Abstract
MicroRNAs (miRNAs) are noncoding RNAs that regulate posttranscriptional gene expression. In this study we characterized the circulating and urinary miRNA pattern associated with reduced glomerular filtration rate, using Affymetrix GeneChip miR 4.0 in 28 patients with chronic kidney disease (CKD). Top miRNA discoveries from the human studies were validated in an Alb/TGFβ mouse model of CKD, and in rat renal proximal tubular cells (NRK52E) exposed to TGFβ1. Plasma and urinary levels of procollagen III N-terminal propeptide and collagen IV were elevated in patients with decreased estimated glomerular filtration rate (eGFR). Expression of 384 urinary and 266 circulatory miRNAs were significantly different between CKD patients with eGFR ≥30 vs. <30 ml·min−1·1.73 m−2. Pathway analysis mapped multiple miRNAs to TGFβ signaling-related mRNA targets. Specifically, Let-7a was significantly downregulated, and miR-130a was significantly upregulated, in urine of patients with eGFR <30; miR-1825 and miR-1281 were upregulated in both urine and plasma of patients with decreased eGFR; and miR-423 was significantly downregulated in plasma of patients with decreased eGFR. miRNA expression in urine and plasma of Alb/TGFβ mice generally resembled and confirmed most, although not all, of the observations from the human studies. In response to TGFβ1 exposure, rat renal proximal tubular cells overexpressed miR-1825 and downregulated miR-423. Thus, miRNA are associated with kidney fibrosis, and specific urinary and plasma miRNA profile may have diagnostic and prognostic utility in CKD.
Keywords: chronic kidney disease, fibrosis, TGFβ
micrornas (miRNAs) are small, noncoding RNAs that exert posttranscriptional control of gene expression. MiRNAs participate in a wide range of biological processes, including cell cycle, apoptosis, cell differentiation, and epithelial-mesenchymal transition (EMT) (2). The latter is the mechanism by which injured tubular epithelial cells transform into mesenchymal cells that contribute to the development of fibrosis (13, 16). Fibrosis is characterized by imbalance in matrix formation and degradation, which leads to excessive accumulation of extracellular matrix (ECM) (9, 21). miRNAs regulate kidney fibrosis through direct repression and/or expression of matrix genes and through transforming growth factor (TGF)β signaling (5, 6, 15, 36). Tissue changes in the expression levels of specific miRNAs are reflected in their plasma levels, suggesting that miRNAs may serve as noninvasive markers of disease processes (24). Furthermore, preliminary evidence indicates that circulating miRNAs could regulate cells and organs distant from the site of origin (35).
In this study, we examined the circulating and urinary miRNA profile associated with decreased glomerular filtration rate (GFR) in patients with chronic kidney disease (CKD), using Affymetrix GeneChip microRNA 4.0 arrays. The pathological significance of top miRNA discoveries from the human studies were validated in an in vitro cell system and in an animal model of CKD.
MATERIALS AND METHODS
Discovery Studies in CKD Patients
Sample collection.
The study was approved in advance by the George Washington University Institutional Review Board for human research, and all subjects provided informed written consent. The electronic medical record system was used to prescreen and select 28 CKD patients with a minimum follow-up of six months. The exclusion criteria were acute kidney injury, active infection, cirrhosis, class III or IV heart failure, human immunodeficiency virus or hepatitis B or C infection, cancer, active or recent immunosuppression, polycystic kidney disease, or pregnancy. Venous blood was collected from all patients and processed immediately to obtain plasma, which was aliquoted and stored at −80°C until analysis. Midday urine samples were obtained and processed within 2 h. Processing involved centrifugation at 2,000 g for 10 min. The supernatant was removed and stored at −80°C until analysis. Clinical data including serum creatinine and 24-h urine protein were recorded. GFR was estimated (eGFR) using the abbreviated Modification of Diet in Renal Disease equation (18).
Measurement of biomarkers of fibrosis.
Procollagen III NH2-terminal propeptide (PIIINP) concentrations in plasma and urine were measured using high-sensitivity ELISA (Cloud-Clone, TX). The inter- and intra-assay coefficients of variation are <12 and <10%, respectively. Plasma and urine concentrations of type IV collagen were measured using a highly sensitive one-step sandwich enzyme immunoassay kit (Echelon Biosciences. UT). The intra- and interassay coefficients of variation were 4.1 and 5.7%, respectively. The urinary concentrations of creatinine were simultaneously measured by the Jaffe method, using the Creatinine Parameter Assay Kit (R&D Systems, MN). The urinary excretion levels of PIIINP and type IV collagen were expressed as micrograms per gram of creatinine. All procedures were performed according to the instructions provided by the manufacturers.
miRNA profiling.
Plasma and urine exosome miRNAs were isolated using Exosome miRNA isolation kits (Norgen Biotek, Thorold, ON, Canada) according to the manufacturer's protocol. In brief, miRNA was extracted from 1 ml of urine and 0.5 ml of plasma that had been stored at −80°C. RNA extractions were eluted in 100 µl of 2H2O and stored at −80°C until use. The quantity and quality of the miRNA extractions were determined by the Agilent Bioanalyzer 2100 with a small miRNA chip for exosomal miRNA (Agilent Technologies, Columbia, MD). miRNA yields were normalized to nanograms of RNA per milliliter of plasma or urine. RNA was labeled with anAffymetrix FlashTag Biotin HSR RNA Labeling Kit (Affymetrix, Santa Clara, CA) according to standard procedures. Labeled RNA was hybridized to Affymetrix GeneChip microRNA 4.0 arrays and run using a Fluidics Station 450 protocol (FS450_002; Affymetrix). Resultant array data will be deposited in NCBI's Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) per GEO guidelines. Affymetrix CEL files were analyzed in Expression Console using RMA+DMBG (Affymetrix) and then exported to Partek Genomics Suite for further analyses (Partek, St. Louis, MO). Only mature human miRNAs were retained for all downstream analyses. Top miRNA discoveries were validated by qRT-PCR.
qRT-PCR.
A TaqMan MicroRNA Reverse Transcription kit and Universal Mater Mix II (Applied Biosystems, Foster City, CA) were used for quantitative RT-PCR assays of selected miRNA, as described by the manufacturer. For RT reactions, 10 ng of RNA, 1× target-specific RT primer, 3.33 U/ml reverse transcriptase, 3.8 units RNAse inhibitor, 0.15 mM dNTPs, and 1× reaction buffer were run in a total reaction volume of 15 µl and incubated at 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min in a thermocycler (Applied Biosystems). Real-time PCR was performed using an Applied Biosystems 7900HT Sequence Detection System in a 10-μl PCR mixture containing 4.5 µl of RT product, 2× TaqMan Universal PCR Master Mix II, 20× TaqMan Assay, and water to adjust the final volume to 10 µl. All reactions were incubated in a 96-well plate at 95°C for 10 min followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min; all were performed in triplicate. Relative miRNA expression levels were compared via the 2−ΔΔCt method (20). U6B small nuclear RNA gene (RNU6B, Applied Biosystems) was used as an endogenous control (28).
Validation Experiments
The top miRNAs from discovery studies were chosen for validation based on statistical significance and biological plausibility.
Alb/TGFβ Mouse Model of CKD
Alb/TGFβ1 mice were established and characterized as previously described (14, 26). Mouse care was performed under a protocol approved in advance by the NIDDK Animal Care and Use Committee and adhered to the Public Health Service Policy on Humane Care and Use of Laboratory Animals (revised 2002) and the NIH Animal Research Advisory Committee Guidelines, which govern the care of animals in the NIH Intramural Research Program. Male C57BL/6J X CBA F1 mice were used as wild-type (WT) controls. Plasma was collected by retroorbital bleeding, and random urine collections were obtained from male mice at 6 wk of age, either carrying the transgene (n = 9) or WT controls (n = 5). Urine and plasma miRNAs were extracted as described above.
In Vitro Cell Model
The rat renal proximal tubular cell line (NRK52E) was obtained from the American Type Culture Collection and grown in DMEM-LG containing 5% FCS. Prior to TGFβ1 stimulation, the medium was changed to DMEM supplemented with 0.5% FCS, and TGFβ1 (R&D Systems) was subsequently added to the medium at 10 ng/ml on the next day for 4 days. The concentration of TGFβ used is based on previous studies (40). Flow cytometry analysis was used to measure expression of E-cadherin, and qRT- PCR was used to determine the expression levels of miRNAs of interest. E-cadherin serves as a pivotal molecule in EMT and fibrosis (39). Briefly, miRNA was purified from NRK-52E cells, using a PureLink miRNA Isolation Kit (Invitrogen, Carlsbad, CA), and potentially contaminating genomic DNA was digested by RNAse-free DNAse according to the manufacturer’s protocol.
Statistical Analyses
Descriptive statistics for selected demographic and clinical characteristics of the study population stratified by eGFR was generated. Student’s t-test was used for two parametric groups. All probabilities were two tailed. Partek Genomics Suite 6.5 (Partek) was used for array statistics and data visualization analyses for differentially expressed and eGFR-correlated miRNAs. For microarray data, one-way ANOVA testing disease status (eGFR ≥ 30 and eGFR < 30 ml·min−1·1.73 m−2) within tissue type (plasma and urine) was performed (covariates included sex and age), as well as Pearson correlation coefficient tests for relationship to eGFR. A Kruskal-Wallis test was used to compare miRNA expression levels between groups, and Spearman’s rank order correlation was used to test associations between miRNA expression levels and clinical parameters. One-way ANOVA was used to determine the differences in collagen levels between groups.
Rather than using a false discovery rate on the initial miRNA data set, all mature human miRNAs that met the unadjusted P < 0.05 cutoff were carried into biological pathway analysis for targeted mRNAs, and more stringent cutoffs were used during the pathway identification process, which filters out most unrelated findings. The relationships between eGFR and miRNA changes within urine and plasma samples were also assessed using Pearson correlation coefficients, filtering at an unadjusted P < 0.05 cutoff.
Ingenuity Pathway Analysis Suite (IPA; Ingenuity, Redwood City, CA) was used to identify the biological pathways containing the mRNA targets of miRNAs that exhibited differential expression or significant association with eGFR. First, mRNA targets of significant miRNAs were determined using IPA’s miRNA Target Filter, which identifies experimentally validated miRNA-mRNA interactions from TarBase, miRecords, and the peer-reviewed biomedical literature, as well as predicted miRNA-mRNA interactions from TargetScan. A conservative filter was applied using only experimentally validated and highly conserved predicted mRNA targets for each miRNA, as identified by TargetScan within the IPA software. Highly conserved pairings are predicted by TargetScan to repress expression of mRNA target to <40% of “normal” levels. These mRNA targets were carried through to Core Pathway Analyses, which identified common pathways containing the mRNAs in our data set. As noted above, P values were assigned to pathways via a Fischer exact test to reduce the risk of false positive findings from the original ANOVA, as pathway components represent interrelated rather than independent elements. Canonical pathways, novel networks, and common upstream regulators were then queried for overlap with targets from our differentially expressed miRNA gene target list.
RESULTS
miRNA Discovery
The baseline demographic and clinical data of the patients are summarized in Table 1. Among the 28 patients studied, 9 had ≥50% increase in serum creatinine during the preceding 6 mo, with a terminal eGFR of <30 ml·min−1·1.73 cm−2 (eGFR<30 group), and the remaining 19 had slower progression of CKD with a terminal creatinine of eGFR ≥30 ml·min−1·1.73 cm−2 (eGFR≥30 group). Patients in the lower eGFR group were younger and had worse proteinuria (P < 0.01) (Table 1).
Table 1.
Clinical characteristics of study participants
| eGFR ≥30 (n = 19) | eGFR <30 (n = 9) | P Value | |
|---|---|---|---|
| Age (years) | 57.4 ± 11.9 | 55.2 ± 12.2 | 0.65 |
| Sex (M/F) | 6/13 | 3/6 | 0.38 |
| Race (AA/W) | 14/2 | 8/1 | 0.41 |
| Body mass index (kg/m2) | 29.9 ± 6.7 | 33.1 ± 7.1 | 0.27 |
| Proteinuria (g/day) | 1.4 ± 1.1 | 3.0 ± 1.5 | <0.01 |
| Serum creatinine (mg/dl) | 1.5 ± 0.4 | 4.0 ± 2.1 | <0.001 |
| BUN | 24.8 ± 8.0 | 47.6 ± 22.3 | <0.001 |
| Total cholesterol (mg/dl) | 182.1 ± 39.4 | 184 ± 50.5 | 0.93 |
| Use of lipid lowering agents | 94% | 44% | <0.01 |
| Systolic blood pressure | 134.2 ± 17 | 136.3 ± 24.4 | 0.8 |
| (mmHg) | 11.8 ± 1.8 | 10.4 ± 2.1 | 0.11 |
| Hemoglobin (%) | 27% | 50% | 0.66 |
| Patients with diabetes (%) | 50.5 ± 22.2 | 18.7 ± 7.4 | <0.001 |
| GFR (ml/min/1.73 m2) | 18.7 ± 7.4 | 50.5 ± 22.2 | <0.001 |
Values are means ± SE P values depict comparisons by Student’s t-test or χ2 test (for categorical values) between diagnosis groups.
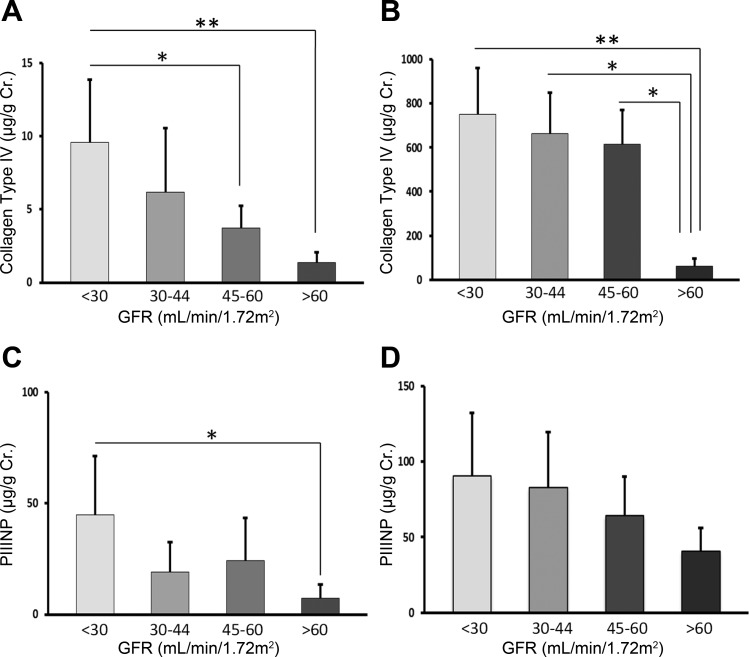
To determine the extent of fibrosis and its association with the decline of kidney function, the plasma and urinary levels of PIIINP and CIV were measured. Fig. 1. There was a significant difference between urine CIV (P = 0.001), plasma CIV (P = 0.002), and urine PIIINP (P = 0.004) levels among the eGFR groups. Although the plasma levels of PIINP tended to increase in those with lower eGFR, the difference was not statistically significant.
Fig. 1.
Urine and plasma levels of collagen type IV (A and B) and procollagen III NH2-terminal propeptide (PIIINP; C and D) in chronic kidney disease (CKD) patients in relation to eGFR levels. The urinary excretion levels of type IV collagen and PIIINP are expressed as micrograms per gram of creatinine. *P < 0.05, ** P < 0.01.
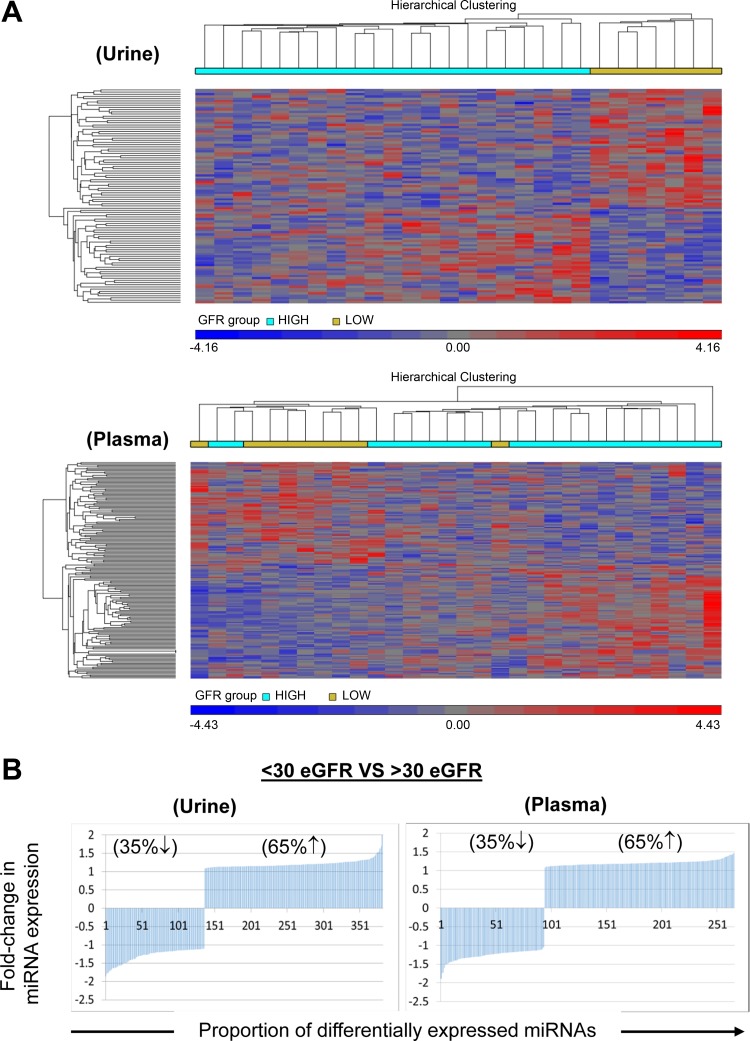
Interrogation of 5,214 human miRNAs in blood and urine samples using the Affymetrix GeneChips revealed that 384 urinary and 266 circulatory miRNAs were differentially expressed between the two eGFR groups. (Fig. 2A) Among these, 248 miRNAs were upregulated and 136 miRNAs were downregulated in urine samples of patients in the eGFR<30 group compared with the eGFR≥30 group (P < 0.05). (Fig. 2B) Similarly, 172 miRNAs were upregulated, and 94 miRNAs were downregulated in plasma samples of patients with eGFR<30 (P < 0.05). The top 10 miRNAs associated with decreased eGFR in plasma and urine are listed in Table 2. A number of miRNAs related to TGFβ were differentially expressed in patients with reduced eGFR, leading us to explore specifically the miRNAs related to TGFβ pathway. IPA mapped 39 urine miRNAs and 15 plasma miRNAs to 6,526 and 3,985 mRNA targets, respectively. As shown in Fig. 3, TGFβ signaling was among the top ranked canonical pathways represented by these mRNAs. Specifically, miR-1281, miR-1825, miR130a-3p, and Let7ap-5p in the urine, and miR-1825p and miR-1281, and miR-423 in the plasma exhibited differential expression. Notably, miR-1825 and miR-1281 were upregulated in both the urine and the plasma of patients with reduced eGFR.
Fig. 2.
Comparative analysis of miRNA expression levels in plasma and urine samples obtained from patients grouped by eGFR. A: heatmap showing unsupervised hierarchical clustering of miRNAs (Pearson correlation, average linkage). Colors represent relative miRNA expression as indicated in the color key for each panel. Brackets on the top margins indicate samples from the same cohort. Samples are in columns, miRNAs in rows. Only miRNAs that survived multiple testing (FDR), and had a fold change >3 or <−3 and P < 0.05 are shown. B: miRNAs with significantly dysregulated expression are graphed for plasma and urine samples. Percentages of up- or downregulated miRNAs are shown with corresponding up or down arrows on each graph.
Table 2.
Top 10 urinary and plasma miRNA discoveries in patients with eGFR <30 ml·min−1·1.73 m−2
| Targeted Genes | Fold Change | P Value | |
|---|---|---|---|
| Urinary miRNAs | |||
| miR-1281 | BMP signaling pathway, TGFβ signaling | 2.2 (↑) | 0.01 |
| miR-1825 | TGFβ signaling, Wnt/β-catenin signaling | 2.1 (↑) | 0.01 |
| miR-1255b-5p | Unknown | 1.9 (↓) | 0.01 |
| MiR-5698 | Unknown | 1.8 (↓) | 0.04 |
| miR-4525 | TGFβ signaling | 1.8 (↓) | 0.02 |
| miR-885-3p | Unknown | 1.8 (↓) | 0.02 |
| miR-6797-3p | Unknown | 1.8 (↑) | 0.03 |
| miR-7846-3p | Unknown | 1.7 (↓) | 0.01 |
| miR-130a-3p | Smad4, TGFβ signaling, Wnt/β-catenin signaling | 1.6 (↑) | 0.01 |
| let7a-5p | TGFβ signaling, Wnt/β-catenin signaling | 1.5 (↓) | 0.03 |
| Circulating miRNAs | |||
| miR-4530 | Unknown | 1.9 (↓) | 0.05 |
| miR-4646-5p | Unknown | 1.7 (↓) | 0.03 |
| miR-423-5p | TGFβ signaling, Wnt/β-catenin signaling | 1.7 (↓) | 0.02 |
| miR-3648 | TGFβ signaling | 1.6 (↓) | 0.01 |
| miR-98-3p | IL-10 | 1.5 (↓) | 0.02 |
| miR-144-5p | TGIF, TGFβ, FGF, VEGF | 1.5 (↑) | 0.01 |
| miR-1825 | TGFβ signaling, Wnt/β-catenin signaling | 1.4 (↑) | 0.01 |
| miR-548ap-5p | TGFβ signaling, BMP signaling pathway | 1.4 (↑) | 0.01 |
| miR-6759-3p | Unknown | 1.4 (↑) | 0.01 |
| miR-3663-3p | Unknown | 1.4 (↑) | 0.04 |
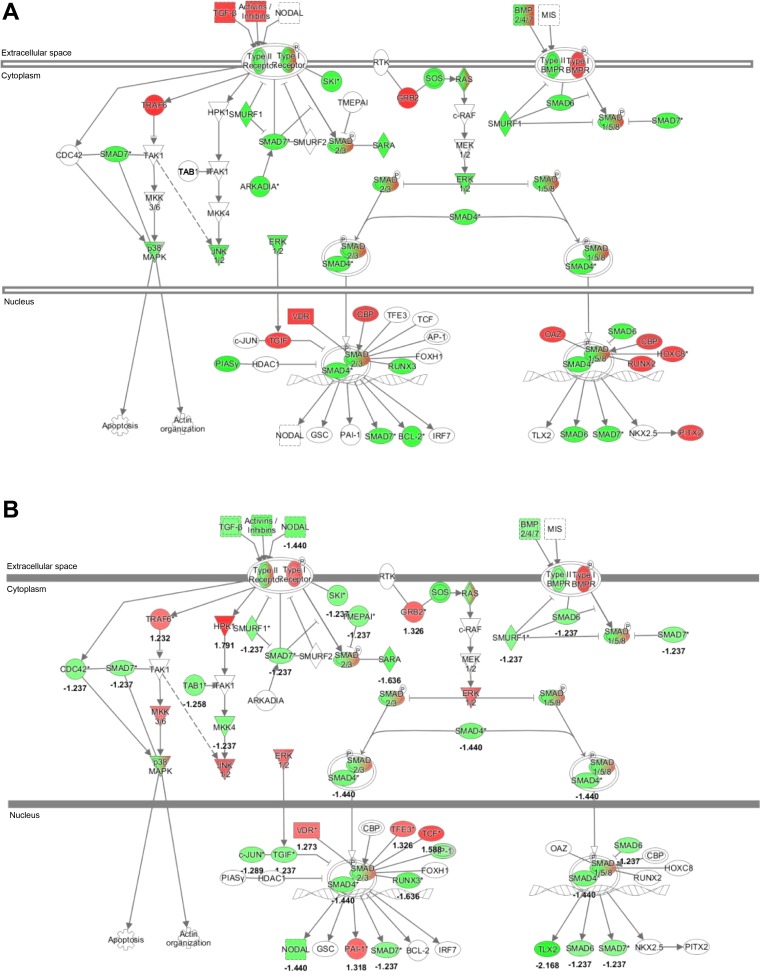
Fig. 3.
Pathway analysis of miRNA discovery. The TGFβ canonical pathway was among the top ranked as determined by Ingenuity Pathways Analysis (IPA) software (Ingenuity, Redwood City, CA). Green indicates predicted downregulation of target transcripts by differentially expressed miRNAs; red indicates predicted upregulation. A, urine; B, plasma. Among the altered miRNAs, expression levels of miR-1825, miR-1281, let-7a, and miR-130a are experimentally-validated.
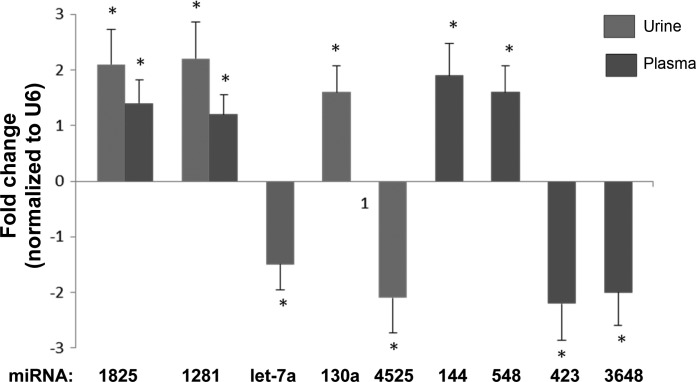
qRT-PCR confirmed that the eGFR<30 group had higher urine levels of miR-1825 (2.1-fold change, P < 0.001), miR-1281 (2.2-fold change, P < 0.001), and miR-130a (1.6-fold change, P < 0.01), and lower urine levels of Let-7a (−1.5-fold change, P < 0.01) compared with those with higher eGFRs. (Fig. 4) Plasma levels of miR-1825 (1.4-fold change, P < 0.05) and miR-1281 (1.6-fold change, P = 0.01) were higher, and that of miR-423-5p (1.7-fold change, P = 0.02) were lower, in the eGFR<30 group compared with their counterparts.
Fig. 4.
qRT-PCR confirmation of the microarray discoveries in CKD patients. Error bars are the SE for each analysis. Selected miRNAs were verified using single-well TaqMan qRT-PCR. The 2−ΔΔCt method was used to calculate relative changes in miRNA expression. Results were normalized to levels of U6 snRNA. Each data point represents mean ± SE. *P < 0.05.
The qRT-PCR analysis of urinary miR-4525 and circulating miR-144, miR-548, miR-423, and miR-3648 from the urine and plasma samples of study participants yielded similar results to those obtained from the microarray analysis, further validating the microarray data (Fig. 4) Specifically, miR-4525 was significantly downregulated in urine of patients with eGFR <30 (2.1-fold change, P < 0.01), miR-144-5p (1.9-fold change, P < 0.02), and miR-548ap-5p (1.6-fold change, P < 0.01) were upregulated in plasma of patients with eGFR <30, and miR-423-5p (2.2-fold change, P < 0.001) and miR-3648 (2.0-fold change, P < 0.01) were significantly downregulated in plasma of patients with eGFR <30.
Strong correlations between, miR-1281 and BUN (r = 0.40, P = 0.04), miR-423 and BUN (r = −0.44, P = 0.02), and miR-423 and serum creatinine (r = −0.6, P = 0.002) were identified. No significant association of miRNAs with the urine albumin/creatinine ratio was noted.
Validation of miRNA Discoveries
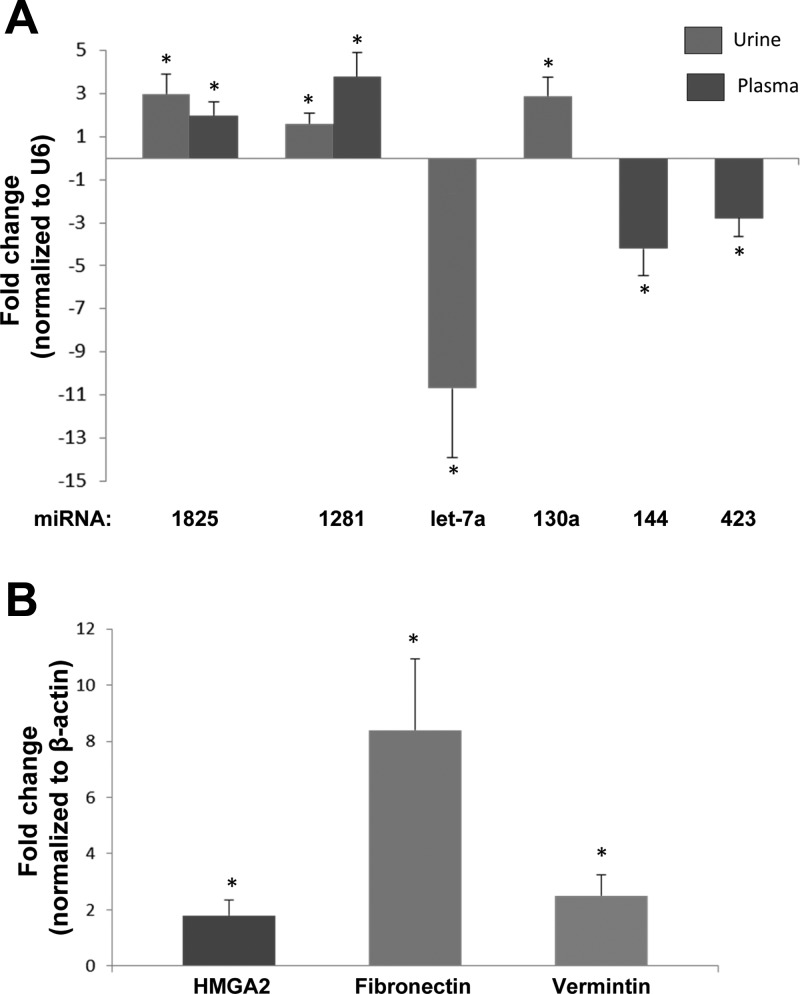
Alb/TGFβ mice overexpress TGF-β and spontaneously develop renal fibrosis and CKD with age. qRT-PCR studies confirmed significant changes in the expression levels of the selected miRNAs in the urine and plasma of Alb/TGF- β1 mice compared with wild-type controls. (Fig. 5) TGFβ1 mice also had significantly higher levels of profibrotic miRNAs, miR-1825 (3.0-fold change, P < 0.001), miR-1281 (1.6-fold change, P < 0.05), and miR-130a (2.9-fold change, P < 0.01), and significantly lower levels of Let-7a (−10.7-fold change, P < 0.001) in the urine. We also found significantly increased levels of miR-1825 (2.0-fold change; P < 0.01), miR-1281 (3.8-fold change, P < 0.01), and miR-130a (3.8-fold change, P < 0.01) in plasma samples from the TGFβ1 mice.
Fig. 5.
A: qRT-PCR verification of microarray discoveries in urine and plasma samples of Alb/TGFβ mice compared with wild-type mice. Error bars are SE for each analysis. Selected miRNAs were verified using single-well TaqMan qRT-PCR. The 2−ΔΔCt method was used to calculate relative changes in miRNA expression. Results were normalized to levels of U6 snRNA. Each data point represents the mean ± SE, P < 0.05. B: qRT-PCR verification of mRNA targets of fibrosis-associated miRNAs. The 2−ΔΔCt method was used to calculate relative changes in mRNA expression. Results were normalized to levels of endogenous β-actin. Each data point represents the mean ± SE. *P < 0.05.
Among other miRNAs, miR-423-5p was also found to be downregulated in the plasma of Alb/TGFβ mice (2.8-fold change, P < 0.001), confirming the observation from the human studies (Fig. 5A). Interestingly, in Alb/TGFβ mouse plasma, miR-144 levels were significantly downregulated (4.2-fold change, P < 0.001), which contrasted its expression levels in human plasma. The remaining miRNAs were either not detected or had no significant change in their expression levels.
We also investigated the expression levels of some potential targets of miRNA from our miRNA profile in the kidney tissues from the Alb/TGFβ mice. Specifically, we measured mRNA expression levels of Hmga2, insulin-like growth factor-1 (Igf1), and IGF1 receptor (Igf1r) genes (all known targets of let-7 miRNA), TGFβ receptor 1 (Tgfbr1) and tissue plasminogen activator (Plat) (both targets of miR-144), EMT marker, fibronectin (Fn1), which is induced by TGFβ1, the mesenchymal marker, vimentin (Vim, a miR-548 target), and Smad4 (miR-130a target).
qRT-PCR analysis of Alb/TGFβ mice kidney mRNA showed significantly increased levels of Hmga2 (1.8-fold change, P = 0.01), fibronectin 1 (8.4-fold change, P < 0.001), and vimentin (2.5-fold change, P < 0.01), compared with normal control mice. No significant change was observed for the expression levels of Tgfb1, Igf1, Igfr11, Plat, or Smad4. (Fig. 5B).
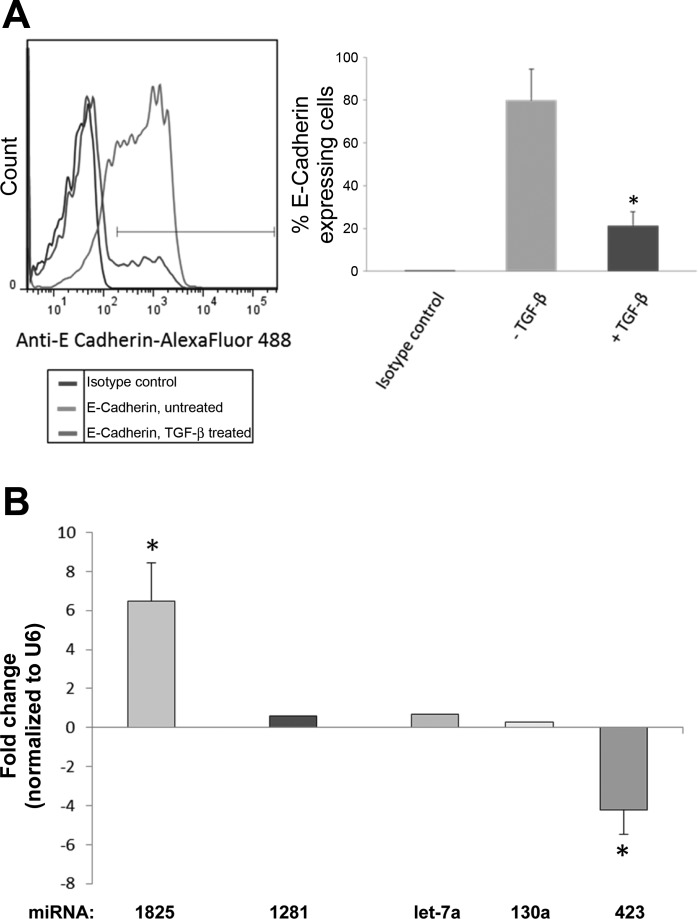
To examine the direct effect of TGFβ on expression of the selected fibrotic miRNAs, we studied the miRNA expression levels in a rat renal proximal tubular cell line (NRK52E) in vitro, after treatment with TGFβ1. EMT phenotype acquisition was confirmed by measuring the expression of E-cadherin in these cells, which was significantly decreased following TGFβ1 treatment, in a time- and dose-dependent manner, peaking at 24 h (Fig. 6). Examination of miRNA expression by qRT- PCR demonstrated upregulation of miR-1825 (6.5-fold change, P = 0.001) and downregulation of miR-423-5p (4.2-fold change, P < 0.001), further validating the finding from human and CKD mouse experiments (Fig. 6). None of the other above miRNA could be detected in NRK52E cells.
Fig. 6.
A: flow cytometry analysis showing expression of E-cadherin in rat renal proximal tubular cell line (NRK52E) with or without TGFβ stimulation. B: real-time PCR of fibrotic miRNAs in NRK52E cells stimulated by TGFβ. Selected miRNAs were verified using single-well TaqMan qRT-PCR. The 2−ΔΔCt method was used to calculate relative changes in miRNA expression. Results were normalized to levels of U6 snRNA. Each data point represents the mean ± SE. *P < 0.05.
DISCUSSION
In this pilot study, we compared the plasma and urinary miRNA profiles in CKD patients with eGFR <30 ml·min−1·1.73 cm−2 and those with eGFR ≥30 ml·min−1·1.73 cm−2 using Affymetrix GeneChip miR 4.0 array and found 384 urinary and 266 circulatory miRNAs were differently expressed. IPA mapped many miRNAs to TGFβ signaling-related mRNA targets. miR-1825 and miR-130a-3p, both related to TGFβ signaling, were upregulated in urine and in plasma of patients with advanced CKD, whereas Let-7a expression, also related to TGFβ signaling, was significantly downregulated in the urine samples. miRNA expression in urine, and plasma of Alb/TGFβ mice generally resembled and confirmed most, although not all, of the observations from the human studies. We also noted that, in response to TGFβ1 challenge, rat renal proximal tubular cells increased expression of miR-1825 and decrease miR-423 expression.
These results suggest that miRNAs are potentially involved in kidney fibrosis and could be a useful biomarker in CKD. Although there is no “direct” evidence linking the identified miRNA with renal fibrosis in humans yet, our study is perhaps novel in being the first to find association between the circulatory and urinary levels of certain miRNAs and advanced stages of CKD. Future mechanistic studies are needed to confirm the involvement of these miRNAs in kidney fibrosis.
Fibrosis is the final common pathway downstream of most kidney injuries, leading to progressive loss of kidney function (27). Fibrosis is characterized by an imbalance between matrix formation and degradation, resulting in excessive accumulation of ECM (9, 21). Increased urinary concentrations of collagen type IV is associated with the decline of kidney function in patients with CKD (1, 8, 25, 29). PIIINP correlates with kidney function and the extent of interstitial fibrosis in kidney biopsies (10, 32). In our study, we found that the urinary levels of PIIINP and collagen IV were elevated in patients with reduced eGFR.
We employed Affymetrix GeneChip miR 4.0 high-density array, in which most miRNA sequences reported in miRBase Release 2.0 are represented for an unbiased discovery of miRNAs associated with CKD progression. Based on our results, 384 urinary and 266 circulatory miRNAs varied by level of kidney function. We found that a number of miRNAs related to TGFβ signaling were dysregulated in patients with advanced CKD. IPA linked a number of miRNA discoveries to the TGFβ pathway.
When IPA was limited to validated targets only, the number of significantly dysregulated miRNAs and their targets were reduced: 13 miRNAs targeting 186 genes in urine, and six miRNAs targeting 200 genes in plasma. As shown in the supplementary Tables S1–S4 supplenentary tables can be found linked to the online version of his paper), the TGFβ pathway continues to be a significant target for both plasma and urine miRNA.
We found that miR-130a-3p was upregulated in the urine, mir-1285 and miR-1825 were upregulated in urine and plasma, whereas Let 7a-5p was downregulated in urine, and miR-423-5P was downregulated in the plasma of patients belonging to the eGFR<30 group. These miRNAs have confirmed/putative targets in the TGFβ pathway (4, 11). miR-130a is involved in cell cycle regulation of granulocytic cells through engagement of Smad4 in the TGFβ pathway (23). Preliminary evidence indicates that miR-130a-3p-regulates epithelial mesenchymal transition in hepatoma cells through inhibition of Smad4 (19).
The TGFβ superfamily consists of highly pleiotropic molecules such as bone morphogenic proteins, growth differentiation factors and glia-derived neurotrophic factors, activins, and inhibins. They are involved in regulation of inflammation, fibrosis, cell apoptosis, and cell proliferation (7). Active TGFβ1 binds to type II TGFβ receptors, leading to phosphorylation of receptor-associated SMADs, which in turn regulate the transcription of target genes. An equilibrium shift of TGFβ/SMAD signaling has been proposed as the underlying mechanism for renal fibrosis (23). Interestingly, TGFβ regulates expression of several microRNAs, some of which are which are positively induced by TGFβ signaling, but others are inhibited, possibly to protect kidneys from progressive fibrosis (23), which is similar to what was observed in our study.
We measured the expression of these miRNAs in Alb/TGFβ transgenic mice, which are transgenic for the murine albumin enhancer/promoter expressing a full-length porcine TGFβ1 gene, likely on the Y chromosome. The transgene is expressed exclusively in the liver, leading to elevated circulating levels of TGFβ1 in as early as 2 wk of age, causing glomerular injury in all males (14, 26). qRT-PCR analysis revealed that the changes in miRNA expression in urine (miR-1825, miR-1281, let7a-5p, and miR-130a) and plasma (miR-1825 and miR-130a) of TGFβ1 mice were similar to those observed in advanced CKD patients. Both the urine and plasma levels of miR-130a were significantly upregulated in advanced CKD patients and also in the Alb/TGFβ transgenic mice. miR-130a targets SMAD4, which is downstream of the TGFβ pathway (11) and involved in renal fibrosis and inflammation (22). Urine samples from CKD patients with decreased eGFR and from Alb/TGFβ mice showed a significant decrease in the expression levels of let-7a miRNA. The members of the let-7 miRNA family regulate cell proliferation and differentiation, and reduced expression of these miRNA can lead to EMT as well as increased cell migration and invasion (3, 33). Reduction in let-7 miRNA levels in endothelium increases expression of TGFβ ligands and receptors, resulting in EMT (4). Downregulation of let-7a in Alb/TGFβ mice in our study points to the existence of a potential negative regulatory mechanism of let-7 expression by TGFβ, as discussed above. This possibility is supported by a previous study, which showed that another member of the let-7 family, let-7d was downregulated in samples from idiopathic pulmonary fibrosis patients, which was sufficient to cause EMT in lung epithelial cells (30).
We noted that the target molecule of let-7a, Hmga2 is overexpressed in the Alb/TGFβ mouse kidney cells. The expression of let-7a is inhibited by TGFβ. Among the genes suppressed by the let-7 family is Hmga2, a structural transcriptional regulator which confers a growth advantage to fibroblasts (41) and a mediator of TGFβ–induced EMT (34). We found a dramatic decrease in let-7a urinary miRNA expression level accompanied by an increase in Hmga2 in Alb/TGFβ mice kidney cells. We did not observe changes in the expression levels of other fibrosis-relevant targets of let-7 such as insulin-like growth factor-1 (Igf1), and IGF1 receptor (Igf1r) (Fig. 5B). Taken together, our results suggest that downregulation of let-7a microRNA may be important in determining the progression of CKD.
We investigated the role of miR-144 in CKD-associated fibrosis. miR-144, one target of which is TGFβ1 according to miRTarBase (12), was previously reported to be downregulated in lung fibrosis (38). Our results showed that miR-144 was downregulated in Alb/TGFβ mouse kidney compared with normal mouse kidney cells. When tested by q-RT-PCR, there was no significant change in the mRNA levels of tissue plasminogen activator (tPA), a potential target of miR-144 in the Alb/TGFβ mice kidney cells. This was in contrast to an increase observed in our advanced CKD patient cohort (GFR<30). The significant downregulation of miR-144 expression in Alb/TGFβ mouse kidney could be explained by the high levels of circulating TGFβ in these mice. Furthermore, recent studies showed that plasma miR-144 is markedly upregulated by erythropoietin (17, 31).
Hager et al. demonstrated that miR-130a is differentially expressed during granulopoiesis and targets Smad4 mRNA, identifying two miR-130a binding sites in the 3-untranslated region of the Smad4 mRNA (11). Overexpression of miR- 130a in HEK-293, A549, and 32Dcl3 cells repressed synthesis of Smad4 protein without affecting Smad4 mRNA level. In our study, although the urinary levels of miR-130a was significantly increased in both the eGFR<30 patients and the in Alb/TGFβ mice, there was no significant changes in the Smad4 mRNA levels in the kidney tissues of Alb/TGFβ mice when examined by q-RT-PCR.
Another one of the fibrosis-associated miRNA from our miR profile, miR-548, has been shown to target and directly bind to the 3′-untranslated region of vimentin mRNA, a marker of the EMT (42). The downregulation of vimentin suppresses the proliferation and invasion of pancreatic cancer cells in vitro and in vivo. In addition, vimentin was inversely correlated with miR-548 expression in pancreatic cancer samples. While the circulating levels of miR-548 expression were not significantly changed in the Alb/TGFβ mice, there was a significant increase in the vimentin expression levels (2.5-fold change, P < 0.01) in the kidney tissues of Alb/TGFβ mice when examined by q-RT-PCR.
We also examined the expression levels of other fibrotic miRNA discovered in human studies in a TGFβ-induced EMT model in renal tubular cells (NRK52E) in vitro. Among the miRNAs tested, only the miR-1825 and miR-423 expression significantly dysregulated in response to TGFβ challenge, suggesting that renal tubular cells are a potential source of circulating and urinary miR-1825, and circulating miR-423. Another miRNA that was identified to be upregulated in advanced CKD by our microarray analysis is miR-1825. Although not much is known about miR-1825, it has been shown to putatively targets member-1 of the discoidin domain family of receptors (DDR1). Since DDR1 is a putative oncogene, it is proposed that miR-1825 might function as a tumor-suppressor, inhibiting DDR1 translation (37).
Our study has a number of strengths. It is the first study exploring the utility of extracellular exosomal miRNA as a biomarker for underlying kidney fibrosis and decreased eGFR. Strengths of the study include 1) unbiased discovery of miRNA using Affymetrix GeneChip microRNA 4.0 arrays to interrogate 30,424 miRNAs, and confirmation of the finding using q-RT-PCR, 2) simultaneous comparison of both plasma and urine miRNA profiles, and 3) validation of the functional significance of miRNAs in a relevant animal model of CKD as well as a cell model. We acknowledge the relatively small sample size, as well as the asymmetrical distribution of eGFR, diabetes, and proteinuria among the study population and the preponderance of patients of African American descent, as potential limitations.
To summarize, this study shows that patients with advanced CKD have a distinct circulating and urine miRNA profile, with a number of these miRNAs linked to kidney fibrosis. We confirmed the pathological significance of the miRNA discovered in human CKD patients in an animal model of CKD and cell-based studies. We found that miR-1825 is consistently linked to progression of CKD, in humans, the animal model of CKD, and a cell model of EMT, which needs further investigation to define its role in kidney fibrosis. Our results provide proof of “association” but not the “mechanism”. Regardless, these findings have their own merit and significance in providing strong miRNA candidates as biological markers of kidney fibrosis and potential targets for future mechanistic and therapeutic studies. If these preliminary findings are confirmed, miRNAs could be used as a surrogate marker of underlying kidney fibrosis and when combined with GFR and proteinuria could improve the prognostication.
GRANTS
D. S. Raj is supported by NIDDK grants 1R01 DK-073665-01A1, 1U01 DK-099924-01, and 1U01 DK-099914-01. J. B. Kopp and S. Srivastav. are supported by the Intramural Research Program, NIDDK, National Institutes of Health, Bethesda, MD. This publication was supported in part by Award Number UL1 TR-001876 from the NIH National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1.Araki S, Haneda M, Koya D, Isshiki K, Kume S, Sugimoto T, Kawai H, Nishio Y, Kashiwagi A, Uzu T, Maegawa H. Association between urinary type IV collagen level and deterioration of renal function in type 2 diabetic patients without overt proteinuria. Diabetes Care 33: 1805–1810, 2010. doi: 10.2337/dc10-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butz H, Rácz K, Hunyady L, Patócs A. Crosstalk between TGFβ signaling and the microRNA machinery. Trends Pharmacol Sci 33: 382–393, 2012. doi: 10.1016/j.tips.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Chang CJ, Hsu CC, Chang CH, Tsai LL, Chang YC, Lu SW, Yu CH, Huang HS, Wang JJ, Tsai CH, Chou MY, Yu CC, Hu FW. Let-7d functions as novel regulator of epithelial-mesenchymal transition and chemoresistant property in oral cancer. Oncol Rep 26: 1003–1010, 2011. doi: 10.3892/or.2011.1360. [DOI] [PubMed] [Google Scholar]
- 4.Chen PY, Qin L, Barnes C, Charisse K, Yi T, Zhang X, Ali R, Medina PP, Yu J, Slack FJ, Anderson DG, Kotelianski V, Wang F, Tellides G, Simons M. FGF regulates TGFβ signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Reports 2: 1684–1696, 2012. doi: 10.1016/j.celrep.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung AC, Huang XR, Meng X, Lan HY. miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol 21: 1317–1325, 2010. doi: 10.1681/ASN.2010020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Y, Yu X, Liu Y, Kriegel AJ, Heng Y, Xu X, Liang M, Ding X. miR-29c is downregulated in renal interstitial fibrosis in humans and rats and restored by HIF- activation. Am J Physiol Renal Physiol 304: F1274–F1282, 2013. doi: 10.1152/ajprenal.00287.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farris AB, Colvin RB. Renal interstitial fibrosis. Curr Opin Nephrol Hypertens 21: 289–300, 2012. doi: 10.1097/MNH.0b013e3283521cfa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furumatsu Y, Nagasawa Y, Shoji T, Yamamoto R, Iio K, Matsui I, Takabatake Y, Kaimori JY, Iwatani H, Kaneko T, Tsubakihara Y, Imai E, Isaka Y, Rakugi H. Urinary type IV collagen in nondiabetic kidney disease. Nephron Clin Pract 117: c160–c166, 2011. doi: 10.1159/000319794. [DOI] [PubMed] [Google Scholar]
- 9.Genovese F, Manresa AA, Leeming DJ, Karsdal MA, Boor P. The extracellular matrix in the kidney: a source of novel non-invasive biomarkers of kidney fibrosis? Fibrogenesis Tissue Repair 7: 4, 2014. doi: 10.1186/1755-1536-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghoul BE, Squalli T, Servais A, Elie C, Meas-Yedid V, Trivint C, Vanmassenhove J, Grünfeld JP, Olivo-Marin JC, Thervet E, Noël LH, Prié D, Fakhouri F. Urinary procollagen III aminoterminal propeptide (PIIINP): a fibrotest for the nephrologist. Clin J Am Soc Nephrol 5: 205–210, 2010. doi: 10.2215/CJN.06610909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Häger M, Pedersen CC, Larsen MT, Andersen MK, Hother C, Grønbæk K, Jarmer H, Borregaard N, Cowland JB. MicroRNA-130a-mediated down-regulation of Smad4 contributes to reduced sensitivity to TGFβ1 stimulation in granulocytic precursors. Blood 118: 6649–6659, 2011. doi: 10.1182/blood-2011-03-339978. [DOI] [PubMed] [Google Scholar]
- 12.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, Jian TY, Lin FM, Chang TH, Weng SL, Liao KW, Liao IE, Liu CC, Huang HD. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res 42, D1: D78–D85, 2014. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest 112: 1776–1784, 2003. doi: 10.1172/JCI200320530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopp JB, Factor VM, Mozes M, Nagy P, Sanderson N, Böttinger EP, Klotman PE, Thorgeirsson SS. Transgenic mice with increased plasma levels of TGF-beta 1 develop progressive renal disease. Lab Invest 74: 991–1003, 1996. [PubMed] [Google Scholar]
- 15.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283: 14910–14914, 2008. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeBleu VS, Taduri G, O’Connell J, Teng Y, Cooke VG, Woda C, Sugimoto H, Kalluri R. Origin and function of myofibroblasts in kidney fibrosis. Nat Med 19: 1047–1053, 2013. doi: 10.1038/nm.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leuenberger N, Jan N, Pradervand S, Robinson N, Saugy M. Circulating microRNAs as long-term biomarkers for the detection of erythropoiesis-stimulating agent abuse. Drug Test Anal 3: 771–776, 2011. doi: 10.1002/dta.370. [DOI] [PubMed] [Google Scholar]
- 18.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D; Modification of Diet in Renal Disease Study Group . A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Ann Intern Med 130: 461–470, 1999. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Li Y, Wang R, Qin S, Liu J, Su F, Yang Y, Zhao F, Wang Z, Wu Q. MiR-130a-3p regulates cell migration and invasion via inhibition of Smad4 in gemcitabine resistant hepatoma cells. J Exp Clin Cancer Res 35: 19, 2016. doi: 10.1186/s13046-016-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408, 2001. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 3: a005058, 2011. doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng XM, Huang XR, Xiao J, Chung AC, Qin W, Chen HY, Lan HY. Disruption of Smad4 impairs TGFβ/Smad3 and Smad7 transcriptional regulation during renal inflammation and fibrosis in vivo and in vitro. Kidney Int 81: 266–279, 2012. doi: 10.1038/ki.2011.327. [DOI] [PubMed] [Google Scholar]
- 23.Meng XM, Tang PM, Li J, Lan HY. TGFβ/Smad signaling in renal fibrosis. Front Physiol 6: 82, 2015. doi: 10.3389/fphys.2015.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med 18: 371–390, 2014. doi: 10.1111/jcmm.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita M, Uchigata Y, Hanai K, Ogawa Y, Iwamoto Y. Association of urinary type IV collagen with GFR decline in young patients with type 1 diabetes. Am J Kidney Dis 58: 915–920, 2011. doi: 10.1053/j.ajkd.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 26.Mozes MM, Böttinger EP, Jacot TA, Kopp JB. Renal expression of fibrotic matrix proteins and of transforming growth factor-beta (TGF-beta) isoforms in TGF-beta transgenic mice. J Am Soc Nephrol 10: 271–280, 1999. [DOI] [PubMed] [Google Scholar]
- 27.Nath KA. Tubulointerstitial changes as a major determinant in the progression of renal damage. Am J Kidney Dis 20: 1–17, 1992. doi: 10.1016/S0272-6386(12)80312-X. [DOI] [PubMed] [Google Scholar]
- 28.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58: 1375–1381, 2009. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 29.Okonogi H, Nishimura M, Utsunomiya Y, Hamaguchi K, Tsuchida H, Miura Y, Suzuki S, Kawamura T, Hosoya T, Yamada K. Urinary type IV collagen excretion reflects renal morphological alterations and type IV collagen expression in patients with type 2 diabetes mellitus. Clin Nephrol 55: 357–364, 2001. [PubMed] [Google Scholar]
- 30.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, Richards T, Selman M, Watkins SC, Pardo A, Ben-Yehudah A, Bouros D, Eickelberg O, Ray P, Benos PV, Kaminski N. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 182: 220–229, 2010. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schumacher YO, Saugy M, Pottgiesser T, Robinson N. Detection of EPO doping and blood doping: the haematological module of the Athlete Biological Passport. Drug Test Anal 4: 846–853, 2012. doi: 10.1002/dta.406. [DOI] [PubMed] [Google Scholar]
- 32.Soylemezoglu O, Wild G, Dalley AJ, MacNeil S, Milford-Ward A, Brown CB, el Nahas AM. Urinary and serum type III collagen: markers of renal fibrosis. Nephrol Dial Transplant 12: 1883–1889, 1997. doi: 10.1093/ndt/12.9.1883. [DOI] [PubMed] [Google Scholar]
- 33.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64: 3753–3756, 2004. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 34.Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol 174: 175–183, 2006. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659, 2007. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 36.Wang B, Ricardo S. Role of microRNA machinery in kidney fibrosis. Clin Exp Pharmacol Physiol 41: 543–550, 2014. doi: 10.1111/1440-1681.12249. [DOI] [PubMed] [Google Scholar]
- 37.Weiner HL, Huang H, Zagzag D, Boyce H, Lichtenbaum R, Ziff EB. Consistent and selective expression of the discoidin domain receptor-1 tyrosine kinase in human brain tumors. Neurosurgery 47: 1400–1409, 2000. doi: 10.1097/00006123-200012000-00028. [DOI] [PubMed] [Google Scholar]
- 38.Xie T, Liang J, Guo R, Liu N, Noble PW, Jiang D. Comprehensive microRNA analysis in bleomycin-induced pulmonary fibrosis identifies multiple sites of molecular regulation. Physiol Genomics 43: 479–487, 2011. doi: 10.1152/physiolgenomics.00222.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng G, Lyons JG, Tan TK, Wang Y, Hsu TT, Min D, Succar L, Rangan GK, Hu M, Henderson BR, Alexander SI, Harris DC. Disruption of E-cadherin by matrix metalloproteinase directly mediates epithelial-mesenchymal transition downstream of transforming growth factor-beta1 in renal tubular epithelial cells. Am J Pathol 175: 580–591, 2009. doi: 10.2353/ajpath.2009.080983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Q, Zeng R, Xu C, Liu L, Chen L, Kou P, Pei G, Bai S, Zhang Y, Li C, Rong S, Han M, Xu G. Erbin inhibits TGFβ1-induced EMT in renal tubular epithelial cells through an ERK-dependent pathway. J Mol Med (Berl) 90: 563–574, 2012. doi: 10.1007/s00109-011-0833-4. [DOI] [PubMed] [Google Scholar]
- 41.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 376: 771–774, 1995. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 42.Zhu S, He C, Deng S, Li X, Cui S, Zeng Z, Liu M, Zhao S, Chen J, Jin Y, Chen H, Deng S, Liu Y, Wang C, Zhao G. MiR-548an, Transcriptionally Downregulated by HIF1/HDAC1, Suppresses Tumorigenesis of Pancreatic Cancer by Targeting Vimentin Expression. Mol Cancer Ther 15: 2209–2219, 2016. doi: 10.1158/1535-7163.MCT-15-0877. [DOI] [PubMed] [Google Scholar]