus elicits antibody and CD8+ T cell responses in a mouse model. Scientific reports. 2017;7:252. doi: 10.1038/s41598-017-00193-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim E, et al. Preventative Vaccines for Zika Virus Outbreak: Preliminary Evaluation. EBioMedicine. 2016;13:315–320. doi: 10.1016/j.ebiom.2016.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzman MG, et al. Induction of neutralizing antibodies and partial protection from viral challenge in Macaca fascicularis immunized with recombinant dengue 4 virus envelope glycoprotein expressed in Pichia pastoris. Am J Trop Med Hyg. 2003;69:129–134. [PubMed] [Google Scholar]
- 14.Kelly EP, Greene JJ, King AD, Innis BL. Purified dengue 2 virus envelope glycoprotein aggregates produced by baculovirus are immunogenic in mice. Vaccine. 2000;18:2549–2559. doi: 10.1016/S0264-410X(00)00032-3. [DOI] [PubMed] [Google Scholar]
- 15.Coller BA, Clements DE, Bett AJ, Sagar SL, Ter Meulen JH. The development of recombinant subunit envelope-based vaccines to protect against dengue virus induced disease. Vaccine. 2011;29:7267–7275. doi: 10.1016/j.vaccine.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mamo T, Poland GA. Nanovaccinology: the next generation of vaccines meets 21st century materials science and engineering. Vaccine. 2012;30:6609–6611. doi: 10.1016/j.vaccine.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 17.Block OK, et al. A tetravalent recombinant dengue domain III protein vaccine stimulates neutralizing and enhancing antibodies in mice. Vaccine. 2010;28:8085–8094. doi: 10.1016/j.vaccine.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Midgley CM, et al. An in-depth analysis of original antigenic sin in dengue virus infection. J Virol. 2011;85:410–421. doi: 10.1128/JVI.01826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams KL, Wahala WM, Orozco S, de Silva AM, Harris E. Antibodies targeting dengue virus envelope domain III are not required for serotype-specific protection or prevention of enhancement in vivo. Virology. 2012;429:12–20. doi: 10.1016/j.virol.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Alwis R, et al. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci USA. 2012;109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allison SL, Stadler K, Mandl CW, Kunz C, Heinz FX. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J Virol. 1995;69:5816–5820. doi: 10.1128/jvi.69.9.5816-5820.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fibriansah G, et al. DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science. 2015;349:88–91. doi: 10.1126/science.aaa8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallichotte EN, et al. A new quaternary structure epitope on dengue virus serotype 2 is the target of durable type-specific neutralizing antibodies. mBio. 2015;6:1461–1469. doi: 10.1128/mBio.01461-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metz SW, et al. Precisely Molded Nanoparticle Displaying DENV-E Proteins Induces Robust Serotype-Specific Neutralizing Antibody Responses. PLoS Negl Trop Dis. 2016;10:5071–5088. doi: 10.1371/journal.pntd.0005071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dejnirattisai W, et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nature immunology. 2015;16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dejnirattisai W, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nature immunology. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith SA, et al. Isolation of dengue virus-specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J Virol. 2014;88:12233–12241. doi: 10.1128/JVI.00247-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lok SM, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, et al. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat Struct Mol Biol. 2013;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, et al. Structure of acidic pH dengue virus showing the fusogenic glycoprotein trimers. J Virol. 2015;89:743–750. doi: 10.1128/JVI.02411-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Sun L, Rossmann MG. Temperature dependent conformational change of dengue virus. Current opinion in virology. 2015;12:109–112. doi: 10.1016/j.coviro.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, et al. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc Natl Acad Sci USA. 2013;110:6795–6799. doi: 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fibriansah G, et al. Structural changes in dengue virus when exposed to a temperature of 37 degrees C. J Virol. 2013;87:7585–7592. doi: 10.1128/JVI.00757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slon Campos JL, et al. Temperature-dependent folding allows stable dimerization of secretory and virus-associated E proteins of Dengue and Zika viruses in mammalian cells. Scientific reports. 2017;7:966. doi: 10.1038/s41598-017-01097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barba-Spaeth G, et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature. 2016;536:48–53. doi: 10.1038/nature18938. [DOI] [PubMed] [Google Scholar]
- 36.Swanstrom JA, et al. Dengue Virus Envelope Dimer Epitope Monoclonal Antibodies Isolated from Dengue Patients Are Protective against Zika Virus. mBio. 2016;7:1123–1131. doi: 10.1128/mBio.01123-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S, et al. Neutralization mechanism of a highly potent antibody against Zika virus. Nature communications. 2016;7:13679–13685. doi: 10.1038/ncomms13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouvinski A, et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520:109–113. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 39.Correia BE, et al. Proof of principle for epitope-focused vaccine design. Nature. 2014;507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jardine J, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henchal EA, McCown JM, Burke DS, Seguin MC, Brandt WE. Epitopic analysis of antigenic determinants on the surface of dengue-2 virions using monoclonal antibodies. Am J Trop Med Hyg. 1985;34:162–169. doi: 10.4269/ajtmh.1985.34.162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).


