Abstract
Long intergenic noncoding RNAs (lincRNAs) have emerged as key regulators of cellular functions and physiology. Yet functional lincRNAs often have low, context-specific and tissue-specific expression. We hypothesized that many human monocyte and adipose lincRNAs would be absent in current public annotations due to lincRNA tissue specificity, modest sequencing depth in public data, limitations of transcriptome assembly algorithms, and lack of dynamic physiological contexts. Deep RNA sequencing (RNA-Seq) was performed in peripheral blood CD14+ monocytes (monocytes; average ~247 million reads per sample) and adipose tissue (average ~378 million reads per sample) collected before and after human experimental endotoxemia, an in vivo inflammatory stress, to identify tissue-specific and clinically relevant lincRNAs. Using a stringent filtering pipeline, we identified 109 unannotated lincRNAs in monocytes and 270 unannotated lincRNAs in adipose. Most unannotated lincRNAs are not conserved in rodents and are tissue specific, while many have features of regulated expression and are enriched in transposable elements. Specific subsets have enhancer RNA characteristics or are expressed only during inflammatory stress. A subset of unannotated lincRNAs was validated and replicated for their presence and inflammatory induction in independent human samples and for their monocyte and adipocyte origins. Through interrogation of public genome-wide association data, we also found evidence of specific disease association for selective unannotated lincRNAs. Our findings highlight the critical need to perform deep RNA-Seq in a cell-, tissue-, and context-specific manner to annotate the full repertoire of human lincRNAs for a complete understanding of lincRNA roles in dynamic cell functions and in human disease.
Keywords: RNA-Seq, lincRNA, de novo assembly, human, inflammation
long noncoding RNAs (lncRNAs) have emerged as key regulators of cellular functions and physiology. LncRNAs are typically over 200 nucleotides in length and, like protein-coding mRNAs, often spliced and polyadenylated at their 3′-end. Compared with mRNAs, lncRNAs are shorter and more tissue specific and have fewer exons and lower expression (7, 17, 53). Through nuclear and cytoplasmic interactions with a variety of proteins and other RNA species, lncRNAs can modulate gene and protein functions in epigenetic and posttranscriptional manners and regulate key biological processes, such as X chromosome inactivation (3), embryogenesis (53), cell pluripotency (27), and cell differentiation (71). Recent work has implicated multiple distinct lncRNAs in cell-specific biology including leukocyte inflammatory functions (e.g., LincRNA-Cox2 and THRIL) (9, 39) as well as white (e.g., Firre and ADINR) and brown (lnc-BATE1) adipocyte differentiation (2, 29, 75).
LncRNAs have rapidly evolved in primates relative to other small noncoding RNAs and protein-coding genes and are often not conserved between human and mouse (70). Thus, it is important to perform unbiased discovery of lncRNAs in human biomaterials to identify candidates relevant to human physiology and disease. Several RNA sequencing (RNA-Seq) initiatives, including the human GENCODE project (30) and the human Bodymap (ArrayExpress E-MTAB-513), have annotated thousands of human lncRNAs in multiple cells and tissues. Other studies have annotated lncRNAs in individual tissues (58, 69). Yet these resources are based on moderate sequencing depth. Furthermore, due to limitations of de novo transcriptome assembly algorithms, it is challenging to reconstruct full-length lncRNA transcripts especially for low expression transcripts not previously annotated. Thus, many human lncRNAs, particularly lowly expressed, tissue-specific, and dynamically expressed transcripts not found in other species, may be absent in current public resources. These deficiencies are particularly concerning because a number of lncRNAs reported to be functional in the literature showed low or context- and tissue-specific expression e.g., lnc-DC (72) and cyrano (71).
Here, we addressed these challenges in peripheral blood CD14+ monocytes (monocytes) and human adipose tissue. We focused on these two tissues because of their central roles in diverse complex metabolic and inflammatory diseases. To avoid any overlap of RNA sequence with protein coding mRNA and because of our interest in intergenic regulatory features in human traits, we limited our interrogation to long intergenic noncoding RNAs (lincRNAs), which comprise approximately two-thirds of all known lncRNAs. We performed deep RNA sequencing (RNA-Seq) in the context of human experimental endotoxemia through low-dose intravenous lipopolysaccharide (LPS) administration, a clinically relevant in vivo inflammatory stress (47, 64), to identify dynamically expressed, tissue-specific and physiologically relevant transcripts. Using these strategies as well as a merger of reads from multiple independent human samples, de novo assembly, and stringent filtering criteria, we report hundreds of monocyte and adipose lincRNAs not previously annotated. A subset of these lincRNAs was validated and replicated for their presence and inflammatory induction in independent human samples. Finally, through interrogation of public genome-wide association data, we provide preliminary evidence of disease association of selective unannotated lincRNAs. Our findings highlight the importance of deep RNA-Seq in a cell-, tissue-, and context-specific manner to annotate the full repertoire of human lncRNAs and to understand their role in dynamic cell functions and disease.
MATERIALS AND METHODS
Human subjects.
Healthy noncigarette-smoking participants (n = 294; ~50% women and 30% African American, aged 18–45 yr) on no medications and with no history of clinical disease were enrolled in the Genetics of Evoked-responses to Niacin and Endotoxemia (GENE) study, a University of Pennsylvania Clinical and Translational Research Center protocol as previously described (22). All subjects underwent low-dose experimental endotoxemia (1 ng/kg intravenous LPS) to evoke a systemic inflammatory response. The study protocol was reviewed and approved by the U.Penn institutional review board (IRB), and all participants provided informed consent. We selected a subset of GENE participants at the extremes of inflammatory responsiveness to LPS based on integration of their body temperature, plasma tumor-necrosis factor alpha (TNF-α), and plasma interleukin-6 (IL-6) responses. Top and bottom 5% responders for each sex/race group were selected, i.e., 10 high- and 10 low-responder European Ancestry subjects (50% women) and 6 high and 6 low-responder African ancestry subjects (50% female) for RNA-Seq of monocytes and adipose tissue. Based on the availability of high-quality RNA [minimum of 300 ng input RNA; RNA integrity number (RIN) >6, average RIN = 7.4], monocytes of 8 high responders and 7 low responders and adipose tissue of 13 high responders and 12 low responders were selected for de novo assembly. Based on our previous experience (42, 62), we selected baseline and 4 h post-LPS samples or baseline and 2 h post-LPS samples for sequencing of adipose and monocyte samples, respectively. The clinical characteristics of study subjects are summarized in Supplementary Table S1. (The online version of this article contains supplemental material.)
Library preparation, sequencing, and read alignment.
Sample handling, library preparation, and sequencing were performed as previously described (40, 42, 76). Briefly, CD14+ monocytes were isolated from buffy coat by positive magnetic selection using Dynabeads CD14 (ThermoFisher, Waltham, MA), and frozen in TRIzol reagent (ThermoFisher) for subsequent RNA extraction via standard protocol. RNA from frozen adipose tissue was extracted using the RNeasy Lipid Tissue total RNA mini kit (Qiagen, Valencia, CA). Extracted RNA samples underwent quality control assessment using the Agilent Bioanalyzer (Agilent, Santa Clara, CA). Poly-A library preparation was performed using the TruSeq Stranded mRNA Sample Preparation Kit (Illumina, San Diego, CA) per standard protocols as described (42). The prepared libraries were sequenced on an Illumina HiSeq 2000 at the U.Penn Next Generation Sequencing Core. Monocyte samples generated on average ~247 million reads per sample, and adipose samples generated on average ~378 million reads per sample. RNA-Seq data were aligned to hg19 reference genome using STAR aligner v2.3.0 (18) with known splice junction annotation from GENCODE v14 (30). To ensure high-quality mapping, mapped reads were retained for downstream analysis only if 1) they were uniquely mapped and 2) reads within a pair were mapped to the same chromosome with expected orientations and the distance between the reads was <500,000 bp. Mapping statistics are summarized in Supplementary Table S2.
LincRNA discovery.
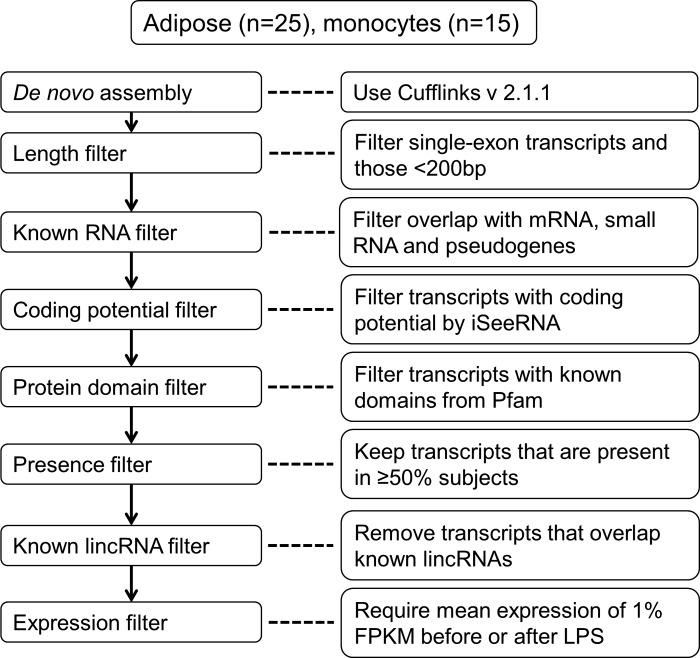
BAM files were merged across samples before de novo assembly to improve the sensitivity of transcriptome reconstruction for low abundance transcripts. As the overall sequencing depth was high in adipose samples, we merged the BAM files in two batches so that the computing time and memory requirement were manageable. Merged BAM files had duplicate reads identified and removed using samtools v0.1.18 (38). We performed de novo transcriptome assembly on the monocyte and two adipose merged BAM files using Cufflinks v2.1.1 (67) requiring a minimum of three fragments per transcript (–min-frags-per-transfrag 3). As the assembled transcripts included both coding and noncoding RNAs, we applied multiple filtering steps to generate a set of lincRNAs. Figure 1 shows the flowchart of our analysis pipeline.
Fig. 1.
Pipeline for discovery of unannotated human peripheral blood CD14+ monocytes and adipose tissue lincRNAs. RNA sequencing (RNA-Seq) data generated from human peripheral blood CD14+ monocytes and adipose tissue were analyzed using the outlined de novo assembly and filtering pipeline. LincRNA, long intergenic noncoding RNA; FPKM, fragments per kilobase of exon per million fragments mapped.
First, the assembled transcripts were size selected to exclude single-exon ones and those <200 bp long since this set of transcripts is more likely to harbor false positive RNA molecules (7). Second, we overlapped the assembled transcriptome with known protein-coding genes, small RNAs, and pseudogenes based on the following annotations: 1) RefSeq genes with accession prefix “NM”; 2) GENCODE v18 coding genes (30); 3) UCSC coding genes; 4) GENCODE v18 microRNA, tRNA, snoRNA, and rRNA; 5) GENCODE v18 pseudogenes, Gerstein group pseudogene database (77), and Vega pseudogene database (74). Transcripts that had an exon overlapping with any of the transcripts from the above annotations were excluded from downstream analysis. Third, we evaluated the coding potential of each transcript using iSeeRNA (65), a support vector machine-based classifier that was designed to separate coding transcripts from noncoding ones. This tool utilized 10 features from three categories: conservation, open reading frame length/proportion, and di- or trinucleotide sequences. Compared with commonly used tools such as PhyloCSF (41), iSeeRNA showed high sensitivity, specificity, and fast running speed that made it an ideal tool for evaluating tens of thousands of transcripts for each sample in our study. Transcripts labeled as “coding” by iSeeRNA were excluded. To identify known protein domains, the remaining transcripts were evaluated by HMMER-3 (25). We translated each transcript in all six possible reading frames and used HMMER-3 to detect any of the known domains from Pfam 27.0 (24). Both Pfam-A and Pfam-B were used, providing a total of 14,831 protein families. Transcripts were excluded if they had a significant Pfam hit. Next, the transcripts assembled from adipose batches were merged by Cuffmerge (67). As lincRNA structures become more complete, it is possible that some of the merged lincRNAs could have coding potential or contain known protein coding domain. Therefore, we performed an additional round of coding potential filtering and known protein coding domain filtering on the merged adipose lincRNA set.
Using multiple data sources, we identified unannotated lincRNAs by eliminating transcripts that overlapped known lincRNA annotations, including 1) RefSeq noncoding genes, 2) GENCODE v18 noncoding RNAs, 3) Ensembl noncoding RNAs, 4) lincRNAs from Cabili et al. (7), 5) MiTranscriptome (33), 6) lincRNAs from Ranzani et al. (58), and 7) lincRNAs from Ballantyne et al. (4). Any transcript not overlapping these existing lincRNA annotations was classified as unannotated lincRNA. To exclude run-on transcripts, we extended each lincRNA by 1 kb on each side and excluded the lincRNA if the extension overlapped existing mRNA or lincRNA annotations. Finally, the expression of unannotated lincRNAs was estimated in monocyte and adipose. To define a set of lincRNAs that were reliably expressed, we applied two expression filters. First, we defined a lincRNA transcript as “present” in one subject if it had fragments per kilobase of exons per million fragments mapped (FPKM) >0 and at least 10 mapped reads before or after LPS administration. Only lincRNA transcripts that were present in at least 50% of the subjects were included. Then we calculated FPKM percentile for each lincRNA based on FPKMs of all lincRNAs and mRNAs in each sample. LincRNAs whose mean expression was above 1 percentile FPKM before or after LPS were considered expressed.
Synteny and conservation of lincRNAs.
We used NCBI HomoloGene release 68 (50) to evaluate synteny of lincRNAs and their neighboring genes in human relative to mouse genomes. The neighboring coding genes of a human lincRNA were first identified and then queried against HomoloGene database for homologous genes. Synteny is defined by a lincRNA having at least one upstream and one downstream homologous gene in mouse. For lincRNAs with synteny in mouse, we evaluated their sequence conservation using BLASTN (8) with the best reciprocal hit strategy. With an E-value cut-off of 1 × 10−10, any hits in mouse genome within the syntenic region were searched against human genome with the same E-value cut-off. Sequences that have significant hits in mouse genome and that could be mapped back to human sequences are best reciprocal hits.
Transposable element in lincRNAs.
Transposable elements (TEs), a class of DNA sequence distributed throughout the human genome (14), are more common in lincRNAs than in protein coding genes (35). TE annotation was obtained from RepeatMasker track (build hg19) from UCSC genome browser (71a). To calculate TE coverage in the genome, we removed gaps from the genome before intersecting TEs. For lincRNAs, we collapsed all exons across each transcript and intersected TEs with the collapsed exons. To test TE coverage in lincRNAs compared with the genome, we applied a sampling approach as described in (36). We randomly sampled 500 times genomic locations matching the size of lincRNA exons using shuffleBed option in Bedtools v2.23.0 (55) and calculated the corresponding TE coverage in each sampled data set. The null distribution was fitted by a normal distribution. P values were calculated from the cumulative density function of the fitted normal distribution.
Chromatin state and histone mark analyses.
ChromHMM tracks from Roadmap Epigenomics (59) were downloaded from the WashU EpiGenome Browser (78). These tracks were created by a multivariate hidden Markov model on multiple chromatin modification data sets to assign chromatin state in the genome (20). The “Adipose Nuclei chromatin state” track was used for adipose lincRNAs, and the “Peripheral Blood Mononuclear Primary cells chromatin state” track was used to compare with monocyte lincRNAs. To examine lincRNA overlap with transcription start site (TSS) states, we extended ± 2.5 kb of each lincRNA start site and counted the number of lincRNAs that overlapped active TSS (state 1) or flanking active TSS (state 2). To examine lincRNA overlap with enhancer states, we took enhancer states (states 6 and 7) and counted the number of lincRNAs that overlapped either state. Bedtools v2.23.0 (55) was used for intersection of genomic coordinates.
H3K4me1 and H3K4me3 ChIP-seq data from adipose and peripheral blood mononuclear cells (PBMCs) were downloaded from WashU EpiGenome Browser in bigWig format. We assessed histone marks profile by counting reads in 10 bp windows within ± 1.5 kb of each lincRNA start site using computeMatrix option from deepTools v1.5.11 (57). Profiles were then visualized using the heatmapper option from deepTools. H3K4me3 heat map was ordered by decreasing mean signal across each region. H3K4me1 heat map was ordered by the same gene orders from the H3K4me3 heat map. The H3K4me1/H3K4me3 ratio was calculated using the mean signal of H3K4me1 and H3K4me3 in each region.
Differential expression of unannotated lincRNAs during endotoxemia.
Differential expression (DE) of unannotated lincRNAs, before and after LPS administration, was tested by Cuffdiff (67) using annotation from RefSeq and unannotated lincRNAs. P values from Cuffdiff output were adjusted by Benjamini-Hochberg method (6) based on the list of expressed unannotated lincRNAs. Due to variation of expression among subjects, we define unannotated lincRNAs as DE if they met the following criteria 1) expressed in at least 80% subjects in either pre-LPS or post-LPS; 2) fold change ≥2; and 3) adjusted P value <0.05. We also highlight three groups of DE lincRNAs with expression ≥5% FPKM, ≥10% FPKM, or ≥20% FPKM in either pre-LPS or post-LPS samples.
LincRNA overlap with trait associated single nucleotide polymorphisms.
Single nucleotide polymorphisms (SNPs) were compiled from genome-wide association study (GWAS) data sets for seven cardiometabolic traits, including plasma levels of triglycerides, total cholesterol, HDL-cholesterol (HDL-C) and LDL-cholesterol (LDL-C) as well as central obesity [waist-hip ratio adjusted for body mass index (BMI)], Type 2 diabetes, and fasting glucose (19, 31, 48, 66). We extended ± 2 kb of each unannotated lincRNA and mapped SNPs to those regions for each trait. To correct for multiple testing, we counted the number of SNPs mapped to unannotated lincRNAs in each trait and adjusted the significance level by dividing 0.05 with the number of SNPs in the unannotated lincRNAs. Linkage disequilibrium (LD) between SNPs was calculated in European subjects from 1000 Genomes phase 3 data (26) and visualized in Haploview (5). In association plots, two neighboring genes of each unannotated lincRNA as well as other genes within the region were plotted using Sushi package (54) from R (56).
Validation and replication of unannotated lincRNAs.
For a small set of unannotated lincRNAs, we performed 1) quantitative (q)PCR validation of their presence and/or LPS modulation in the same GENE samples that had undergone RNA-Seq, 2) qPCR replication of their presence and LPS modulation in different GENE samples, and 3) qPCR in human macrophages and adipocytes to provide additional biological and clinical context for these unannotated lincRNAs. All qPCR assays in our studies were performed using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) on QuantStudio 7 Flex Real-Time PCR System (Applied Biosystem). For each gene target (lincRNA and mRNA), the built-in analysis software of QuantStudio Flex PCR determines cycle threshold based on target abundance and amplification. We uses these default thresholds in all our analyses.
THP1 and primary macrophage were cultured as previously described (52, 76). Briefly, THP1 cells were cultured in RPMI medium containing 10% FBS and differentiated into macrophages by treatment with 100 nM phorbol-12-myristate-13-acetate for 3 days. For human primary macrophage, PBMCs were cultured in RPMI medium containing 20% FBS and 100 ng/ml human macrophage colony-stimulating factor for 7 days to differentiate into macrophage. For M1-phenotype polarization, THP-1 and primary macrophages were treated with 20 ng/ml IFN-γ and 100 ng/mL LPS for 18 h. For human adipocyte differentiation, human adipose stromal cells (ASC) were extracted from freshly isolated subcutaneous adipose tissues and incubated in differentiation media containing 1.7 μM insulin, 250 nM dexamethasone, 500 μM isobutylmethylxanthine, and 2 μM PPAR-γ agonist GW347845 as previously described (4). Mature human adipocytes were subsequently treated with 100 ng/ml LPS for 4 h. For qPCR analysis, total RNA was extracted using RNeasy Plus mini kit (Qiagen). We reverse transcribed 1 μg RNA using High Capacity cDNA kit (Applied Biosystems). Expression levels of lincRNA were assessed by Applied Biosystems QuantStudio 7 Flex Real-Time PCR System. Primers used for lincRNA qPCR analysis are listed in Supplementary Table S3.
We also leveraged independent resources, from Chen et al. (10) for PBMCs and from GTEx (26a) and our own FFAME study (the fenofibrate and omega-3 fatty acid modulation of endotoxemia trial of healthy individuals) data (21, 23) for adipose tissue, to replicate the presence of unannotated lincRNAs in public RNA-Seq data sets. To increase detection sensitivity when using RNA-Seq of moderate depth, we merged RNA-Seq data as described above. Specifically, for Personal omics (10) raw PBMC RNA-Seq data at nine time points of the participant then in good health [Gene Expression Omnibus (GEO) accession number GSE32874, time points 5–8 and 17–21; on average 145 million reads per sample] were downloaded, aligned and filtered and BAM files merged as described above. For GTEx adipose, the five samples with highest sequencing depth (SRA accession number SRR599313, SRR661589, SRR661735, SRR664795, SRR818821; on average 119 million reads per sample) and for FFAME adipose, 14 independent samples (GSE87426, on average 45 million reads per sample), the raw RNA-Seq data were downloaded, aligned, filtered, and merged. LincRNA coverage in these independent PBMC and adipose sample RNA-Seq data was visualized to directly compare with our deeply sequenced GENE monocyte and adipose samples.
Statistical analysis.
Bioinformatics analyses of RNA-Seq data were detailed above. For qPCR validation and replication, data were summarized in mean ± SE. P values were calculated by Student’s t-test. Statistical significance was defined as P < 0.05.
Ethics approval and consent to participate.
The study protocol was approved by the University of Pennsylvania IRB (protocol #805670) and Columbia University’s IRB (protocol #AAAQ6510); all participants provided informed consent.
Availability of data and materials.
GENE adipose RNA-Seq data are available from the National Center for Biotechnology Information (NCBI) GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE76404. GENE monocyte RNA-Seq data are available from NCBI GEO under accession number GSE87290. FFAME adipose RNA-Seq data are available from NCBI GEO under accession number GSE87426.
RESULTS
Discovery and characteristics of unannotated lincRNAs.
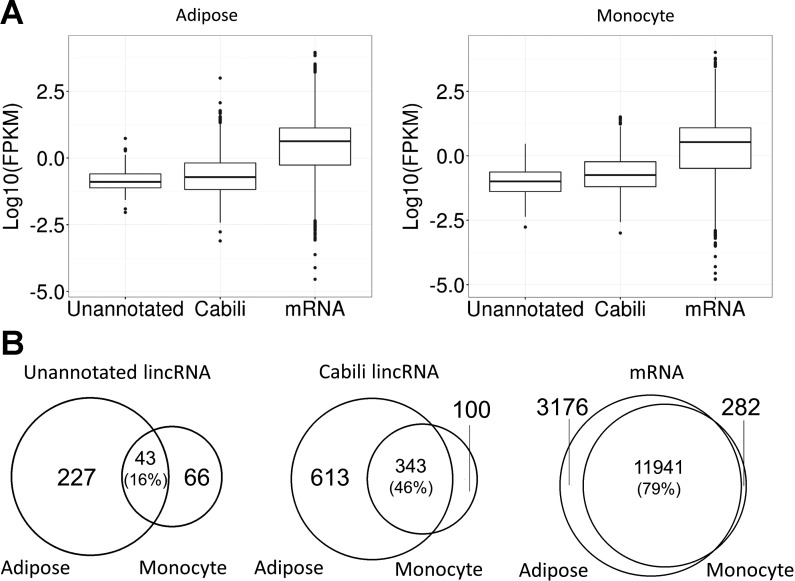
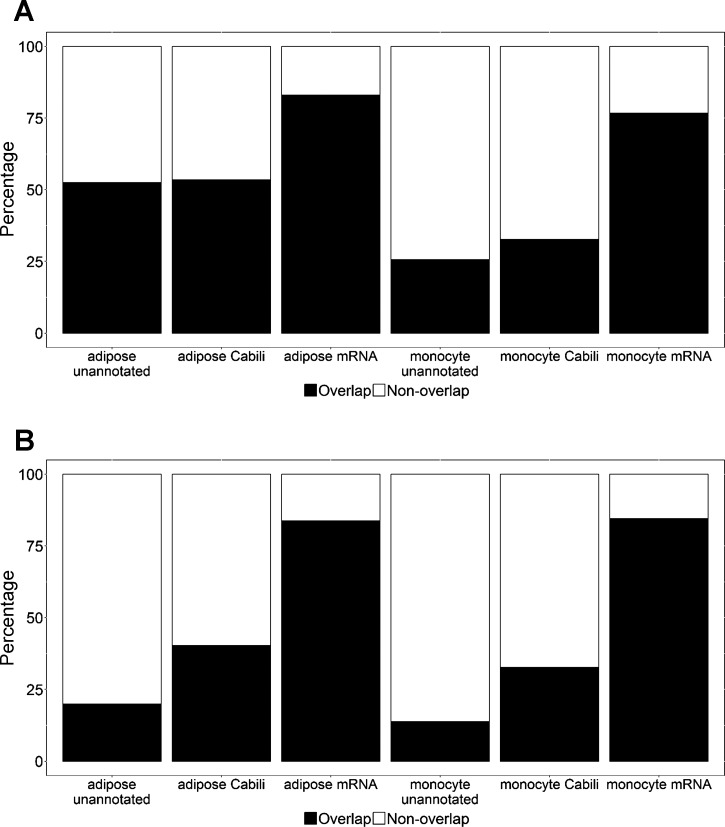
The average mapping rates after filtering was 92.1% in monocyte (227.6 million reads per sample) and 92.7% in adipose (349.7 million per sample) (Supplementary Table S2). Using our pipeline, we found a total of 2,892 and 11,920 lincRNAs present in monocytes and adipose, respectively. After extensive filtering (Fig. 1) we identified 109 unannotated lincRNAs in monocytes (100 pre-LPS, 104 post-LPS, Supplementary Tables S4 and S9) and 270 unannotated lincRNAs in adipose (252 pre-LPS, 246 post-LPS, Supplementary Tables S5 and S10). Unannotated lincRNAs had significantly lower expression than the Cabili stringent set of known lincRNAs (7) in monocytes (Kolmogorov-Smirnov test; P = 6.01 × 10−4) and adipose (Kolmogorov-Smirnov test, P < 2.01 × 10−9) (Fig. 2A). Only 39% unannotated lincRNAs in monocytes and 16% of unannotated lincRNAs in adipose were present in both tissues compared with 77% of Cabili lincRNAs in monocytes and 36% of Cabili lincRNAs in adipose that were present in both tissues. In comparison, expression of protein coding genes is much less tissue specific, with 98% monocyte mRNAs and 79% adipose mRNAs present in both tissues (Fig. 2B).
Fig. 2.
LincRNA expression and tissue overlap. A: boxplot showing expression of unannotated lincRNAs, Cabili set lincRNAs (7), and mRNAs. Unannotated lincRNAs had lower expression than Cabili set lincRNAs in adipose tissue (left, Kolmogorov–Smirnov test, P = 2.01 × 10−9) and in monocytes (right, Kolmogorov–Smirnov test, P = 6.01 × 10−4). B: tissue specificity of unannotated lincRNAs, Cabili set lincRNAs, and mRNAs. Venn diagram shows the number of unannotated lincRNAs (left), Cabili set lincRNAs (middle), and mRNAs (right) expressed in adipose tissue and monocytes. The percentage of overlap based on the number of adipose lincRNAs are shown in parentheses. LincRNAs, especially unannotated lincRNAs, had higher tissue specificity than mRNAs.
Unannotated human lincRNAs have limited synteny and conservation with mouse.
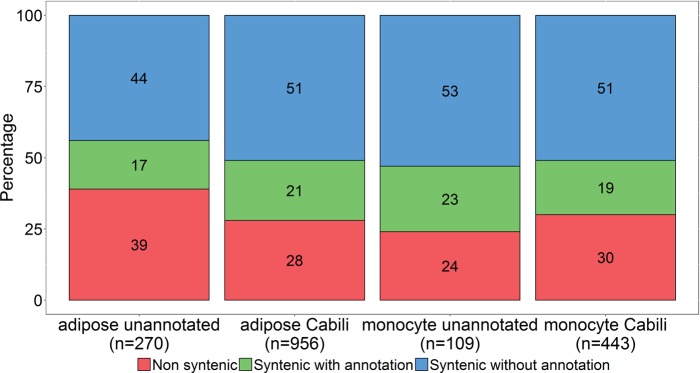
LincRNAs are known to have rapid sequence evolution and limited sequence conservation across vertebrate species (71). Thus, synteny (i.e., conserved neighboring protein coding genes) across species is used as one metric of genomic conservation. We evaluated synteny of unannotated lincRNAs relative to mouse and compared this to the pattern of synteny for known lincRNAs described by Cabili et al. (7). In monocytes, 76% of unannotated lincRNAs were syntenic, of which 30% had mouse lincRNA annotation (49), while for known (7) monocyte lincRNAs, 70% were syntenic in mouse and 26% of these had lincRNAs annotated in mouse (49). We found 61% unannotated lincRNAs in adipose were syntenic, of which 27% had lincRNA annotation in mouse GENCODE annotation M4 (49). Among previously known (7) adipose lincRNAs, 72% were syntenic and 29% of them had lincRNA transcripts annotated in mouse (Fig. 3). Thus, like known lincRNAs, unannotated lincRNAs have limited synteny between human and mouse.
Fig. 3.
Synteny of lincRNAs. The percentages of lincRNAs that are nonsyntenic, syntenic with annotation in mouse, or syntenic without annotation are shown for adipose unannotated lincRNAs, adipose Cabili set lincRNAs, monocyte unannotated lincRNAs, and monocyte Cabili set lincRNAs. Numbers in each bar show the percentage.
For syntenic lincRNAs, we evaluated their sequence conservation. LincRNA sequences in human were searched against mouse genome and the mapped sequences in mouse were then reciprocally searched against human. According to this approach, 20% of unannotated syntenic lincRNAs and 23% of known syntenic lincRNAs were conserved in monocytes, while 23% of unannotated syntenic lincRNAs and 26% of known (7) syntenic lincRNAs were conserved in adipose. In monocytes, the length of conserved sequence averaged 415 bp (range 58–2,678 bp) and 483 bp (range 71–2,332 bp) in unannotated and known lincRNAs, respectively. In adipose, the conserved sequence averaged 532 bp (range 125–2,173 bp) and 459 bp (range 84–1,517 b p) in unannotated and known lincRNAs, respectively. In summary, relative to known human lincRNAs, unannotated human monocyte and adipose lincRNAs had a very similar pattern of synteny (~70%), annotated expression (26–30%), and sequence conservation (20–26%) with mouse.
TEs are prevalent in unannotated lincRNAs.
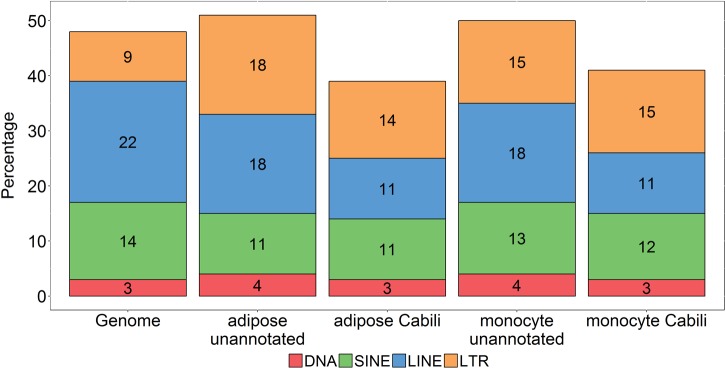
Because of the proposed role of transposable elements in lincRNA function (35), we examined the exonic coverage of four classes of TEs: DNA repeat elements (DNA), short interspersed nuclear elements (SINE), long interspersed nuclear elements (LINE), and long terminal repeat elements (LTR) in our unannotated lincRNAs as well as in the Cabili set of known lincRNAs. The TE coverage in both monocyte and adipose unannotated lincRNAs was higher than in known lincRNAs (Fig. 4). Relative to the whole genome, the total TE coverage in unannotated lincRNAs was similar, while the LTR class was more prevalent in unannotated lincRNAs, consistent with recent reports of enrichment of LTRs in lincRNAs (36). The LTR class, comprising ERV1, ERVK, ERVL, and ERVL-MaLR elements, has been shown to regulate transcription by functioning as alternative promoters (11). Overall, compared with the whole genome, retroviral elements showed 1.6-fold greater coverage in monocyte unannotated lincRNAs (P = 9.16 × 10−5) and 1.9-fold greater coverage in adipose unannotated lincRNAs (P = 8.78 × 10−20). When tested individually, ERVL-MaLR and ERV1 were significantly enriched in monocyte unannotated lincRNAs, while all retroviral elements showed a significant enrichment in adipose unannotated lincRNAs (Table 1). In both monocyte and adipose unannotated lincRNAs, ERVL-MaLR and ERV1 were the most abundant retroviral elements.
Fig. 4.
Prevalence of transposable elements in lincRNAs. The percentages of lincRNA bases that overlap each of the 4 categories of transposable elements are shown for adipose unannotated lincRNAs, adipose Cabili set lincRNAs, monocyte unannotated lincRNAs, and monocyte Cabili set lincRNAs. Whole genome coverage of transposable elements are shown for comparison. DNA, DNA repeat elements; SINE, short interspersed nuclear elements; LINE, long interspersed nuclear elements; LTR, long terminal repeat elements. Numbers within each bar show the percentage.
Table 1.
LTR element coverage in unannotated lincRNAs
| Adipose Tissue |
Monocytes |
|||||||
|---|---|---|---|---|---|---|---|---|
| LTR family | Total Bases That Overlap LTR, n | Total Length of lincRNAs, bp | % lincRNA Bases Occupied by LTR | P Value | Total Bases That Overlap LTR, n | Total Length of lincRNAs, bp | % lincRNA Bases Occupied by LTR | P Value |
| ERV1 | 66,921 | 1,059,097 | 6.32 | 1 × 10−7 | 25,967 | 473,006 | 5.49 | 4.82 × 10−3 |
| ERVL | 38,023 | 3.59 | 5.26 × 10−5 | 11,905 | 2.52 | 0.21 | ||
| ERVK | 11,951 | 1.13 | 1.83 × 10−4 | 4,070 | 0.86 | 0.057 | ||
| ERVL-MaLR | 68,920 | 6.51 | 4.85 × 10−8 | 27,288 | 5.77 | 8.38 × 10−3 | ||
Coverage of ERV1, ERVL, ERVK, and ERVL-MaLR from the long terminal repeat (LTR) class is shown for unannotated lincRNAs in adipose tissue and monocytes. Total length of long intergenic noncoding RNAs (lincRNAs) is the projection of all lincRNA exons. P value for comparison of LTR coverage in unannotated lincRNAs compared with the whole genome is calculated with a sampling approach described in materials and methods.
Unannotated lincRNAs overlap enhancer states more than promoters.
Some lincRNAs may overlap histone promoter marks (7), while others may be characterized by their localization at enhancer marks (37). Using enhancer states defined as per ChromHMM tracks from Roadmap Epigenomics, we found monocyte and adipose unannotated lincRNAs had similar overlap with enhancer states as the Cabili set of known lincRNAs, and both were less than mRNAs (Fig. 5A); e.g., for adipose unannotated lincRNAs, 52.6% overlapped enhancer states compared with 53.6% for the Cabili set of known lincRNAs and 82.9% for mRNAs. Monocytes had similar pattern of TSS overlap as adipose (Fig. 5B); e.g., 20% of adipose unannotated lincRNAs overlapped (±2.5 kb) TSS compared with 40% of known lincRNAs and 84% of mRNAs.
Fig. 5.
LincRNA overlap with enhancer and transcription start site (TSS) states. The percentage of lincRNAs and mRNAs that overlap enhancer states (A) and TSS states (B) within ± 2.5 kb of TSS) are shown in each set. Chromatin states are defined by ChromHMM tracks from Roadmap Epigenomics in adipose nuclei and PBMC. Both unannotated and Cabili set lincRNAs had less overlap with enhancer states and TSS states than mRNAs. Unannotated lincRNAs had similar overlap with enhancer states but less overlap with TSS states compared with Cabili set lincRNAs. PBMC, peripheral blood mononuclear cells.
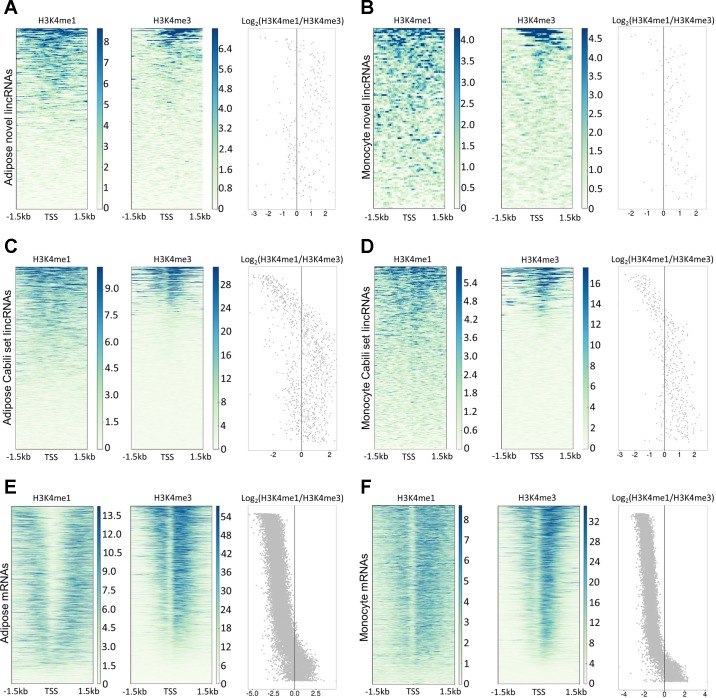
The pattern of limited TSS overlap and moderate enhancer overlap for unannotated lincRNAs in monocytes and adipose suggests that unannotated lincRNAs might be enriched for enhancer RNAs. To explore this possibility, we visualized H3K4me1 and H3K4me3 ChIP-seq data in a ± 1.5 kb window of lincRNA TSSs and calculated the H3K4me1/H3K4me3 ratio (Fig. 6). A high H3K4me1/H3K4me3 ratio suggests enhancer function, while a lower ratio suggests promoters (46, 60). Using a previously proposed cut-off H3K4me1/H3K4me3 ratio of 1.2 (32a), we found that the proportion of putative enhancer RNAs was significantly higher at unannotated lincRNAs than at known lincRNAs in monocytes (68.8 vs. 50.8%, P = 0.0007) and adipose (58.6 vs. 47.8%, P = 0.002) and was markedly greater than for mRNAs in monocytes (8.8%) and adipose (10.3%). As expected, mRNAs were predominantly associated with promoter states in both monocytes and adipose tissue.
Fig. 6.
Histone modification of unannotated lincRNAs, Cabili set lincRNAs, and mRNAs. Histone modifications of H3K4me1 and H3K4me3 in ± 1.5 kb of transcription start site (TSS) are shown for adipose unannotated lincRNAs (A), monocyte unannotated lincRNAs (B), adipose Cabili set lincRNAs (C), monocyte Cabili set lincRNAs (D), adipose mRNAs (E), and monocyte mRNAs (F). Dot plot shows the Log2(H3K4me1/H3K4me3) ratio of each lincRNA. LincRNAs are ordered by decreasing H3K4me3 signal.
Endotoxemia induces unannotated inflammatory lincRNAs in monocytes and adipose.
Table 2 summarizes the number of DE unannotated lincRNAs (fold change ≥2, false discovery rate-adjusted P value <0.05) expressed above the 5th, 10th, and 20th percentile FPKMs; the expression level before and after LPS, fold change and P values of all DE unannotated lincRNAs are presented in Supplementary Tables S6–S8. Consistent with a more direct impact of endotoxemia on circulating cells, there were many more DE lincRNAs in monocytes than adipose, e.g., at the 10th percentile FPKM, 51 DE unannotated lincRNAs in monocytes compared with 5 in adipose. Of these 51 DE unannotated monocyte lincRNAs, 15 had pre-LPS expression below 10th percentile but were markedly induced after LPS stimulation. Such context-specific inflammatory lincRNAs are unlikely to be detected when profiling circulating monocytes at baseline in noninflammatory states even when using deep RNA-Seq.
Table 2.
Experimental endotoxemia modulates expression of unannotated lincRNAs
| Adipose Tissue | High Responder (n = 13) |
Low Responder (n = 12) |
||||
|---|---|---|---|---|---|---|
| FPKM percentile | 5th | 10th | 20th | 5th | 10th | 20th |
| DE lincRNAs, n | 9 | 5 | 2 | 0 | 0 | 0 |
| Monocytes | High Responder (n = 8) |
Low Responder (n = 7) |
||||
|---|---|---|---|---|---|---|
| FPKM percentile | 5th | 10th | 20th | 5th | 10th | 20th |
| DE lincRNAs, n | 61 | 51 | 26 | 4* | 2 | 1 |
The number of endotoxemia-induced differentially expressed (DE) unannotated lincRNAs with fold change ≥2, false discovery rate-adjusted P value <0.05 and presence in ≥80% of participants is shown at the 5th, 10th, and 20th percentile fragments per kilobase of exon per million fragments mapped (FPKM). The number of DE unannotated lincRNAs that are expressed above each percentile in either pre-LPS or post-LPS samples are shown in adipose tissue and monocytes. LPS, lipopolysaccharide.
In monocytes 4 of 4 unannotated lincRNA DE in low responders were also DE in the high-responder samples.
Validation and replication of unannotated lincRNAs.
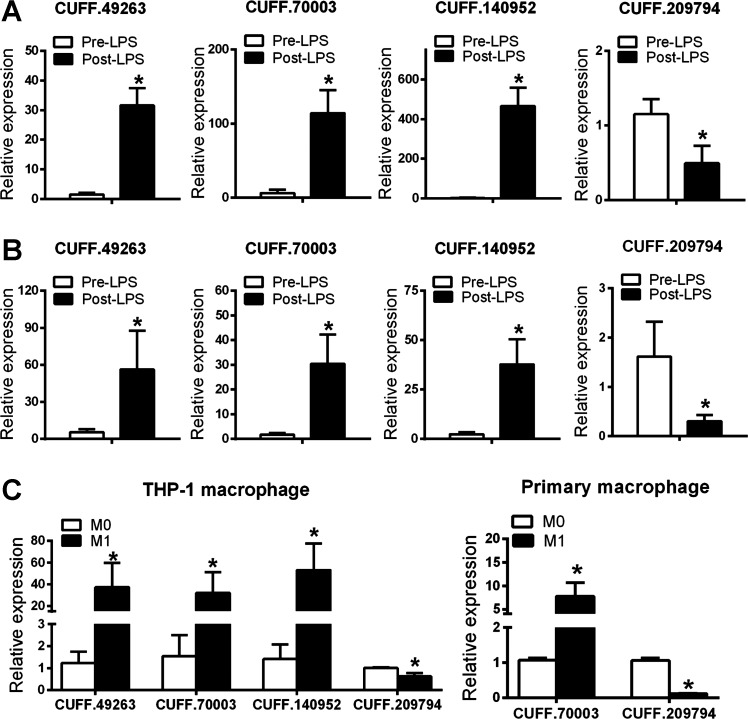
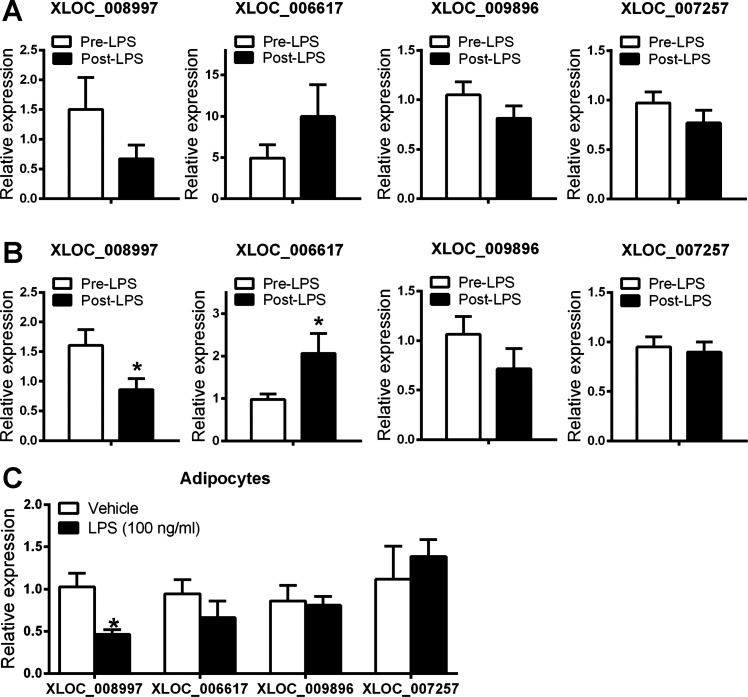
Based on expression level and higher proportion of subjects with expression, as well as LPS induction, we chose a target set of unannotated monocyte and adipose lincRNAs for further examination (Figs. 7 and 8). First, using qPCR we validated both expression and LPS modulation of monocyte lincRNAs in the same GENE samples in which we had discovered these lincRNAs using deep RNA-Seq (Fig. 7A). Adipose lincRNAs were also validated and exhibited the same trends of LPS modulation in the same GENE samples (Fig. 8A). Second, using qPCR we replicated their presence and LPS modulation in monocyte and adipose of independent GENE study participants (Figs. 7B and 8B). Third, also using qPCR, we determined the cellular sources for these monocyte and adipose lincRNAs in human cells of specific relevance to leukocyte and metabolic biology. A number of unannotated monocyte lincRNAs DE between pre-LPS and post-LPS were also expressed in human THP-1 macrophages and primary macrophages and showed similar consistent expression pattern in response to inflammatory stimuli (Fig. 7C). In human ASC-derived adipocytes, all adipose unannotated lincRNAs were detected (Fig. 8C).
Fig. 7.
Validation and replication of selected unannotated monocyte lincRNAs by quantitative (q)PCR. qPCR quantification of selected unannotated lincRNAs in monocytes from the same GENE subjects as for RNA-Seq discovery (n = 7 per group) (A), independent GENE participants (n = 8 per group) (B), and in human THP-1 and primary macrophage (C). Data are presented as means ± SE. *P < 0.05 compared with pre-LPS or M0.
Fig. 8.
Validation and replication of selected unannotated adipose lincRNAs by qPCR. qPCR quantification of selected unannotated lincRNAs in adipose samples from the same GENE subjects for RNA-Seq (n = 9 per group) (A), independent GENE participants (n = 8 per group) (B), and in human adipose stromal cell-derived adipocytes (C). Note that XLOC_008997 and XLOC_006617 were modulated, whereas XLOC_009896 and XLOC_007257 were not modulated by endotoxin in vivo in the original GENE discovery sample. Data are presented as means ± SE. *P < 0.05 compared with pre-LPS or vehicle.
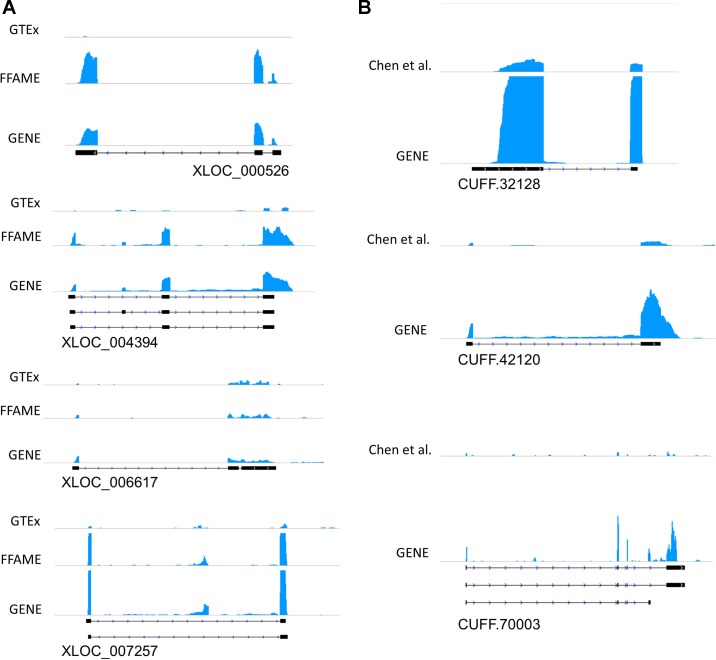
We also leveraged independent resources to visualize and replicate the presence of unannotated lincRNAs in existing public RNA-Seq data sets. Fig. 9 shows examples of lincRNAs illustrative of different levels of expression; Fig. 10 shows coverage plots for individual sample data. The lowest expression transcripts would not have been identified if assembly was done in individual samples but were detected when we merged reads across samples. The coverage plots of public data sets aligned well with our GENE study exons from de novo assembly. CUFF.32128, a higher expression unannotated lincRNA in monocytes, is one example with clear coverage in public data. Other unannotated lincRNAs, however, have less coverage because of limited sequencing depth in the data available from GTEx, FFAME study, and Chen et al. (10). For example, even with merger of the five samples with the highest sequencing depth from GTEx adipose tissue the exon structures of unannotated lincRNAs could not always be demonstrated. This observation illustrates the need for deep RNA-Seq to detect tissue-specific lincRNAs, especially for those with lower expression.
Fig. 9.
Coverage plots of unannotated lincRNAs in public RNA-Seq data sets compared with GENE study RNA-Seq data. RNA-Seq coverage for illustrative unannotated lincRNAs in adipose (A) and monocytes (B). In adipose tissue, each plot shows the coverage from the 1) GENE study, 2) FFAME study, and 3) GTEx. In monocytes, each plot shows the coverage from the 1) GENE study and 2) PBMC from Chen et al. (10). Exon structures are shown below each coverage plot. PBMC, peripheral blood mononuclear cells.
Fig. 10.
RNA-Seq coverage plots of validated unannotated lincRNAs. A–D: RNA-Seq coverage plots of 8 subjects are shown for 4 validated monocyte unannotated lincRNAs. E–H: RNA-Seq coverage plots of 13 subjects are shown for 4 validated adipose unannotated lincRNAs. Each row shows the coverage in pre-LPS and post-LPS on the same scale.
Trait-associated SNPs in unannotated lincRNAs.
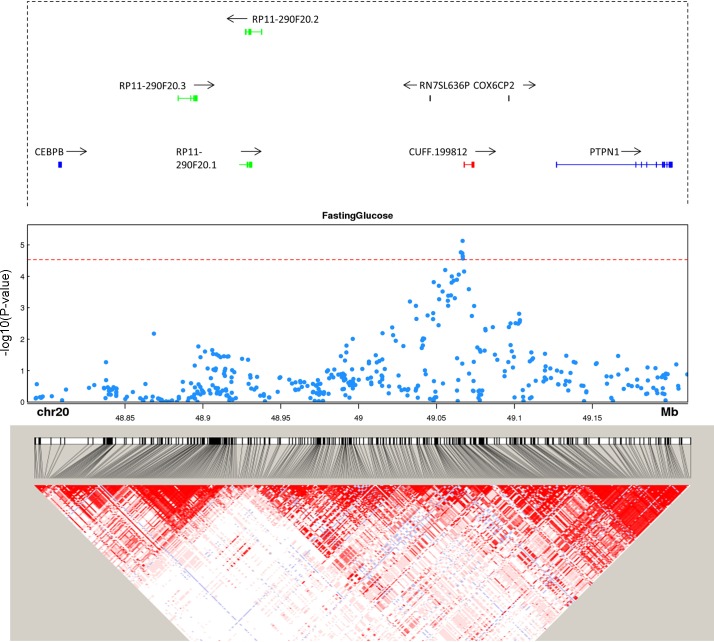
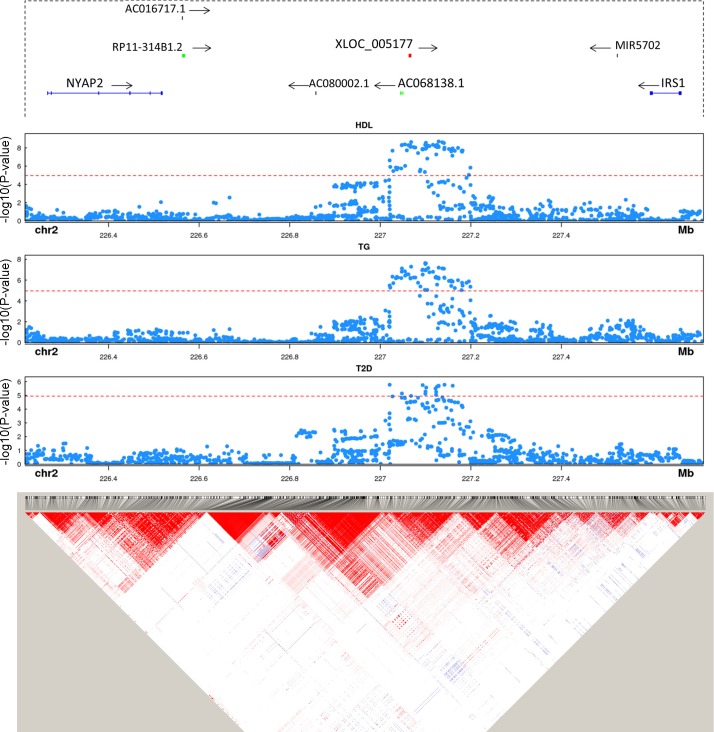
Most complex trait-associated SNPs identified through GWAS lie in intergenic regions (32), and recent work (7, 34), including from our group (4), demonstrates that many trait-associated SNPs reside in noncoding RNAs. To determine if our unannotated adipose and monocyte lincRNAs may harbor trait-associated SNPs, we compiled a list of SNPs associated with seven cardiometabolic traits that might function via adipose or inflammatory leukocytes in mediating trait association. These include central obesity (waist-hip ratio adjusted for BMI), Type 2 diabetes, fasting glucose, plasma triglycerides, plasma total cholesterol, plasma HDL-cholesterol, and plasma LDL-cholesterol. These SNPs were then mapped to ± 2 kb of each unannotated lincRNA. Six of the unannotated lincRNAs harbor trait-associated SNPs. In two instances (CUFF.199812, Fig. 11; XLOC_005177, Fig. 12) the top signals were in the lincRNA and did not have strong LD with SNPs in neighboring coding genes, suggesting a potential functional role in the locus-trait association through cis actions on local candidate protein coding genes or through well described lincRNA trans function in nucleus or cytoplasm.
Fig. 11.
Trait-associated single nucleotide polymorphisms (SNPs) overlap unannotated lincRNA CUFF.199812 at genome-wide association study (GWAS) loci. Gene annotation, trait-associated SNPs, and linkage disequilibrium (LD) structure are shown for CUFF.199812. Genes from GENCODE v19 are plotted in each locus. Protein coding genes are shown in blue, unannotated lincRNAs in red, other lincRNAs in green. All other genes, e.g., COX6CP2, are shown in black. P value cut-off is corrected for the number of SNPs mapped to lincRNAs. LD heat map color scheme is the default D′/LOD scheme in Haploview.
Fig. 12.
Trait-associated SNPs overlap unannotated lincRNA XLOC_005177 at GWAS loci. Gene annotation, trait-associated SNPs, and LD structure are shown for XLOC_005177. Genes from GENCODE v19 are plotted in each locus. Protein coding genes are shown in blue, unannotated lincRNAs in red, other lincRNAs in green. All other genes, e.g., MIR5702, are shown in black. P value cut-off is corrected for the number of SNPs mapped to lincRNAs. LD heat map color scheme is the default D′/LOD scheme in Haploview.
PTPN1 is a protein tyrosine phosphatase known to be a negative regulator of insulin signaling (16). Deletion of PTPN1 in mice has been shown to alleviate diet-induced endoplasmic reticulum stress (16) and to improve glucose homeostasis (15). CUFF.199812 is a monocyte unannotated lincRNA located upstream of PTPN1 and downstream of CEBP-β. Top SNPs associated with fasting glucose are located near the 5′ of the unannotated lincRNA while having no strong LD with SNPs in PTPN1 (Fig. 11). At baseline PTPN1 and CUFF.199812 expression have weak correlation (r = 0.23), while the change of expression induced by LPS had much stronger correlation (r = −0.57). This suggests that the glucose-associated SNPs at this locus might modulate PTPN1 expression through regulation of CUFF.199812.
XLOC_005177 is an adipose unannotated lincRNA located downstream of IRS1. SNPs near IRS1 have been associated with insulin resistance (45), Type 2 diabetes (61), and lipid levels (66). The trait-associated SNPs overlap XLOC_005177 and are in high LD with each other (Fig. 12). However, there is almost no LD between the trait-associated SNPs and SNPs in IRS1. In contrast to a weak baseline correlation (r = 0.012), the correlation of LPS-induced change in expression of IRS1 and XLOC_005177 was much stronger (r = 0.57), suggesting that the top obesity-associated SNPs at the locus might modulate IRS1 expression through regulation of XLOC_005177 expression.
DISCUSSION
We hypothesized that many human monocyte and adipose lincRNAs would be absent in current public annotations because of lincRNA tissue specificity, modest sequencing depth of public data sets, limitations of current transcriptome assembly algorithms, and lack of interrogation in physiological contexts. Here, through deep RNA-Seq, use of a physiological inflammatory stress, sample merging across multiple human samples for de novo assembly, and stringent filtering criteria, we identified hundreds of unannotated human monocyte and adipose lincRNAs. Most unannotated lincRNAs are not conserved between human and rodents and are tissue specific, while many have features of regulated expression and contain TE domains. Specific subsets have enhancer RNA characteristics, are expressed only during inflammatory stress, or overlap GWAS SNPs for complex human traits. These findings underscore the importance of tissue-specific deep RNA-Seq applied in appropriate physiological contexts to discover the full repertoire of human lincRNAs with potential functional roles in human dynamic cell biology and disease.
Largely driven by RNA-Seq technology, there has been an explosion of interest in the role of lncRNAs in cell biology and pathophysiologies. LncRNAs are shorter, have lower expression, and are more tissue specific than protein-coding mRNAs, yet there are now many examples of low-abundance, tissue-selective lncRNAs with critical functional roles in development, differentiation, and disease (9, 71, 72). Indeed, on the basis of predominantly cell and rodent studies, there are rapidly emerging sets of lncRNAs with important functions in regulation of adipocyte differentiation and adipose functions (2, 29, 75), as well as in leukocyte homeostasis and response to inflammatory stresses (9, 39). However, relative to protein-coding genes, lncRNAs have rapidly evolved in primates, and they are often not conserved between humans and classical model organisms such as mouse and zebrafish. Therefore, unlike protein-coding genes, sequence conservation with mouse is not a sensitive strategy for prioritization and functional follow-up of most human lncRNAs; most human functional lincRNAs are likely not conserved at the primary sequence level, and many are not conserved at their genomic position (syntenic) in rodents. Discovery and functional studies using humans and human systems as the model system are required for a complete understanding of important lncRNAs in human physiology and disease. In keeping with recent evolutionary perspectives on lncRNA contributions to species- and tissue-specific functions (71, 73), we found that the majority of unannotated adipose and monocyte lincRNAs are neither syntenic nor conserved in mouse. Furthermore, the lack of synteny did not confer a reduced likelihood of functional features including tissue enrichment, promoter marks, or evidence of enhancer RNA features.
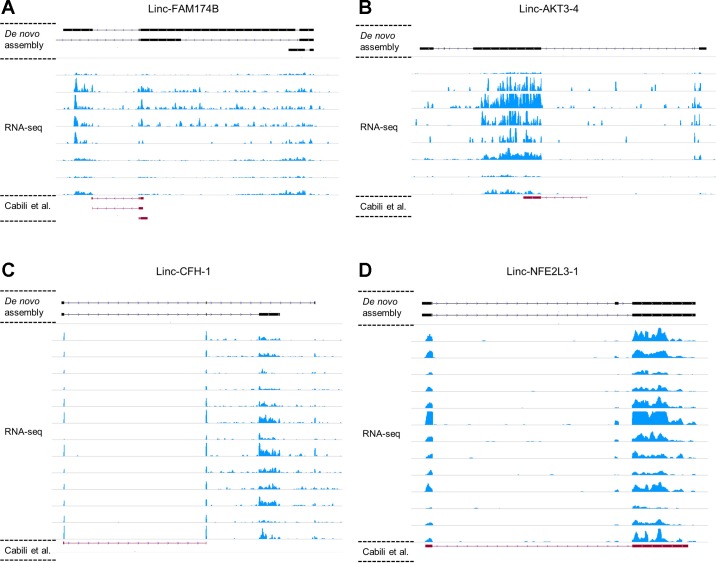
Beyond species specificity, there are several reasons that current public human lincRNA annotations may be lacking. First, the combination of lower expression and tissue specificity means that deep RNA-Seq of each cell and tissue type is required to identify the true diversity of human lncRNAs. Notably, Cap Analysis of Gene Expression (CAGE) from FANTOM5 (12, 43), a technology that identifies promoters, failed to annotate >95% of the unannotated lincRNAs we identified in the same tissue and cell type even for ones we validated by qPCR (data not shown). Second, technical challenges with transcript assembly have yet to be overcome (28, 68), a fact apparent even for well-studied functional lncRNAs where a lack of consistency is apparent between the annotated exon structures in public data and the observed reads in our raw RNA-Seq in adipose and monocytes (e.g., Fig. 13). Thus, the combination of short sequence lengths, low expression, and limitations of transcriptome assembly algorithms means that many low-expression lncRNAs are not annotated even when RNA-Seq reads are present at the locus. In this context, our combination of deep RNA-Seq and merging of within-tissue reads across multiple individuals before de novo assembly enhanced discovery. Third, and of specific clinical relevance, we identified dozens of unannotated monocyte lincRNAs that were induced during in vivo inflammatory stress. Indeed, many endotoxemia-induced lincRNAs were not detected at baseline and likely would not be identified even with deep RNA-Seq in the absence of inflammatory stress. LPS induction of lincRNAs can be mediated by direct signaling of LPS and its receptors, such as TLR4 (44) and caspase-11 (63), and by indirect actions of multiple downstream cytokine-induced inflammatory response (e.g., TNF-α, IL-6) triggered by LPS signaling. Using these strategies, we identified hundreds of unannotated lincRNAs in both tissues even after stringent filtering. Validation and replication of a subset of lincRNAs by qPCR confirmed their presence as did targeted interrogation of public RNA-Seq data of lower sequencing depth, in which unannotated transcripts were discernible despite failing to pass expression filters in the original analysis.
Fig. 13.
Comparison of known lincRNA exon structure and RNA-Seq coverage. Examples of Cabili set lincRNA exon structures are compared with monocyte RNA-Seq data from 8 subjects at baseline (A, B) and adipose RNA-Seq data from 13 subjects at baseline (C, D). Red, exons from Cabili set lincRNAs (red, bottom of each plot); black, exon structures assembled in our study (black, top of each plot). With deep RNA-Seq, de novo assembly is able to identify longer isoforms and exons that are not annotated in public data set.
Our sets of unannotated lincRNAs had several features suggesting their functional significance. First, although modest relative to mRNAs, positional and sequence conservation were similar on average to well-annotated lincRNAs in public data sets. Second, in both adipose and monocyte unannotated lincRNAs, ERVL-MaLR and ERV1 were the most abundant retroviral elements, and such endogenous retroviral LTRs have been shown to regulate transcription and can be functional domains in lincRNAs (11). Third, although unannotated lincRNAs had less overlap with predicted TSS than mRNA and known lincRNAs, they actually had higher rates of overlap with predicted enhancer consistent with their possible role as enhancer RNA molecules. Fourth, several unannotated lincRNAs, induced during physiological stress (endotoxemia) in monocytes and adipose samples, were validated by qPCR and replicated in independent samples, and the directionality of response to inflammatory stimulation was consistent in THP-1-derived macrophages and primary macrophages. Last, several unannotated lincRNAs overlapped SNPs associated with cardiometabolic traits suggesting potential roles in modulating human pathophysiologies. Overall, such features provide complementary support that aids in the prioritization of these unannotated lincRNAs for further functional and clinical interrogation.
This work has several strengths but also limitations. To our knowledge, these are the deepest RNA-Seq and de novo assembly resources for adipose and monocyte lincRNAs that include men and women of both European and African ancestry. Yet, for adipose tissue these data are restricted to the subcutaneous adipose depot. Here, we focused on multi-exon intergenic lncRNA (lincRNAs); future work is required to characterize and unravel trait association for novel anti-sense and other lncRNAs that overlap protein-coding genes. We have validated and replicated only a small subset of prioritized unannotated lincRNAs, although these data provide strong preliminary support for their biological relevance and a prioritization strategy for these unannotated lincRNAs. Mechanistic studies are required to establish functional importance of these less abundant lincRNAs particularly in defining clinical roles for unannotated lincRNAs that overlap trait-associated SNPs.
GRANTS
This work is supported by National Institutes of Health (NIH) Grant R01-HL-113147 (to M. P. Reilly and M. Li) and NIH R01-108600 (to M. Li). X. Zhang is supported by American Diabetes Association Fellowship 1-16-PDF-137. H. Zhang is supported by American Heart Association Postdoctoral Fellowship 15POST25620017 and NIH Grant K99-HL-130574. M. P. Reilly is also supported by R01-HL-111694 and K24-HL-107643. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
C.X., M.L., and M.P.R. conceived and designed research; C.X. and X.Z. analyzed data; C.X. prepared figures; C.X. and M.L. drafted manuscript; C.X., M.L., and M.P.R. edited and revised manuscript; C.X., X.Z., H.Z., J.F.F., Y.W., C.H., M.L., and M.P.R. approved final version of manuscript; X.Z., H.Z., and J.F.F. performed experiments.
Supplementary Material
REFERENCES
- 2.Alvarez-Dominguez JR, Bai Z, Xu D, Yuan B, Lo KA, Yoon MJ, Lim YC, Knoll M, Slavov N, Chen S, Chen P, Lodish HF, Sun L. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab 21: 764–776, 2015. doi: 10.1016/j.cmet.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet 12: 429–442, 2011. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 4.Ballantyne RL, Zhang X, Nuñez S, Xue C, Zhao W, Reed E, Salaheen D, Foulkes AS, Li M, Reilly MP. Genome-wide interrogation reveals hundreds of long intergenic noncoding RNAs that associate with cardiometabolic traits. Hum Mol Genet 25: 3125–3141, 2016. doi: 10.1093/hmg/ddw154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265, 2005. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 6.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B 57: 289–300, 1995. [Google Scholar]
- 7.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927, 2011. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics 10: 421, 2009. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LAJ, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341: 789–792, 2013. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, Cheng Y, Clark MJ, Im H, Habegger L, Balasubramanian S, O’Huallachain M, Dudley JT, Hillenmeyer S, Haraksingh R, Sharon D, Euskirchen G, Lacroute P, Bettinger K, Boyle AP, Kasowski M, Grubert F, Seki S, Garcia M, Whirl-Carrillo M, Gallardo M, Blasco MA, Greenberg PL, Snyder P, Klein TE, Altman RB, Butte AJ, Ashley EA, Gerstein M, Nadeau KC, Tang H, Snyder M. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148: 1293–1307, 2012. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448: 105–114, 2009. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M, Itoh M, Andersson R, Mungall CJ, Meehan TF, Schmeier S, Bertin N, Jørgensen M, Dimont E, Arner E, Schmidl C, Schaefer U, Medvedeva YA, Plessy C, Vitezic M, Severin J, Semple C, Ishizu Y, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer JA, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake JA, Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Burroughs AM, Califano A, Cannistraci CV, Carbajo D, Chen Y, Chierici M, Ciani Y, Clevers HC, Dalla E, Davis CA, Detmar M, Diehl AD, Dohi T, Drabløs F, Edge AS, Edinger M, Ekwall K, Endoh M, Enomoto H, Fagiolini M, Fairbairn L, Fang H, Farach-Carson MC, Faulkner GJ, Favorov AV, Fisher ME, Frith MC, Fujita R, Fukuda S, Furlanello C, Furino M, Furusawa J, Geijtenbeek TB, Gibson AP, Gingeras T, Goldowitz D, Gough J, Guhl S, Guler R, Gustincich S, Ha TJ, Hamaguchi M, Hara M, Harbers M, Harshbarger J, Hasegawa A, Hasegawa Y, Hashimoto T, Herlyn M, Hitchens KJ, Ho Sui SJ, Hofmann OM, Hoof I, Hori F, Huminiecki L, Iida K, Ikawa T, Jankovic BR, Jia H, Joshi A, Jurman G, Kaczkowski B, Kai C, Kaida K, Kaiho A, Kajiyama K, Kanamori-Katayama M, Kasianov AS, Kasukawa T, Katayama S, Kato S, Kawaguchi S, Kawamoto H, Kawamura YI, Kawashima T, Kempfle JS, Kenna TJ, Kere J, Khachigian LM, Kitamura T, Klinken SP, Knox AJ, Kojima M, Kojima S, Kondo N, Koseki H, Koyasu S, Krampitz S, Kubosaki A, Kwon AT, Laros JF, Lee W, Lennartsson A, Li K, Lilje B, Lipovich L, Mackay-Sim A, Manabe R, Mar JC, Marchand B, Mathelier A, Mejhert N, Meynert A, Mizuno Y, de Lima Morais DA, Morikawa H, Morimoto M, Moro K, Motakis E, Motohashi H, Mummery CL, Murata M, Nagao-Sato S, Nakachi Y, Nakahara F, Nakamura T, Nakamura Y, Nakazato K, van Nimwegen E, Ninomiya N, Nishiyori H, Noma S, Noma S, Noazaki T, Ogishima S, Ohkura N, Ohimiya H, Ohno H, Ohshima M, Okada-Hatakeyama M, Okazaki Y, Orlando V, Ovchinnikov DA, Pain A, Passier R, Patrikakis M, Persson H, Piazza S, Prendergast JG, Rackham OJ, Ramilowski JA, Rashid M, Ravasi T, Rizzu P, Roncador M, Roy S, Rye MB, Saijyo E, Sajantila A, Saka A, Sakaguchi S, Sakai M, Sato H, Savvi S, Saxena A, Schneider C, Schultes EA, Schulze-Tanzil GG, Schwegmann A, Sengstag T, Sheng G, Shimoji H, Shimoni Y, Shin JW, Simon C, Sugiyama D, Sugiyama T, Suzuki M, Suzuki N, Swoboda RK, ’t Hoen PA, Tagami M, Takahashi N, Takai J, Tanaka H, Tatsukawa H, Tatum Z, Thompson M, Toyodo H, Toyoda T, Valen E, van de Wetering M, van den Berg LM, Verado R, Vijayan D, Vorontsov IE, Wasserman WW, Watanabe S, Wells CA, Winteringham LN, Wolvetang E, Wood EJ, Yamaguchi Y, Yamamoto M, Yoneda M, Yonekura Y, Yoshida S, Zabierowski SE, Zhang PG, Zhao X, Zucchelli S, Summers KM, Suzuki H, Daub CO, Kawai J, Heutink P, Hide W, Freeman TC, Lenhard B, Bajic VB, Taylor MS, Makeev VJ, Sandelin A, Hume DA, Carninci P, Hayashizaki Y; FANTOM Consortium and the RIKEN PMI and CLST (DGT) . A promoter-level mammalian expression atlas. Nature 507: 462–470, 2014. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384, 2011. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delibegovic M, Bence KK, Mody N, Hong EG, Ko HJ, Kim JK, Kahn BB, Neel BG. Improved glucose homeostasis in mice with muscle-specific deletion of protein-tyrosine phosphatase 1B. Mol Cell Biol 27: 7727–7734, 2007. doi: 10.1128/MCB.00959-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delibegovic M, Zimmer D, Kauffman C, Rak K, Hong EG, Cho YR, Kim JK, Kahn BB, Neel BG, Bence KK. Liver-specific deletion of protein-tyrosine phosphatase 1B (PTP1B) improves metabolic syndrome and attenuates diet-induced endoplasmic reticulum stress. Diabetes 58: 590–599, 2009. doi: 10.2337/db08-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789, 2012. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21, 2013. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Mägi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparsø T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proença C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Böttcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jørgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martínez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orrù M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvänen AC, Tanaka T, Thorand B, Tichet J, Tönjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, DIAGRAM Consortium, GIANT Consortium, Global BPgen, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Ríos M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF, Anders Hamsten on behalf of Procardis Consortium, MAGIC investigators, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42: 105–116, 2010. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216, 2012. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson JF, Mulvey CK, Patel PN, Shah RY, Doveikis J, Zhang W, Tabita-Martinez J, Terembula K, Eiden M, Koulman A, Griffin JL, Mehta NN, Shah R, Propert KJ, Song WL, Reilly MP. Omega-3 PUFA supplementation and the response to evoked endotoxemia in healthy volunteers. Mol Nutr Food Res 58: 601–613, 2014. doi: 10.1002/mnfr.201300368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferguson JF, Patel PN, Shah RY, Mulvey CK, Gadi R, Nijjar PS, Usman HM, Mehta NN, Shah R, Master SR, Propert KJ, Reilly MP. Race and gender variation in response to evoked inflammation. J Transl Med 11: 63, 2013. doi: 10.1186/1479-5876-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferguson JF, Xue C, Hu Y, Li M, Reilly MP. Adipose tissue RNASeq reveals novel gene-nutrient interactions following n-3 PUFA supplementation and evoked inflammation in humans. J Nutr Biochem 30: 126–132, 2016. doi: 10.1016/j.jnutbio.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Res 42, D1: D222–D230, 2014. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39, Suppl: W29–W37, 2011. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR;. A global reference for human genetic variation. Nature 526: 68–74, 2015. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585, 2013. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300, 2011. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510, 2010. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21: 198–206, 2014. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–1774, 2012. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, Thorleifsson G, Zillikens MC, Speliotes EK, Mägi R, Workalemahu T, White CC, Bouatia-Naji N, Harris TB, Berndt SI, Ingelsson E, Willer CJ, Weedon MN, Luan J, Vedantam S, Esko T, Kilpeläinen TO, Kutalik Z, Li S, Monda KL, Dixon AL, Holmes CC, Kaplan LM, Liang L, Min JL, Moffatt MF, Molony C, Nicholson G, Schadt EE, Zondervan KT, Feitosa MF, Ferreira T, Lango Allen H, Weyant RJ, Wheeler E, Wood AR, Estrada K, Goddard ME, Lettre G, Mangino M, Nyholt DR, Purcell S, Smith AV, Visscher PM, Yang J, McCarroll SA, Nemesh J, Voight BF, Absher D, Amin N, Aspelund T, Coin L, Glazer NL, Hayward C, Heard-Costa NL, Hottenga JJ, Johansson A, Johnson T, Kaakinen M, Kapur K, Ketkar S, Knowles JW, Kraft P, Kraja AT, Lamina C, Leitzmann MF, McKnight B, Morris AP, Ong KK, Perry JR, Peters MJ, Polasek O, Prokopenko I, Rayner NW, Ripatti S, Rivadeneira F, Robertson NR, Sanna S, Sovio U, Surakka I, Teumer A, van Wingerden S, Vitart V, Zhao JH, Cavalcanti-Proença C, Chines PS, Fisher E, Kulzer JR, Lecoeur C, Narisu N, Sandholt C, Scott LJ, Silander K, Stark K, Tammesoo ML, Teslovich TM, Timpson NJ, Watanabe RM, Welch R, Chasman DI, Cooper MN, Jansson JO, Kettunen J, Lawrence RW, Pellikka N, Perola M, Vandenput L, Alavere H, Almgren P, Atwood LD, Bennett AJ, Biffar R, Bonnycastle LL, Bornstein SR, Buchanan TA, Campbell H, Day IN, Dei M, Dörr M, Elliott P, Erdos MR, Eriksson JG, Freimer NB, Fu M, Gaget S, Geus EJ, Gjesing AP, Grallert H, Grässler J, Groves CJ, Guiducci C, Hartikainen AL, Hassanali N, Havulinna AS, Herzig KH, Hicks AA, Hui J, Igl W, Jousilahti P, Jula A, Kajantie E, Kinnunen L, Kolcic I, Koskinen S, Kovacs P, Kroemer HK, Krzelj V, Kuusisto J, Kvaloy K, Laitinen J, Lantieri O, Lathrop GM, Lokki ML, Luben RN, Ludwig B, McArdle WL, McCarthy A, Morken MA, Nelis M, Neville MJ, Paré G, Parker AN, Peden JF, Pichler I, Pietiläinen KH, Platou CG, Pouta A, Ridderstråle M, Samani NJ, Saramies J, Sinisalo J, Smit JH, Strawbridge RJ, Stringham HM, Swift AJ, Teder-Laving M, Thomson B, Usala G, van Meurs JB, van Ommen GJ, Vatin V, Volpato CB, Wallaschofski H, Walters GB, Widen E, Wild SH, Willemsen G, Witte DR, Zgaga L, Zitting P, Beilby JP, James AL, Kähönen M, Lehtimäki T, Nieminen MS, Ohlsson C, Palmer LJ, Raitakari O, Ridker PM, Stumvoll M, Tönjes A, Viikari J, Balkau B, Ben-Shlomo Y, Bergman RN, Boeing H, Smith GD, Ebrahim S, Froguel P, Hansen T, Hengstenberg C, Hveem K, Isomaa B, Jørgensen T, Karpe F, Khaw KT, Laakso M, Lawlor DA, Marre M, Meitinger T, Metspalu A, Midthjell K, Pedersen O, Salomaa V, Schwarz PE, Tuomi T, Tuomilehto J, Valle TT, Wareham NJ, Arnold AM, Beckmann JS, Bergmann S, Boerwinkle E, Boomsma DI, Caulfield MJ, Collins FS, Eiriksdottir G, Gudnason V, Gyllensten U, Hamsten A, Hattersley AT, Hofman A, Hu FB, Illig T, Iribarren C, Jarvelin MR, Kao WH, Kaprio J, Launer LJ, Munroe PB, Oostra B, Penninx BW, Pramstaller PP, Psaty BM, Quertermous T, Rissanen A, Rudan I, Shuldiner AR, Soranzo N, Spector TD, Syvanen AC, Uda M, Uitterlinden A, Völzke H, Vollenweider P, Wilson JF, Witteman JC, Wright AF, Abecasis GR, Boehnke M, Borecki IB, Deloukas P, Frayling TM, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, North KE, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, Hirschhorn JN, Assimes TL, Wichmann HE, Thorsteinsdottir U, van Duijn CM, Stefansson K, Cupples LA, Loos RJ, Barroso I, McCarthy MI, Fox CS, Mohlke KL, Lindgren CM; MAGIC . Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet 42: 949–960, 2010. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 106: 9362–9367, 2009. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.IIott NE, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L, Goodhead I, Hertz-Fowler C, Heger A, Hall N, Donnelly LE, Sims D, Lindsay MA. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun 5: 3979, 2014. doi: 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47: 199–208, 2015. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, Chu LW, Zhang Z, Zhao H, Zheng SL, Isaacs WB, Xu J. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis 32: 1655–1659, 2011. doi: 10.1093/carcin/bgr187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470, 2013. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol 13: R107, 2012. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187, 2010. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079, 2009. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci USA 111: 1002–1007, 2014. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin J, Hu Y, Nunez S, Foulkes AS, Cieply B, Xue CY, Gerelus M, Li WJ, Zhang HR, Rader DJ, Musunuru K, Li MY, Reilly MP. Transcriptome-wide analysis reveals modulation of human macrophage inflammatory phenotype through alternative splicing. Arterioscl Throm Vas 36: 1434–1437, 2016. doi: 10.1161/ATVBAHA.116.307573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282, 2011. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Ferguson JF, Xue C, Ballantyne RL, Silverman IM, Gosai SJ, Serfecz J, Morley MP, Gregory BD, Li M, Reilly MP. Tissue-specific RNA-Seq in human evoked inflammation identifies blood and adipose LincRNA signatures of cardiometabolic diseases. Arterioscler Thromb Vasc Biol 34: 902–912, 2014. doi: 10.1161/ATVBAHA.113.303123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, Abugessaisa I, Fukuda S, Hori F, Ishikawa-Kato S, Mungall CJ, Arner E, Baillie JK, Bertin N, Bono H, de Hoon M, Diehl AD, Dimont E, Freeman TC, Fujieda K, Hide W, Kaliyaperumal R, Katayama T, Lassmann T, Meehan TF, Nishikata K, Ono H, Rehli M, Sandelin A, Schultes EA, ’t Hoen PA, Tatum Z, Thompson M, Toyoda T, Wright DW, Daub CO, Itoh M, Carninci P, Hayashizaki Y, Forrest AR, Kawaji H; FANTOM Consortium . Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 16: 22, 2015. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine 42: 145–151, 2008. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu CT, Bielak LF, Prokopenko I, Amin N, Barnes D, Cadby G, Hottenga JJ, Ingelsson E, Jackson AU, Johnson T, Kanoni S, Ladenvall C, Lagou V, Lahti J, Lecoeur C, Liu Y, Martinez-Larrad MT, Montasser ME, Navarro P, Perry JR, Rasmussen-Torvik LJ, Salo P, Sattar N, Shungin D, Strawbridge RJ, Tanaka T, van Duijn CM, An P, de Andrade M, Andrews JS, Aspelund T, Atalay M, Aulchenko Y, Balkau B, Bandinelli S, Beckmann JS, Beilby JP, Bellis C, Bergman RN, Blangero J, Boban M, Boehnke M, Boerwinkle E, Bonnycastle LL, Boomsma DI, Borecki IB, Böttcher Y, Bouchard C, Brunner E, Budimir D, Campbell H, Carlson O, Chines PS, Clarke R, Collins FS, Corbatón-Anchuelo A, Couper D, de Faire U, Dedoussis GV, Deloukas P, Dimitriou M, Egan JM, Eiriksdottir G, Erdos MR, Eriksson JG, Eury E, Ferrucci L, Ford I, Forouhi NG, Fox CS, Franzosi MG, Franks PW, Frayling TM, Froguel P, Galan P, de Geus E, Gigante B, Glazer NL, Goel A, Groop L, Gudnason V, Hallmans G, Hamsten A, Hansson O, Harris TB, Hayward C, Heath S, Hercberg S, Hicks AA, Hingorani A, Hofman A, Hui J, Hung J, Jarvelin MR, Jhun MA, Johnson PC, Jukema JW, Jula A, Kao WH, Kaprio J, Kardia SL, Keinanen-Kiukaanniemi S, Kivimaki M, Kolcic I, Kovacs P, Kumari M, Kuusisto J, Kyvik KO, Laakso M, Lakka T, Lannfelt L, Lathrop GM, Launer LJ, Leander K, Li G, Lind L, Lindstrom J, Lobbens S, Loos RJ, Luan J, Lyssenko V, Mägi R, Magnusson PK, Marmot M, Meneton P, Mohlke KL, Mooser V, Morken MA, Miljkovic I, Narisu N, O’Connell J, Ong KK, Oostra BA, Palmer LJ, Palotie A, Pankow JS, Peden JF, Pedersen NL, Pehlic M, Peltonen L, Penninx B, Pericic M, Perola M, Perusse L, Peyser PA, Polasek O, Pramstaller PP, Province MA, Räikkönen K, Rauramaa R, Rehnberg E, Rice K, Rotter JI, Rudan I, Ruokonen A, Saaristo T, Sabater-Lleal M, Salomaa V, Savage DB, Saxena R, Schwarz P, Seedorf U, Sennblad B, Serrano-Rios M, Shuldiner AR, Sijbrands EJ, Siscovick DS, Smit JH, Small KS, Smith NL, Smith AV, Stančáková A, Stirrups K, Stumvoll M, Sun YV, Swift AJ, Tönjes A, Tuomilehto J, Trompet S, Uitterlinden AG, Uusitupa M, Vikström M, Vitart V, Vohl MC, Voight BF, Vollenweider P, Waeber G, Waterworth DM, Watkins H, Wheeler E, Widen E, Wild SH, Willems SM, Willemsen G, Wilson JF, Witteman JC, Wright AF, Yaghootkar H, Zelenika D, Zemunik T, Zgaga L, Wareham NJ, McCarthy MI, Barroso I, Watanabe RM, Florez JC, Dupuis J, Meigs JB, Langenberg C; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Multiple Tissue Human Expression Resource (MUTHER) Consortium . A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 44: 659–669, 2012. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol 14: R131, 2013. doi: 10.1186/gb-2013-14-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehta NN, McGillicuddy FC, Anderson PD, Hinkle CC, Shah R, Pruscino L, Tabita-Martinez J, Sellers KF, Rickels MR, Reilly MP. Experimental endotoxemia induces adipose inflammation and insulin resistance in humans. Diabetes 59: 172–181, 2010. doi: 10.2337/db09-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan JA, Lindgren CM, Muller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJF, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WHL, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JRB, Platou CGP, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stancakova A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 44: 981– 990, 2012. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mudge JM, Harrow J. Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm Genome 26: 366– 378, 2015. doi: 10.1007/s00335-015-9583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.NCBI. HomoloGene. 2014. [Google Scholar]
- 52.Park EK, Jung HS, Yang HI, Yoo MC, Kim C, Kim KS. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm Res 56: 45–50, 2007. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 53.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, Schier AF. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22: 577–591, 2012. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phanstiel DH, Boyle AP, Araya CL, Snyder MP. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30: 2808–2810, 2014. doi: 10.1093/bioinformatics/btu379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842, 2010. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R Core Team R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2015. [Google Scholar]
- 57.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42, W1: W187–W191, 2014. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M, De Francesco R, Geginat J, Bodega B, Abrignani S, Pagani M. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol 16: 318–325, 2015. doi: 10.1038/ni.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330, 2015. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robertson AG, Bilenky M, Tam A, Zhao Y, Zeng T, Thiessen N, Cezard T, Fejes AP, Wederell ED, Cullum R, Euskirchen G, Krzywinski M, Birol I, Snyder M, Hoodless PA, Hirst M, Marra MA, Jones SJM. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res 18: 1906–1917, 2008. doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rung J, Cauchi S, Albrechtsen A, Shen L, Rocheleau G, Cavalcanti-Proença C, Bacot F, Balkau B, Belisle A, Borch-Johnsen K, Charpentier G, Dina C, Durand E, Elliott P, Hadjadj S, Järvelin MR, Laitinen J, Lauritzen T, Marre M, Mazur A, Meyre D, Montpetit A, Pisinger C, Posner B, Poulsen P, Pouta A, Prentki M, Ribel-Madsen R, Ruokonen A, Sandbaek A, Serre D, Tichet J, Vaxillaire M, Wojtaszewski JFP, Vaag A, Hansen T, Polychronakos C, Pedersen O, Froguel P, Sladek R. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet 41: 1110–1115, 2009. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 62.Shah R, Lu Y, Hinkle CC, McGillicuddy FC, Kim R, Hannenhalli S, Cappola TP, Heffron S, Wang X, Mehta NN, Putt M, Reilly MP. Gene profiling of human adipose tissue during evoked inflammation in vivo. Diabetes 58: 2211–2219, 2009. doi: 10.2337/db09-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514: 187–192, 2014. . [DOI] [PubMed] [Google Scholar]
- 64.Suffredini AF, Fromm RE, Parker MM, Brenner M, Kovacs JA, Wesley RA, Parrillo JE. The cardiovascular response of normal humans to the administration of endotoxin. N Engl J Med 321: 280–287, 1989. doi: 10.1056/NEJM198908033210503. [DOI] [PubMed] [Google Scholar]
- 65.Sun K, Chen X, Jiang P, Song X, Wang H, Sun H. iSeeRNA: identification of long intergenic non-coding RNA transcripts from transcriptome sequencing data. BMC Genomics 14, Suppl 2: S7, 2013. doi: 10.1186/1471-2164-14-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707–713, 2010. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578, 2012. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515, 2010. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, Sarkar MK, Li B, Ding J, Voorhees JJ, Kang HM, Nair RP, Chinnaiyan AM, Abecasis GR, Elder JT. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol 16: 24, 2015. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 154: 26–46, 2013. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550, 2011. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71a.UCSC Genome Browser. [Google Scholar]
- 72.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344: 310–313, 2014. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 73.Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res 24: 616–628, 2014. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res 36, Database: D753–D760, 2008. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiao T, Liu L, Li H, Sun Y, Luo H, Li T, Wang S, Dalton S, Zhao RC, Chen R. Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPα. Stem Cell Reports 5: 856–865, 2015. doi: 10.1016/j.stemcr.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang H, Xue C, Shah R, Bermingham K, Hinkle CC, Li W, Rodrigues A, Tabita-Martinez J, Millar JS, Cuchel M, Pashos EE, Liu Y, Yan R, Yang W, Gosai SJ, VanDorn D, Chou ST, Gregory BD, Morrisey EE, Li M, Rader DJ, Reilly MP. Functional analysis and transcriptomic profiling of iPSC-derived macrophages and their application in modeling Mendelian disease. Circ Res 117: 17–28, 2015. doi: 10.1161/CIRCRESAHA.117.305860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Z, Carriero N, Zheng D, Karro J, Harrison PM, Gerstein M. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics 22: 1437–1439, 2006. doi: 10.1093/bioinformatics/btl116. [DOI] [PubMed] [Google Scholar]
- 78.Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, Koebbe BC, Nielsen C, Hirst M, Farnham P, Kuhn RM, Zhu J, Smirnov I, Kent WJ, Haussler D, Madden PAF, Costello JF, Wang T. The Human Epigenome Browser at Washington University. Nat Methods 8: 989–990, 2011. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GENE adipose RNA-Seq data are available from the National Center for Biotechnology Information (NCBI) GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE76404. GENE monocyte RNA-Seq data are available from NCBI GEO under accession number GSE87290. FFAME adipose RNA-Seq data are available from NCBI GEO under accession number GSE87426.