Abstract
Pulmonary fibrosis is a severe condition with no cure and limited therapeutic options. A better understanding of its pathophysiology is needed. Recent studies have suggested that pulmonary fibrosis may be driven by accelerated aging-related mechanisms. Sirtuins (SIRTs), particularly SIRT1, SIRT3, and SIRT6, are well-known mediators of aging; however, limited data exist on the contribution of sirtuins to lung fibrosis. We assessed the mRNA and protein levels of all seven known sirtuins in primary lung fibroblasts from patients with idiopathic pulmonary fibrosis (IPF) and systemic sclerosis-associated interstitial lung disease (SSc-ILD) in comparison with lung fibroblasts from healthy controls. These unbiased tests revealed a tendency for all sirtuins to be expressed at lower levels in fibroblasts from patients compared with controls, but the greatest decrease was observed with SIRT7. Similarly, SIRT7 was decreased in lung tissues of bleomycin-challenged mice. Inhibition of SIRT7 with siRNA in cultured lung fibroblasts resulted in an increase in collagen and α-smooth muscle actin (α-SMA). Reciprocally, overexpression of SIRT7 resulted in lower basal and TGF-β-induced levels of COL1A1, COL1A2, COL3A1, and α-SMA mRNAs, as well as collagen and α-SMA proteins. Induced changes in SIRT7 had no effect on endogenous TGF-β mRNA levels or latent TGF-β activation, but overexpression of SIRT7 reduced the levels of Smad3 mRNA and protein. In conclusion, the decline in SIRT7 in lung fibroblasts has a profibrotic effect, which is mediated by changes in Smad3 levels.
Keywords: SIRT7, pulmonary fibrosis, idiopathic pulmonary fibrosis, systemic sclerosis, aging
pulmonary fibrosis, a pathologic process with high morbidity and mortality, complicates a variety of pulmonary and systemic diseases (53). Idiopathic pulmonary fibrosis (IPF) and systemic sclerosis-associated interstitial lung disease (SSc-ILD) are two of the most common and severe diffuse parenchymal lung diseases. IPF has a median survival of approximately three years (47), and currently available medications merely slow the decline of lung function without affecting mortality (21). SSc-ILD is the leading cause of death in systemic sclerosis (SSc), an autoimmune disease that results in progressive fibrosis in multiple organs (36, 50, 56). SSc-ILD and IPF may manifest different histologic patterns, but both diseases are thought to share similar pathophysiological features, such as epithelial and endothelial cell injury, aberrant cell death processes, oxidative and endoplasmic reticulum stress, coagulation abnormalities, immune dysregulation, and exaggerated transforming growth factor-β (TGF-β) activation and signaling, all of which result in excessive extracellular matrix (ECM) deposition by activated myofibroblasts, the hallmark of fibrosis (36, 58).
Numerous molecular mediators of pulmonary fibrosis have been identified, including cell surface proteins, intracellular proteins, and soluble molecules, particularly cytokines (58). Pulmonary fibrosis is mediated by multiple pathways, but despite extensive research over the past several decades, its precise cellular and molecular mechanisms remain to be elucidated. New molecules that may play key regulatory roles in the complex networks involved in fibrosis are being identified and studied with the ultimate goal of manipulating pathways to attenuate or prevent its development.
Recent studies (2, 55, 64, 66) have established a connection between fibrosis and sirtuins, a family of histone deacetylases that require NAD+ for their catalytic activity. Sirtuin 1 (SIRT1) mRNA and protein levels have been shown to be decreased in the skin and dermal fibroblasts of patients with SSc, as well as in mouse models of bleomycin-induced skin fibrosis (64, 66). Recently, SIRT3 was reported to be decreased in lung tissue from patients with IPF and SSc (55), as well as skin biopsies and fibroblasts from patients with SSc (2).
Sirtuins, best known for their role in aging, have been shown to prolong mean and maximal life span in many species across all taxonomic groups (yeast, worms, flies, mice, primates) in response to caloric restriction or activation with molecules, such as resveratrol (5, 6, 12, 22, 27, 33, 49, 63). SIRT6 and SIRT7 knockout mice age more quickly and die at a younger age (43, 62) and, in human populations, polymorphisms within the SIRT3 gene have been associated with longevity (3, 4). While sirtuins have been shown to directly increase life span in lower organisms, their role in aging in mammals is more complex and likely related to diverse genomic, cellular, and metabolic processes involved in stress responses, DNA repair, genomic stability, gene expression, cell cycle regulation, proteostasis, and cellular senescence (15, 20, 25, 29, 35).
Sirtuins may be directly relevant to IPF and SSc-ILD, which are now often being considered as aging-related conditions similar to cardiovascular disease, cancer, and neurodegenerative diseases. The incidence and prevalence of IPF, which has a median age at diagnosis of 66 yr, have been shown to increase with age (48). SSc, although not traditionally viewed as a disease of aging, has recently been described as an accelerated aging phenotype (36), with a peak incidence between 45 and 64 yr of age (17) and a significantly higher prevalence of interstitial lung disease in those greater than 65 yr of age (44).
Little published data exist on the role of sirtuins in pulmonary fibrosis, and most studies have focused on SIRT1 and SIRT3 (2, 55, 64, 66). We began to study a connection between sirtuins and lung fibrosis by assessing, in an unbiased fashion, the expression of all seven sirtuins in primary lung fibroblasts from patients with IPF, SSc-ILD, and from healthy controls. We found a significant decline in SIRT7 expression in pulmonary fibroblasts from patients with IPF and SSc-ILD compared with healthy controls. We also began addressing mechanistic implications of this observed difference by investigating the possible effects of SIRT7 on collagen and α-smooth muscle actin (α-SMA) production and its interplay with the profibrotic cytokine TGF-β in primary lung fibroblast cell cultures.
MATERIALS AND METHODS
Patients and controls.
This study was reviewed and approved by the University of Maryland Institutional Review Board. All participants contributing tissue samples to the study consented in writing. In all patients, lung tissues remaining after diagnostic biopsies or lung transplantation were used as a source of primary pulmonary fibroblasts. Normal lungs initially intended but ultimately not used for transplantation were used as a source of normal human lung fibroblasts (NHLF). Other NHLF were commercially purchased from Lonza (Walkersville, MD).
Primary fibroblast cell cultures.
Primary adult pulmonary fibroblasts were expanded in cell culture from the lung tissues of patients with IPF and SSc and from healthy controls. Some of the fibroblast cultures were a gift of Dr. Carol Feghali-Bostwick (Medical University of South Carolina). Additional NHLF, adult normal human dermal fibroblast (NHDF) cultures, and neonatal human dermal fibroblasts were commercially purchased from Lonza. Fibroblasts were cultured in DMEM (Corning Cellgro, Manassas, VA) supplemented with 10% bovine calf serum (BCS; HyClone Laboratories, GE Healthcare Life Sciences, Logan, UT), 2 mM l-glutamine (Gemini Bio-Products, West Sacramento, CA), 1 mM sodium pyruvate, MEM nonessential amino acids solution, and antibiotic-antimycotic (10,000 units/ml penicillin, 10,000 μg/ml streptomycin, and 25 μg/ml amphotericin B), all from GIBCO Life Technologies (Thermo Fisher Scientific, Waltham, MA). Fibroblast cultures were maintained in T75 culture flasks (Nest Biotechnology, Rahway, NJ) in a humidified atmosphere with 5% CO2 at 37°C. For experiments, fibroblast cultures were incubated with DMEM medium containing the same supplements and 0.5% BCS for at least 12 h before testing. Fibroblasts were passaged by washing with filtered PBS, trypsinizing with 2–5 ml 0.25% trypsin-EDTA (GIBCO), and reconstituting cells in DMEM with 10% BCS before transfer to new T75 flasks, dishes, or six-well plates (Nest). In all experiments, fibroblast cell cultures were tested in passages four to seven.
In vivo experiments.
Experiments were performed in wild-type female C57BL/6 mice aged 10–12 wk (The Jackson Laboratory, Bar Harbor, ME). The animals were treated in accordance with a research protocol reviewed and approved by the University of Maryland Institutional Animal Care and Use Committee. Animals were maintained in sterile microisolator cages with sterile rodent feed and water. Daily maintenance of mice was performed at the Baltimore VA Medical Center Research Animal Facility/University of Maryland Animal Facility, which is approved by the Association for Assessment and Accreditation of Laboratory Animal Care. To model pulmonary inflammation and fibrosis, a single dose of 0.075 U of bleomycin (Sigma-Aldrich, St. Louis, MO), diluted in 50 ml of sterile PBS, was delivered to mouse lungs on day 0, as previously reported (37, 38, 46). Briefly, a minor anterior midline neck incision was made to make the trachea visible, a MicroSprayer (Penn-Century, Wyndmoor, Philadelphia, PA) was inserted intratracheally, and the bleomycin solution was instilled. Control mice were instilled with 50 ml of sterile PBS with no additives. On day 14 after bleomycin challenge, mice were euthanized by CO2 asphyxiation followed by cervical dislocation. Immediately postmortem, the lungs were rapidly dissected, snap-frozen in liquid nitrogen, thawed in 0.5 ml of PBS containing a protease inhibitor cocktail (Sigma), and further homogenized.
Transient transfection of primary fibroblast cell cultures.
For SIRT7 overexpression, transfections were performed with an untagged human SIRT7-encoding plasmid, GFP-tagged human SIRT7 plasmid (SIRT7-GFP), and noncoding vehicle control plasmid with a similar backbone, all from Origene (Rockville, MD). For SIRT7 silencing, transfections were performed with human control and SIRT7 siRNA from Santa Cruz Biotechnology (Dallas, TX) and Qiagen (Valencia, CA).
Primary lung fibroblast cultures were grown to ~80% confluency in T75 flasks. Transfections with recombinant plasmids and silencing RNA were performed by electroporation using the Nucleofector II system and the Amaxa basic nucleofector kit for primary mammalian fibroblasts from Lonza. For each transfection, 0.5–1 million cells were electroporated with 1–3 μg of plasmid or 30–50 pmol of siRNA using program A023 and transferred into six-well plates. Fresh media were added 18–24 h later. Cell lysates for subsequent assays, such as Western blot analysis and mRNA analysis, were collected 48–120 h after transfection. For all transfections with SIRT7-encoding plasmids, overexpression of SIRT7 protein was confirmed by Western blot analysis. In some experiments, transfected cells were stimulated by recombinant human (rh) TGF-β1 (R&D Systems, Minneapolis, MN) at a concentration of 5 ng/ml in DMEM with 0.5% BCS 48 h after transfection, and cell lysates were collected 12–72 h later.
Isolation of cytoplasmic and nuclear cell lysates.
NHLF and primary lung fibroblasts from patients with IPF and SSc-ILD were seeded on 100-mm Falcon Corning tissue culture plates (VWR, Radnor, PA) at a density of 1–2 million cells per plate. When the cells reached confluency, cytoplasmic and nuclear cell lysates were isolated using the nuclear extract kit (Active Motif, Carlsbad, CA), according to the manufacturer’s instructions.
RNA isolation, cDNA synthesis, and real-time PCR.
Total RNA was isolated from fibroblasts or homogenized mouse lung tissue using TRIzol reagent (Ambion, Life Technologies, Carlsbad, CA). RNA was isolated by phase separation with 0.2 ml chlorophorm per 1 ml TRIzol followed by precipitation overnight at −20°C with isopropanol. After washing with ethanol, RNA was dissolved in PCR-certified water (Tecknova, Hollister, CA), and RNA concentration and purity were determined using the Nanodrop 2000 Spectrophotometer (Thermo Scientific). Complementary cDNA was synthesized from 1 to 3 μg of RNA using the first-strand cDNA synthesis kit from GE Healthcare. For fibroblast transfections, 1 μg of purified RNA was first treated with DNase (Promega, Madison, WI), and DNase-treated RNA was then reverse transcribed into cDNA using the SuperScript first-strand synthesis kit from Invitrogen Life Technologies (Carlsbad, CA). RT-qPCR was performed with SYBR Green PCR Master Mix (Life Technologies, Thermo Fisher Scientific), primers for human SIRTs 1–7, COL1A1, COL1A2, COL3A1, α-SMA, CTGF, SMAD2, SMAD3, SMAD7, TGFβ-R1, TGFβ-R2, TGF-β, and 18S, primers for mouse SIRT1, SIRT7, and 18S, and the Applied Biosystems StepOne Plus PCR system. All primers were obtained from Qiagen (Valencia, CA).
Plasminogen activator inhibitor-1/luciferase assay for active TGF-β.
PAIL cells (mink lung epithelial cells stably transfected with an expression construct containing a truncated plasminogen activator inhibitor-1 promoter fused to the firefly luciferase reporter gene) were a gift of Dr. Daniel B. Rifkin (New York University School of Medicine). The cells were maintained and used as previously described (1). Stimulation with rhTGF-β (R&D Systems) was used as a positive control, revealing a dose-dependent increase in luminescence in the concentration range 0.05–1.00 ng/ml. Luminescence was measured using a luciferase assay system (Promega), according to the manufacturer’s recommendations.
Immunoblotting, immunohistochemistry, and immunocytochemistry.
Western blots were performed with primary rabbit antibodies to SIRTs 1–3 and 5–7, phosphorylated and total Smad2/3, GAPDH, β-actin, and MEK1/2 from Cell Signaling Technology (Danvers, MA), HDAC2 and α-SMA from Abcam (Cambridge, MA), and collagen type I from Rockland Antibodies and Assays (Limerick, PA).
For Western blot analysis, fibroblast cultures in six-well plates were washed with ice-cold PBS and lysed with 100–200 μl Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA) containing mercaptoethanol (Sigma-Aldrich) in a 1:20 ratio. Samples were reduced and denatured by boiling for 10 min, and electrophoresis was performed using polyacrylamide gels: 10% Bolt Bis-Tris gels (Invitrogen, Thermo Fisher Scientific) for SIRT1–7 primary antibodies and 7.5% Tris-glycine gels (Bio-Rad Laboratories) for collagen I, Smad2/3, or SIRT7 antibodies. Protein bands were transferred to PVDF membranes (Bio-Rad Laboratories). Membranes were blocked at room temperature for 1–2 h and incubated overnight at 4°C with primary antibodies dissolved in TBS (Quality Biological, Gaithersburg, MD) with 3% BSA (Gemini Bio-Products) and 0.1% Tween-20 (Sigma-Aldrich). Membranes were probed with 1:1,000 dilutions of SIRT1–3, 5–7, 1:5,000 dilution of collagen type I and α-SMA, and 1:1,000 dilution of Smad2/3 primary antibodies, followed by secondary HRP-conjugated goat anti-rabbit antibody (Millipore, Temecula, CA) at 1:2,000 dilution for 1 h at room temperature. Membrane protein bands were developed with SuperSignal West Pico Chemiluminescent (Thermo Fisher Scientific) or Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA) and Blu-Ray X-ray Western blotting film (Next Day Science, Columbia, MD). Membranes were stripped with Re-Blot Plus (Millipore) for 25 min and incubated with GAPDH at 1:10,000 dilution or β-actin at 1:2,000 dilution to determine relative amounts of total protein for whole cell lysates, MEK1/2 at 1:2,000 dilution for cytoplasmic cell fractions (68), and HDAC2 at 1:2,000 dilution for nuclear cell lysates (13). Gel images were scanned using LabScan software (GE Healthcare) and protein densities measured with ImageJ (52).
For immunohistochemical staining, 5-µm sections of formalin-fixed, paraffin-embedded, human lung tissues were deparaffinized and processed for antigen retrieval (Dako, Carpinteria, CA). Staining for SIRT7 was performed using a rabbit polyclonal antibody from Abcam (Cambridge, MA) followed by alkaline phosphatase-labeled goat anti-rabbit antibody and chromogen both from Vector Laboratories (Burlingame, CA), resulting in a brown appearance of SIRT7-positive structures.
For immunocytochemistry of cultured fibroblasts, cells were transfected with NULL or SIRT7-encoding plasmids, or scrambled or SIRT7 siRNA, and seeded at a density of 15–20,000 cells per well in eight-well chamber slides (Falcon, Becton Dickinson Labware, Franklin Lakes, NJ). Media were changed to 0.5% DMEM 24 h later. For plasmid transfections, cells were stimulated with rhTGF-β1 48 h after transfection and staining for α-SMA performed 48 h later; for siRNA transfections, staining was performed 48 h after transfection. Cells were fixed by incubating with methanol at −20°C for 20 min, washed with PBS, and blocked for 1 h at room temperature with 5% BSA in 0.1% Tween 20 in PBS. Cells were incubated overnight with primary antibody to α-SMA (Abcam) at 1:100 dilution, washed, incubated with secondary FITC antibody to α-SMA (Santa Cruz) at 1:1,000 dilution for 1 h, and visualized with a fluorescent microscope.
Statistical analyses.
Data are reported as mean values ± SD. Differences between two groups were assessed using the Student’s two-tailed unequal variance t-test or the Mann-Whitney U-test, as indicated for specific experiments. P values less than 0.05 were considered statistically significant.
RESULTS
Expression levels of sirtuins in lung tissues and cultured pulmonary fibroblasts.
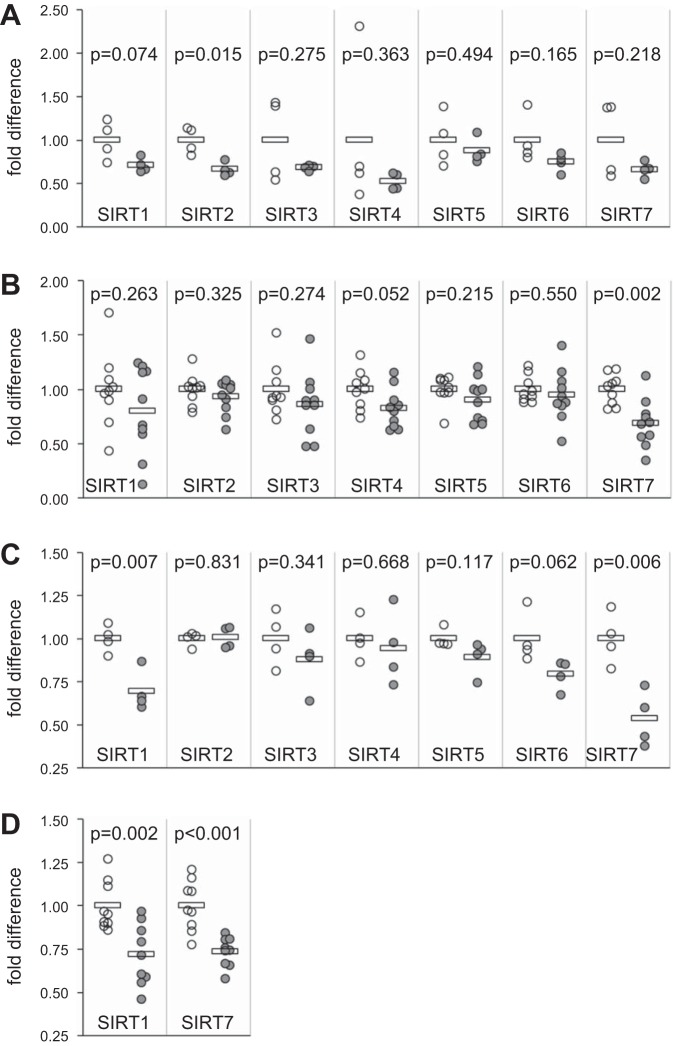
Initial experiments were performed to determine whether mRNA levels of SIRTs 1–7 in lung tissues differed between patients with IPF and healthy controls. Lung tissue samples were obtained from four patients with IPF and four healthy controls, RNA was isolated, and SIRT 1–7 levels were determined by quantitative PCR (qPCR). All seven sirtuin mRNA levels tended to be decreased in patients with IPF compared with controls, but only the decrease in SIRT2 mRNA reached significance (P = 0.015, Fig. 1A).
Fig. 1.
Sirtuin mRNA expression in whole lung tissue and pulmonary fibroblasts. RNA was isolated and reverse transcribed into cDNA, and sirtuin mRNA expression was determined by RT-quantitative PCR (qPCR) for normal and idiopathic pulmonary fibrosis (IPF) lung tissue (A), fibroblasts cultured from lung explants of patients with IPF (B) and systemic sclerosis-associated interstitial lung disease (SSc-ILD; C), and lung tissue from PBS- and bleomycin-treated mice (D). For each SIRT, mRNA levels were normalized to 18S rRNA levels and further normalized to the average value of all control samples. Donors included 12 patients with IPF, four patients with SSc-ILD, and 13 healthy controls. Open circles represent control samples, and gray circles represent patients or bleomycin-treated mice. Bars represent averages for each group. P values were calculated using the Student’s t-test. A: fold differences for mRNA from four IPF lung tissue samples compared with four normal lung samples. B and C: mRNA-fold differences in pulmonary fibroblasts from 10 patients with IPF (B) and four patients with SSc-ILD (C) compared with control fibroblasts. D: sirtuin mRNA levels in lung tissue from nine control mice (open circles) and nine mice treated with bleomycin (gray circles).
The possibility was considered that substantial differences in lung tissue architecture and cellular composition between patient donors and controls might mask cell-specific differences in sirtuin expression. Therefore, further experiments focused on measuring SIRT1–7 levels in pulmonary fibroblasts from patients with IPF and SSc-ILD compared with controls. RNA isolation followed by quantitative PCR for SIRTs 1–7 was performed in fibroblasts from 10 patients with IPF, 4 patients with SSc-ILD, and 9 healthy controls. All sirtuin mRNA levels tended to be decreased in patients with IPF (Fig. 1B) and SSc-ILD (Fig. 1C) compared with controls. This decrease reached significance for SIRT7 in patients with IPF (P = 0.002) and SSc-ILD (P = 0.006), and for SIRT1 in patients with SSc-ILD (P = 0.007). We also assessed sirtuin levels in the bleomycin mouse model of pulmonary fibrosis. SIRT1 and SIRT7 mRNA levels were measured from homogenized lung tissues of nine bleomycin-treated mice and nine PBS-treated control mice. Similar to the findings in fibroblast cultures from patients with pulmonary fibrosis, SIRT1 and SIRT7 mRNA levels were significantly lower in lung tissues from bleomycin-challenged mice compared with control mice (P = 0.002 for SIRT1, P < 0.001 for SIRT7, Fig. 1D).
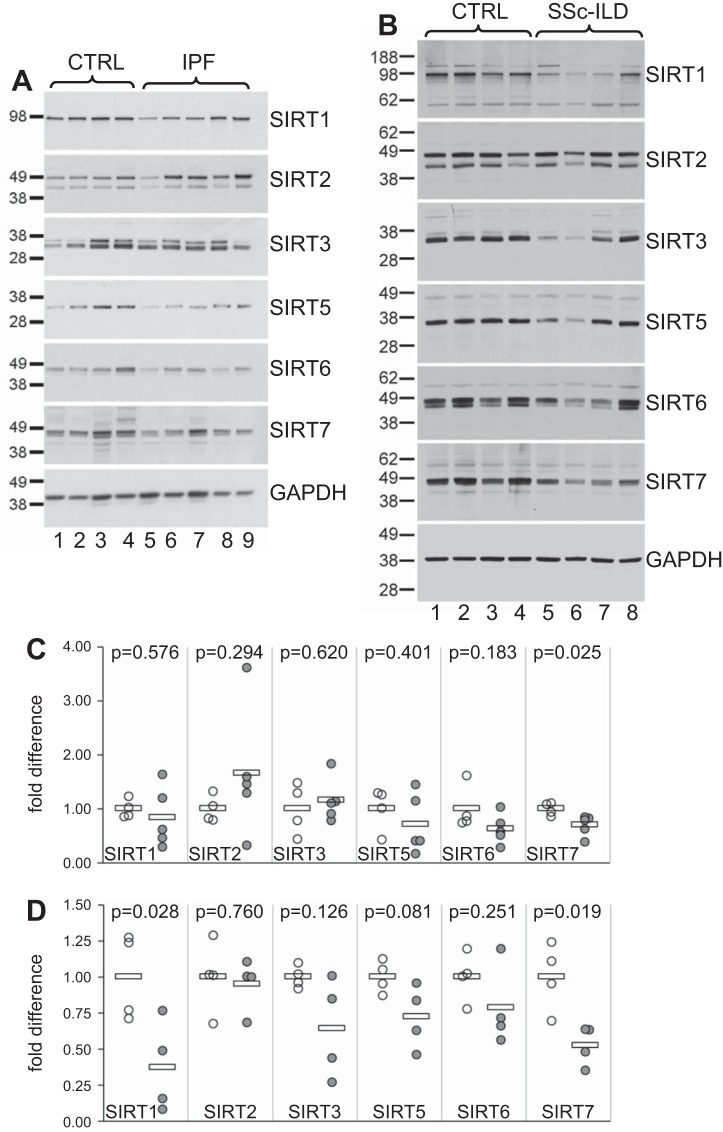
We next compared sirtuin protein expression in pulmonary fibroblasts from patients with pulmonary fibrosis and healthy donors. Primary pulmonary fibroblasts from five patients with IPF, four patients with SSc-ILD, and eight healthy controls were tested for SIRT1–3 and SIRT5–7 protein levels by Western blot analysis (Fig. 2, A and B). SIRT1, SIRT5, SIRT6, and SIRT7 protein levels were decreased in fibroblasts from patients with IPF compared with controls (Fig. 2, A and C), and the decrease in SIRT7 reached significance (P = 0.025). In SSc-ILD, all seven sirtuins were decreased in patients compared with controls (Fig. 2, B and D), and this decrease was significant for SIRT1 (P = 0.028) and SIRT7 (P = 0.019). Given the significant decline in SIRT7 mRNA and protein levels both in SSc-ILD and IPF, our next experiments focused on SIRT7, including SIRT1, in some experiments for comparison.
Fig. 2.
Sirtuin protein expression in pulmonary fibroblasts. Primary pulmonary fibroblasts from patients with IPF, SSc-ILD, and healthy controls were propagated for 4–6 passages in cell culture and Western blot analysis for SIRTs 1–3 and 5–7 performed on cell lysates. A: SIRT protein expression in lung fibroblasts from four control donor fibroblasts (lanes 1–4) compared with five patients with IPF (lanes 5–9). B: SIRT protein expression in four healthy donor controls (lanes 1–4) compared with four patients with SSc-ILD (lanes 5–8). C and D: densitometry of protein bands for each sample normalized to GAPDH and further normalized to the average value of all control samples for patients with IPF (C) and SSc-ILD (D). Open circles represent control fibroblasts, and gray circles represent fibroblast samples from patients with IPF (C) or SSc-ILD (D). Horizontal bars indicate average values. P values were calculated using the Student’s t-test.
Expression levels of sirtuins in adult and neonatal dermal fibroblasts.
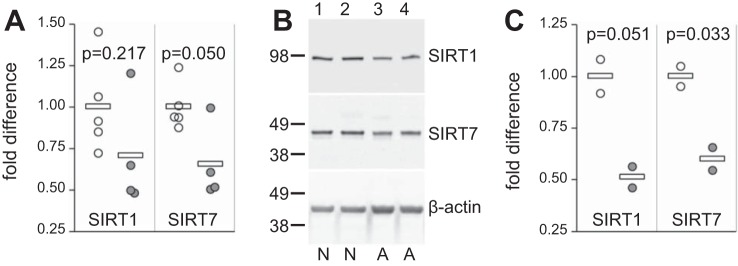
Previous studies have reported that sirtuins decrease with age (8, 24, 39) and mitigate aging-related disorders (54). We investigated possible aging-related changes in SIRT1 and SIRT7 in dermal fibroblasts by comparing SIRT1 and SIRT7 mRNA and protein levels in five neonatal and four adult dermal fibroblast cultures. Both SIRT1 and SIRT7 mRNA levels were decreased in adult compared with neonatal dermal fibroblasts with a notable decrease in SIRT7 (P = 0.05, Fig. 3A). SIRT1 and SIRT7 protein levels were compared by Western blot analysis between two neonatal and two adult dermal fibroblast cultures (Fig. 3B). Similar to the mRNA findings, both SIRT1 and SIRT7 protein levels were decreased in adult compared with neonatal dermal fibroblasts, reaching significance for SIRT7 (P = 0.033, Fig. 3C).
Fig. 3.
Sirtuin expression in neonatal and adult dermal fibroblasts. Primary dermal fibroblasts were isolated, propagated for four or five passages in cell culture, and mRNA (A) and protein (B and C) levels for SIRT1 and SIRT7 were tested. Donors included five neonatal controls (○) and four adult healthy volunteers (gray circles). A: SIRT1 and SIRT7 mRNA levels normalized to 18S and further normalized to the average of all neonatal samples. B: SIRT1 and SIRT7 protein levels in two neonatal cultures (N samples, lanes 1 and 2) and two adult cultures (A samples, lanes 3 and 4). C: densities of SIRT bands in B normalized to β-actin and further normalized to the average of neonatal samples. Horizontal bars in A and C indicate average values. P values were calculated using the Student’s t-test.
In light of these data, the possibility was considered that decreased SIRT7 levels observed in Figs. 1 and 2 could be driven by either age differences between patients and controls, disease status, or their combination. The median [1st quartile, 3rd quartile] age of patients with IPF who provided pulmonary fibroblasts for the experiments shown in Fig. 1B was 62 [58, 67] years, whereas ages of controls in the same data set were 24 [19, 35] years; this age difference was significant (P < 0.05 by Mann-Whitney U-test). Sample sizes in other data subsets in Figs. 1 and 2 were too small for reliable calculations of differences, but controls tended to be younger than patients. Therefore, the confounding effect of age on SIRT7 differences between patients and controls shown in Figs. 1 and 2 cannot be excluded. However, while several sirtuins are known to decrease with age (8, 32, 55), only SIRT1 and SIRT7, among all sirtuins tested, showed significant declines in patients compared with controls (Figs. 1, B and C and 2, C and D), suggesting that disease status rather than age alone drove the observed decline in SIRT7. Moreover, SIRT7 showed a significant decline in age-matched bleomycin-challenged mice (Fig. 1D), further suggesting an association between SIRT7 and fibrosis, rather than an effect of age alone. To definitively assess the effect of SIRT7 levels on fibrosis, experiments in cell culture were performed as described below.
SIRT7 expression in lung tissues.
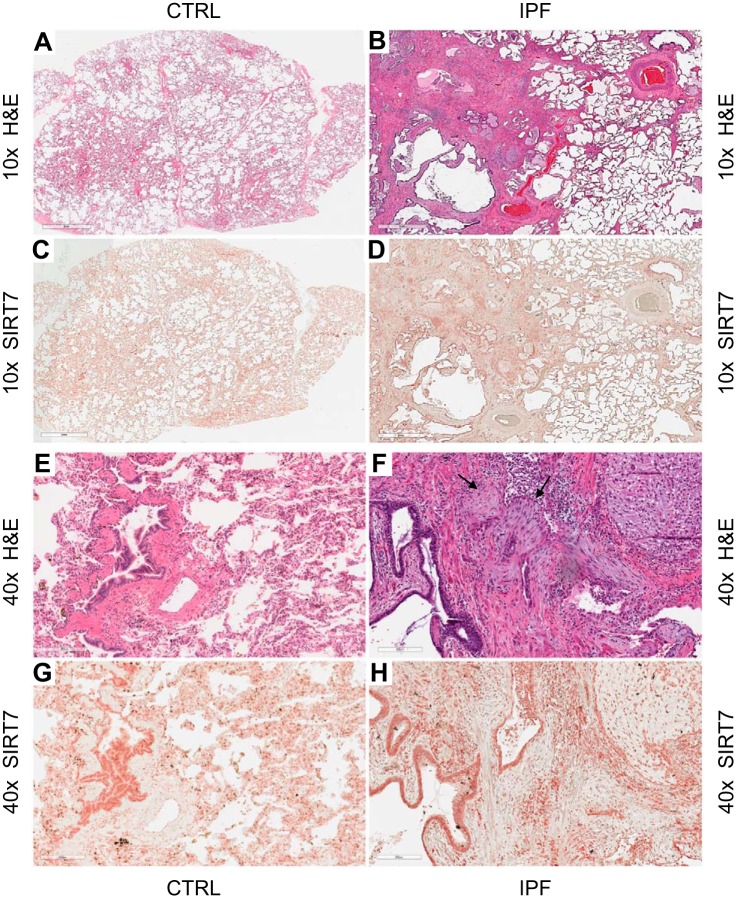
To gain broader insight into the expression of SIRT7 in the lungs of patients and controls, serial lung sections from six patients with IPF and three healthy controls were stained with hematoxylin and eosin (H&E) and SIRT7 antibody. At ×10 objectives with H&E staining, healthy donor lung had the usual appearance of alveoli separated by thin septae of connective tissue (Fig. 4A), while IPF lung exhibited the typical usual interstitial pneumonia (UIP) pattern of distorted lung architecture and areas of relatively preserved alveolar tissue next to areas of dense fibrosis and cystic change (Fig. 4B). Immunohistochemistry for SIRT7 showed widespread cellular expression of SIRT7 both in healthy lung (Fig. 4C) and IPF tissue (Fig. 4D). At ×40 magnification with H&E staining, normal lung parenchyma was seen in control lung (Fig. 4E) and fibroblastic foci (arrows) in IPF lung (Fig. 4F). SIRT7 stained most prominently in areas of dense cellularity, such as bronchial tissue (Fig. 4, G and H) and inflammatory aggregates (Fig. 4D), but it could also be found in mesenchymal cells within fibroblastic foci (Fig. 4H). These observations revealed fairly ubiquitous, yet geographically heterogeneous, and cell type-dependent expression of SIRT7 in normal and fibrotic lungs.
Fig. 4.
Histologic and immunohistochemical analyses of lung tissues. Serial lung sections from a healthy donor (CTRL; A, C, E, and G) and from a patient with IPF (B, D, F, and H) were stained with hematoxylin and eosin (H&E) or SIRT7, as indicated. Digital images were acquired with ×10 (A–D) or ×40 (E–H) objectives. SIRT7-positive cells stain brown, with greater intensity in the nucleus. These experiments were repeated in three healthy control subjects and six patients with IPF with similar results. Arrows in F indicate fribroblastic foci.
Subcellular distribution of SIRT7 in lung fibroblasts.
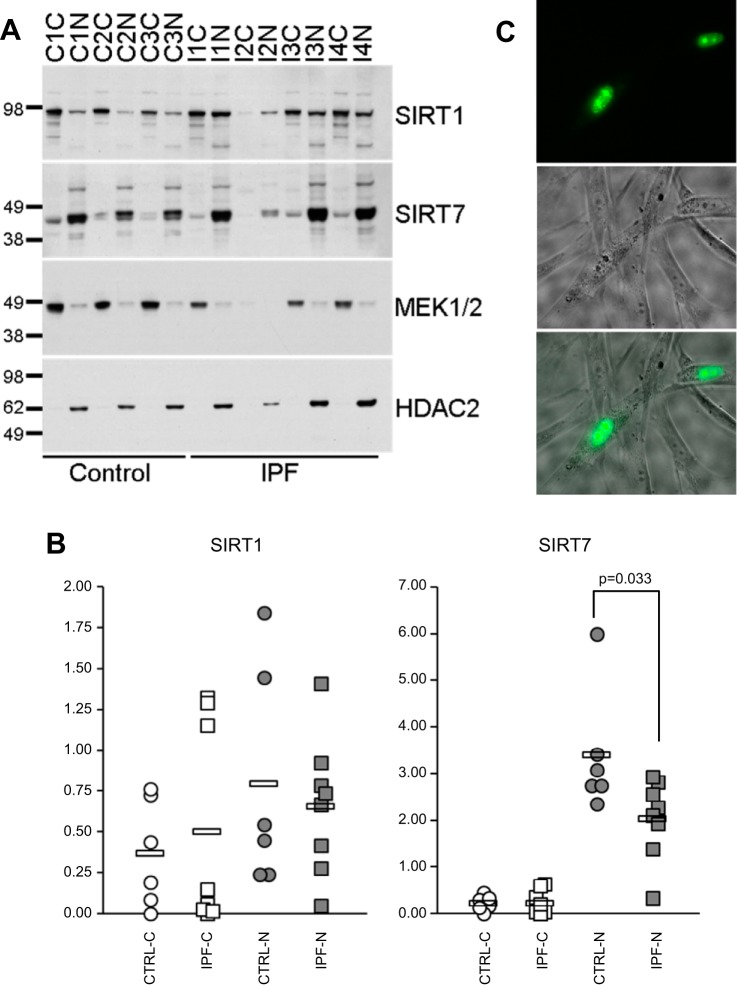
SIRT7 has been described primarily as a nucleolar protein (9, 10, 16, 18, 28, 31, 40, 45, 51, 59, 60, 62), although its presence in the cytoplasm of fibroblasts has been reported as well (30, 67). To determine the location of SIRT7 in primary lung fibroblasts, cytoplasmic and nuclear fractions from cultured fibroblasts obtained from six patients with IPF, two patients with SSc-ILD, and six healthy controls were isolated. Levels of SIRT7, and SIRT1 for comparison, were determined by Western blot analysis (Fig. 5A). As previously shown in different cell types (57), SIRT1 was present in the cytoplasm and nucleus of fibroblasts both in patients with IPF and controls. Compared with control samples, IPF samples tended to have more SIRT1 in the cytoplasm and less SIRT1 in the nucleus (Fig. 5B), but these differences were not significant. Consistent with prior reports, SIRT7 was found predominantly in the nucleus (Fig. 5, A–C). Nuclear fractions had significantly more SIRT7 for all samples combined (P < 0.001), for control samples (P = 0.005), and for IPF samples (P = 0.003). Unlike SIRT1, both control and IPF samples showed minimal SIRT7 in the cytoplasm and, when compared with controls, IPF samples showed a significant decrease of SIRT7 in the nucleus (P = 0.033, Fig. 5B). Transfection of normal human lung primary fibroblasts with a SIRT7-GFP-encoding plasmid and visualization of cells under a fluorescent microscope 24–48 h later corroborated the predominantly nuclear and nucleolar localization of SIRT7 (Fig. 5C). Combined, these data show that SIRT7 is ubiquitously expressed in diverse pulmonary cell types, localized predominantly in the nucleus in lung fibroblasts, and declines in the nucleus in patients with pulmonary fibrosis compared with healthy controls.
Fig. 5.
Nuclear and cytoplasmic distribution of SIRT1 and SIRT7 in primary fibroblasts. Primary lung fibroblasts from six patients with IPF, two patients with SSc-ILD, and six healthy controls were cultured for 4–6 passages, cytoplasmic and nuclear fractions were isolated, and protein levels of SIRT1 and SIRT7 were measured by Western blot analysis in each cellular compartment. A: Western blots showing SIRT1 and SIRT7 protein levels in the cytoplasm (C) and nucleus (N) for three healthy controls (C1–C3) and four patients with IPF (I1–I4). MEK1/2 and histone deacetylases 2 (HDAC2) antibodies were used to control for sample loading of the cytoplasmic and nuclear fractions, respectively. B: SIRT1 and SIRT7 densities for cytoplasmic and nuclear fractions of all samples tested, including three controls and four patients with IPF shown in A. The density of each sample was subtracted from the background density and normalized to MEK1/2 for cytoplasmic fractions and HDAC2 for nuclear fractions. Circles indicate control samples, and squares indicate patient samples. Open circles and squares indicate cytoplasmic fractions, while gray circles and squares indicate nuclear fractions. Horizontal bars indicate average values. P values were calculated using the Mann-Whitney U-test; the significant difference in SIRT7 levels between control and patient fibroblast nuclei is indicated. C: normal human lung fibroblasts overexpressing SIRT7-GFP and viewed under a fluorescent microscope with ×40 objective 24 h after transfection. Top subpanel shows the green fluorescence image, middle subpanel shows phase contrast image, and bottom subpanel shows fluorescent and phase contrast images merged. Note that SIRT7-GFP is localized to the nucleus with greater intensity in the nucleoli.
SIRT7 silencing increases collagen and α-SMA levels in primary fibroblasts.
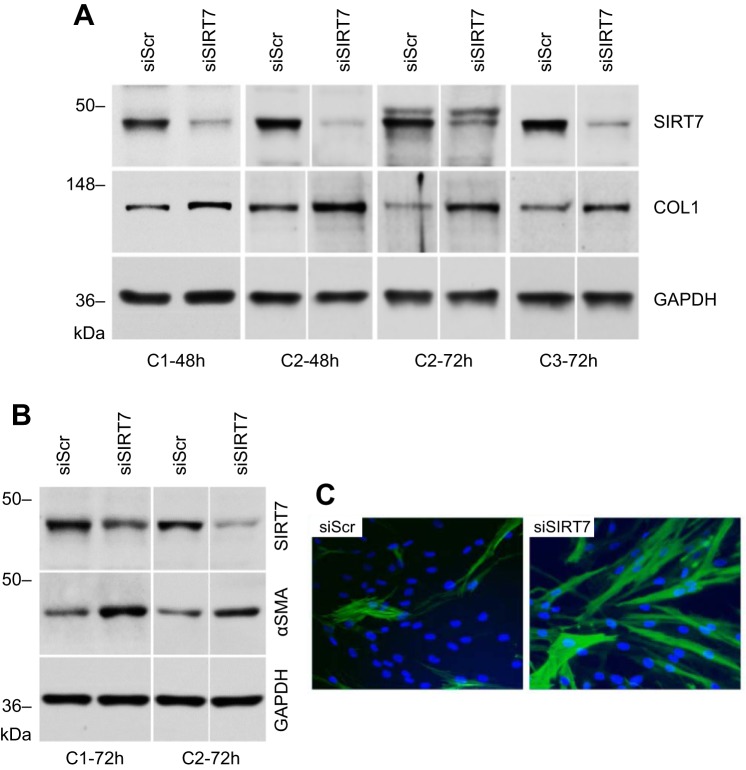
To investigate a possible causative role of decreased SIRT7 levels in pulmonary fibrosis, subsequent experiments assessed the effects of artificially manipulated SIRT7 expression on levels of the fibrosis-related molecules collagen and α-SMA. A robust attenuation of SIRT7 expression was achieved by inhibition with siRNA (Fig. 6A). Electroporation of primary lung fibroblasts with SIRT7 siRNA, when compared with control siRNA, resulted in modest (up to 1.3-fold), increases in COL1A, COL1A2, and COL3A1 mRNA levels 48 to 72 h after transfection and a more substantial, up to 2.5-fold, increase in collagen type I protein after 48 to 72 h (Fig. 6A). SIRT7 silencing increased α-SMA protein levels by approximately twofold 72 h after transfection (Fig. 6B). SIRT7 silencing also showed an increase in α-SMA protein 48 h after transfection when assessed by immunocytochemistry (Fig. 6C).
Fig. 6.
Effect of SIRT7 silencing on collagen and α-SMA protein levels. A: normal adult lung fibroblasts from three healthy control donors (C1–C3) were transfected with scrambled control or SIRT7 siRNAs, as indicated, and Western blotting for collagen type I protein (COL1) performed after 48 or 72 h. B: α-SMA protein levels for two healthy control donors (C1, C2) 72 h after silencing with scrambled or SIRT7 siRNAs. Noncontiguous gels are demarcated by white spaces. C: immunofluorescent staining for α-SMA in fibroblasts from a healthy control donor (different from the two controls shown in B) 48 h after transfection with scrambled or SIRT7 siRNAs. Note the increases in collagen and α-SMA in SIRT7-silenced fibroblast cultures.
Overexpression of SIRT7 suppresses collagen and α-SMA levels and attenuates TGF-β-induced increases in collagen.
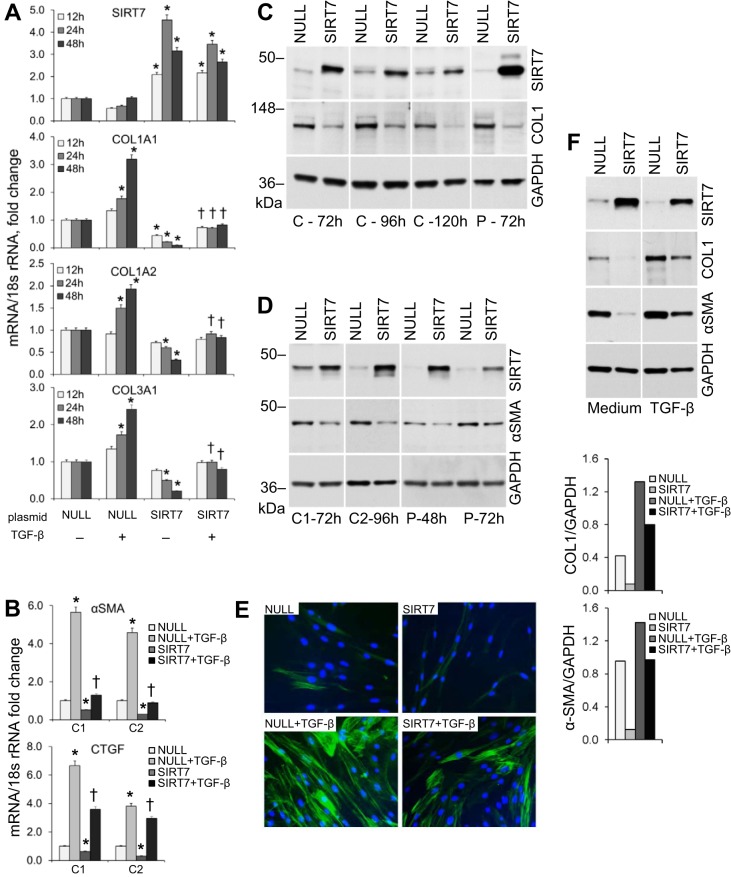
When compared with transfection with a noncoding (NULL) plasmid containing a similar backbone, transfection of NHLF with a SIRT7-encoding plasmid resulted in decreases of COL1A1 mRNA levels by three- to 10-fold, COL1A2 mRNA by two- to threefold, and COL3A1 by two- to five-fold (Fig. 7A). Overexpression of SIRT7 similarly attenuated TGF-β-driven increases in COL1A1 and, to a lesser degree, COL1A2 and COL3A1 (Fig. 7A). Overexpression of SIRT7 also decreased mRNA levels of α-SMA and another fibrosis-associated molecule, connective tissue growth factor (CTGF), by approximately two- to threefold. Additionally, SIRT7 overexpression suppressed TGF-β-induced increases in α-SMA mRNA levels by four- to five-fold and CTGF mRNA levels by up to twofold (Fig. 7B).
Fig. 7.
Effect of SIRT7 overexpression on the levels of fibrosis-associated molecules. A: RT-qPCR for SIRT7, COL1A1, COL1A2, and COL3A1 mRNA levels in cultured primary pulmonary fibroblasts from a healthy control at indicated times after stimulation with rhTGF-β1. Before stimulation, cells were transfected with equal amounts of a SIRT7-encoding or control noncoding (NULL) plasmid; both plasmids had a similar backbone. Cells were stimulated with TGF-β 48 h after transfection. Means ± SD of duplicate measurements are shown; the experiment was repeated in fibroblast cultures from two different donors on two separate occasions. The mRNA levels of each target were normalized to the levels of 18S rRNA and further normalized to the NULL sample without TGF-β at each time point. Significant differences (P < 0.05) from NULL-transfected cells without TGF-β stimulation are indicated by asterisks, and significant differences from NULL-transfected cells with TGF-β are indicated by daggers. B: RT-qPCR for α-SMA and connective tissue growth factor (CTGF) in cultured primary pulmonary fibroblasts from two healthy controls (C1, C2) transfected with NULL or SIRT7 plasmids and stimulated with rhTGF-β1 48 h later. Analyses were performed 24 h after TGF-β stimulation. Significant differences (P < 0.05) are indicated. C: changes in collagen type I protein levels in cultured primary pulmonary fibroblasts from a healthy control (C) and patient with IPF (P) following overexpression of SIRT7. Fibroblasts were electroporated with a control noncoding (NULL) or SIRT7-encoding plasmid and Western blots for SIRT7, COL1, and GAPDH performed at the indicated times. Note the decreases in collagen levels in SIRT7-overexpressing cultures. These experiments were performed in primary cultures from four different healthy donors on at least four separate occasions and two donors with IPF with similar results. Noncontiguous gels are demarcated by white spaces. D: α-SMA protein levels in two healthy controls (C1 and C2) and a patient with IPF (P) at indicated times after transfection with NULL or SIRT7-encoding plasmids. Note the decreases in α-SMA in cultures overexpressing SIRT7. E: immunofluorescent staining for α-SMA in normal lung fibroblasts transfected with NULL or SIRT7 plasmids and stimulated with rhTGF-β 48 h later. Cells were fixed, and staining was performed 48 h after TGF-β stimulation. SIRT7 overexpression appears to slightly decrease α-SMA expression both with and without TGF-β stimulation. F: effects of SIRT7 overexpression combined with rhTGF-β1 stimulation on COL1 and α-SMA protein levels in healthy control lung fibroblasts. Cells were electroporated with NULL- or SIRT7-encoding plasmids and, after 48 h, stimulated with rhTGF-β or cultured with no stimulation (medium) for an additional 72 h. Note that the stimulating effect of TGF-β on collagen and α-SMA is suppressed by SIRT7. F, bottom: densities for COL1 and α-SMA normalized to GAPDH are shown.
Similar to its effect on collagen mRNA levels, overexpression of SIRT7 in primary lung fibroblast cultures resulted in a decrease, up to three-fold, in collagen type I protein 72–120 h after transfection of fibroblasts from normal healthy donors and from patients with IPF. This experiment was performed in primary fibroblast cell cultures from four different control donors and three donors with IPF with consistent results (representative gels are shown in Fig. 7C). Overexpression of SIRT7 also reduced α-SMA protein levels by 2–2.5-fold in fibroblasts from three healthy controls and from two patients with IPF (representative gels are shown in Fig. 7D), an effect confirmed immunocytochemically (Fig. 7E). In addition to its observed independent effects on basal collagen and α-SMA levels, SIRT7 overexpression also attenuated TGF-β-induced increases in collagen and α-SMA (Fig. 7F), an effect that was observed in three different NHLF cultures from separate donors.
SIRT7 regulates the fibrotic phenotype of lung fibroblasts by decreasing Smad3 levels.
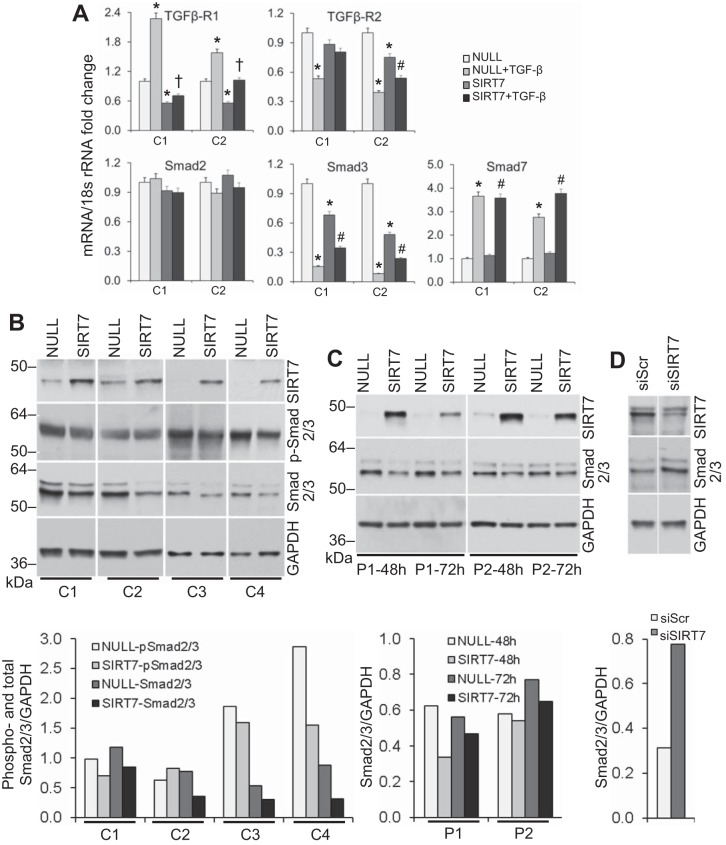
We considered the possibility that SIRT7 suppresses collagen and α-SMA levels by downregulating TGF-β activity or the levels of TGFβ or TGFβ receptors. NHLF overexpressing SIRT7 did not show a significant change in TGF-β1 mRNA levels, although TGFβ-R1 and, to a lesser extent, TGFβ-R2 levels were decreased in two separate SIRT7-overexpressing NHLF cultures (Fig. 8A). To address the possibility that SIRT7 may play a role in activating latent TGF-β protein, experiments with PAIL cells, a genetically manipulated cell line that expresses luciferase in response to active TGF-β (1), were performed. NHLF were transfected with the noncoding (NULL) or SIRT7-encoding plasmids, or with scrambled control siRNA or SIRT7 siRNA, and then cocultured with PAIL cells. Whereas recombinant active TGF-β increased luciferase activity in concentrations as low as 0.03 ng/ml in control experiments, no change in luminescence was observed in cocultures of PAIL cells with SIRT7-transfected fibroblasts. This observation suggested that SIRT7 does not act by modulating TGF-β activity. We also considered that SIRT7 may exert its antifibrotic effect by affecting Smad2, Smad3, or Smad7 levels. RT-qPCR analyses showed decreases in mRNA levels for Smad3, but not Smad2, in two NHLF cultures 72 h after transfection with SIRT7 vs. NULL plasmids (Fig. 8A). While both NULL- and SIRT7-transfected cultures showed significant increases in Smad7 mRNA levels in response to TGF-β stimulation, SIRT7 overexpression had no significant effect on Smad7 levels (Fig. 8A).
Fig. 8.
Effect of SIRT7 overexpression on TGF-β pathway-related molecules. A: RT-qPCR for Smad2, Smad3, Smad7, TGFβ-R1, and TGFβ-R2 in cultured primary pulmonary fibroblasts from two healthy controls (C1 and C2) transfected with NULL or SIRT7 plasmids and stimulated with rhTGF-β1 48 h later. Analyses were performed 24 h after TGF-β stimulation. *Significant difference (P < 0.05) from NULL-transfected cells without TGF-β stimulation. †Significant difference from NULL-transfected cells with TGF-β. #Significant differences from SIRT7-transfected cells without TGF-β. Note the decreases in SMAD3 and TGFβ-R1 in fibroblast cultures overexpressing SIRT7. B: changes in the expression levels of phosphorylated and total Smad2/3 measured by Western blot analysis in cultured primary lung fibroblasts from four different healthy controls (C1–C4) following overexpression of SIRT7. Fibroblasts were electroporated with a control noncoding (NULL) or SIRT7-encoding plasmid, and Western blots for the indicated targets were performed 72 h later. Noncontiguous gels are demarcated by white spaces. B, bottom: densities for phosphorylated and total Smad2/3 normalized to GAPDH are shown. C: changes in total Smad2/3 levels at indicated times in two patients with IPF (P1, P2) following overexpression of SIRT7. The SIRT7 and GAPDH bands for patient 1 are the same as those for the patient shown in Fig. 7D. C, bottom: total Smad2/3 densities normalized to GAPDH are shown. Note the decreases in Smad2/3 levels in both normal and patient SIRT7-overexpressing cultures. D: changes in total Smad2/3 levels in NHLF following SIRT7 silencing with siRNA. Fibroblasts were electroporated with scrambled (Scr) or SIRT7 siRNA (siSIRT7), and analyses were performed 72 h later. Noncontiguous gels are demarcated by white spaces. SIRT7 and GAPDH bands are the same as those for donor 2 at 72 h shown in Fig. 6A. D, bottom: Densities for total Smad2/3 relative to GAPDH are shown.
To evaluate the effect of SIRT7 on Smad2/3 protein levels, four different NHLF cultures were transfected with NULL or SIRT7-encoding plasmids on separate occasions, overexpression of SIRT7 was confirmed, and phosphorylated and total levels of Smad2/3 protein were measured by Western blot analysis after 72 h. All four cultures showed a decrease in the levels of total Smad2/3 protein, and three out of four showed a decrease in basal Smad2/3 phosphorylation (Fig. 8B). Similar to its effect in NHLF cultures, SIRT7 overexpression resulted in a decrease in total Smad2/3 protein levels in fibroblast cultures from two patients with IPF (Fig. 8C). Reciprocally, SIRT7 depletion with siRNA resulted in increased total Smad2/3 levels in NHLF (Fig. 8D). Taken together with the data on SIRT7’s effects on Smad2 and Smad3 mRNA levels, these results suggest that decreases in Smad3 levels contribute to the antifibrotic effect of SIRT7.
DISCUSSION
To our knowledge, this is the first report to address the expression of all seven sirtuins in pulmonary fibrosis associated with IPF and SSc-ILD. Given their role in aging, sirtuins (particularly SIRT1 and SIRT3) have recently been described in connection with skin fibrosis in scleroderma and in murine models of pulmonary and skin fibrosis (2, 55, 64, 66). Our work builds on these studies by identifying SIRT7, the least studied mammalian sirtuin, as a novel regulator of fibrosis in human lung fibroblasts. Furthermore, our data suggest that SIRT7 exerts its antifibrotic effect by reducing total Smad3 levels, which is in agreement with the proposed mechanism for the antifibrotic effects of SIRT1 and SIRT3 (2, 11, 23, 55).
We undertook a comprehensive evaluation of sirtuin expression in pulmonary fibrosis by investigating mRNA and protein levels of all seven sirtuins in patients with IPF and SSc-ILD. Our results indicate that, similar to other sirtuins, SIRT7 mRNA levels tend to be lower in lung tissue samples from patients with pulmonary fibrosis compared with healthy controls (Fig. 1A). In primary cell cultures, SIRT7 mRNA levels were significantly lower in lung fibroblasts derived from patients with IPF (Fig. 1B) and SSc-ILD (Fig. 1C) than in control fibroblasts. In the mouse model of bleomycin-induced pulmonary fibrosis, SIRT7 mRNA levels were also significantly decreased compared with PBS-challenged mice (Fig. 1D). Consistent with these mRNA findings, SIRT7 protein levels were significantly decreased in cultured primary lung fibroblasts from patients with IPF and SSc-ILD (Fig. 2) compared with fibroblasts derived from healthy control lungs. To our knowledge, this is the first report of decreased SIRT7 expression in patients with pulmonary fibrosis.
Sirtuins are best known for their role in aging (5, 14, 15, 19, 22, 25–27, 29, 33), and their activity and levels, particularly for SIRT1, the most studied sirtuin, have been shown to decrease with age in multiple tissues and cell types (7, 8, 24, 30, 32, 39, 41, 42, 61, 65). We corroborated these findings in fibroblast cultures by showing a decrease in SIRT1 and SIRT7 mRNA and protein levels in adult compared with neonatal dermal fibroblasts (Fig. 3). Multiple aging-related mechanisms, such as genomic instability and mitochondrial dysfunction have been associated with decreased sirtuin levels and SIRT7 loss, in particular (34, 41, 51, 62). Interestingly, while both SIRT1 and SIRT3 have been implicated in fibrosis, among all of the sirtuins, only SIRT6 and SIRT7-deficient animal models have shown progeric phenotypes, lending further support to the role of SIRT7 in aging-related processes.
We next aimed to characterize the location of sirtuin 7 in whole lung tissue and primary adult human lung fibroblasts. Our goal was to determine the extent and expression by cell type of SIRT7 in healthy and fibrotic lung tissue and investigate possible differences in SIRT7 subcellular localization that may exist between patients with pulmonary fibrosis and controls. Immunohistochemical staining with SIRT7 of serial lung sections from patients with IPF and healthy controls showed widespread SIRT7 expression in multiple cell types, including epithelial, mesenchymal, and inflammatory cells (Fig. 4). Because of significant differences in cellular and ECM composition between normal and fibrotic lung tissue, it is difficult to quantitate differences in SIRT7 expression between patients with IPF and controls based on immunohistochemical analyses. We then focused on the subcellular localization of SIRT7, and SIRT1 for comparison, in lung fibroblasts from patients with pulmonary fibrosis and healthy controls. Consistent with most prior reports (9, 10, 16, 18, 28, 31, 40, 45, 51, 59, 60, 62), SIRT7 was found predominantly in the nucleus (Fig. 5). Compared with fibroblasts from healthy controls, fibroblasts from patients with IPF and SSc-ILD showed a significant decrease of SIRT7 in the nucleus. A decrease of SIRT7 in the nucleus and reciprocal increase in the cytoplasm have been observed during replicative senescence in TIG3 fibroblasts and in response to DNA-damaging agents (30, 67), and SIRT7 movement from the nucleolus into the nucleoplasm has been shown to be induced by genotoxic, low-glucose, and hyperosmolar stress (9, 10, 62). Although we did not detect an increase of SIRT7 in the cytoplasm, our observed decrease of SIRT7 in the nucleus in fibroblasts from patients compared with controls suggests a change in pulmonary fibrosis that may be related to premature aging or cellular stress.
To address the possible antifibrotic role of SIRT7, our next experiments investigated the effects of SIRT7 silencing and overexpression on collagen, α-SMA, and CTGF expression in cultured human lung fibroblasts. SIRT7 silencing increased collagen type I and α-SMA proteins (Fig. 6). Reciprocally, SIRT7 overexpression significantly decreased COL1A1, COL1A2, COL3A1, α-SMA, and CTGF mRNA levels and collagen I and α-SMA proteins (Fig. 7). Furthermore, SIRT7 overexpression attenuated TGF-β-induced increases in collagen and α-SMA mRNA and protein levels (Fig. 7). Combined, these observations suggest that SIRT7 is protective against fibrosis in adult human lung fibroblasts and corroborate the antifibrotic effects of SIRT1 and SIRT3 (2, 55, 64).
To assess the molecular mechanisms of SIRT7’s antifibrotic effect, we considered the possibility that SIRT7 may regulate collagen levels by participating in the activation, transcription, and profibrotic signaling pathways of TGF-β. SIRT7 overexpression in NHLF did not have a significant effect on steady-state TGF-β mRNA levels, as quantified by RT-qPCR or activate TGF-β, given the absence of luminescence in coculture with PAIL cells. We did, however, observe a decrease in TGFβ-R1 and Smad3 mRNA levels in NHLF, as well as total Smad2/3 protein levels in both normal lung and IPF fibroblast cultures overexpressing SIRT7 (Fig. 8), suggesting that SIRT7 may exert its antifibrotic effect, at least in part, by attenuating levels of these signaling mediators.
Taken together, these results suggest that the antifibrotic effect of SIRT7 occurs, in part, by decreasing Smad3 levels, which is in agreement with other sirtuins described in pulmonary, renal, and cardiac fibrosis (2, 11, 23, 55), and set the stage for further mechanistic work to identify molecular processes mediating SIRT7’s independent effects on fibroblasts and interaction with TGF-β. SIRT7 may become a new target of therapeutic manipulation in pulmonary fibrosis.
GRANTS
This study was supported in part by the VA Fellowship in Advanced Geriatrics (A. E. Wyman) and VA Merit Award I01BX002499 (to S. P. Atamas) with the use of resources and facilities at the VA Maryland Health Care System, Baltimore VA Medical Center, as well as National Heart, Lung, and Blood Institute Grant R01HL-126897 (S. P. Atamas).
DISCLOSURES
No conflicts of interest, financial or otherwise are declared by the authors.
AUTHOR CONTRIBUTIONS
A.E.W. and S.P.A. conceived and designed research; A.E.W., Z.N., R.F., V.L., N.G.S., N.W.T., and S.P.A. performed experiments; A.E.W. and S.P.A. analyzed data; A.E.W. and S.P.A. interpreted results of experiments; A.E.W. and S.P.A. prepared figures; A.E.W. and S.P.A. drafted manuscript; A.E.W., N.G.S., N.W.T., and S.P.A. edited and revised manuscript; A.E.W., Z.N., R.F., V.L., N.G.S., N.W.T., and S.P.A. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Carol Feghali-Bostwick for generously providing primary lung fibroblast cultures from patients with SSc-ILD, IPF, and from healthy controls. We thank Dr. Daniel B. Rifkin for providing PAIL cells.
REFERENCES
- 1.Abe M, Harpel JG, Metz CN, Nunes I, Loskutoff DJ, Rifkin DB. An assay for transforming growth factor-beta using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal Biochem 216: 276–284, 1994. doi: 10.1006/abio.1994.1042. [DOI] [PubMed] [Google Scholar]
- 2.Akamata K, Wei J, Bhattacharyya M, Cheresh P, Bonner MY, Arbiser JL, Raparia K, Gupta MP, Kamp DW, Varga J. SIRT3 is attenuated in systemic sclerosis skin and lungs, and its pharmacologic activation mitigates organ fibrosis. Oncotarget 7: 69,321–69,336, 2016. doi: 10.18632/oncotarget.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albani D, Ateri E, Mazzuco S, Ghilardi A, Rodilossi S, Biella G, Ongaro F, Antuono P, Boldrini P, Di Giorgi E, Frigato A, Durante E, Caberlotto L, Zanardo A, Siculi M, Gallucci M, Forloni G. Modulation of human longevity by SIRT3 single nucleotide polymorphisms in the prospective study “Treviso Longeva (TRELONG)”. Age (Dordr) 36: 469–478, 2014. doi: 10.1007/s11357-013-9559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, Greco V, Maggiolini M, Feraco E, Mari V, Franceschi C, Passarino G, De Benedictis G. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics 85: 258–263, 2005. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem 73: 417–435, 2004. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 6.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell 6: 759–767, 2007. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 7.Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol 5: e201, 2007. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 153: 1448–1460, 2013. doi: 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S, Blank MF, Iyer A, Huang B, Wang L, Grummt I, Voit R. SIRT7-dependent deacetylation of the U3-55k protein controls pre-rRNA processing. Nat Commun 7: 10734, 2016. doi: 10.1038/ncomms10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S, Seiler J, Santiago-Reichelt M, Felbel K, Grummt I, Voit R. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol Cell 52: 303–313, 2013. doi: 10.1016/j.molcel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Chen T, Li J, Liu J, Li N, Wang S, Liu H, Zeng M, Zhang Y, Bu P. Activation of SIRT3 by resveratrol ameliorates cardiac fibrosis and improves cardiac function via the TGF-β/Smad3 pathway. Am J Physiol Heart Circ Physiol 308: H424–H434, 2015. doi: 10.1152/ajpheart.00454.2014. [DOI] [PubMed] [Google Scholar]
- 12.Colman RJ, Beasley TM, Kemnitz JW, Johnson SC, Weindruch R, Anderson RM. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat Commun 5: 3557, 2014. doi: 10.1038/ncomms4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 370: 737–749, 2003. doi: 10.1042/bj20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell 123: 655–667, 2005. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 15.Fontana L, Partridge L, Longo VD. Extending healthy life span—from yeast to humans. Science 328: 321–326, 2010. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 20: 1075–1080, 2006. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furst DE, Fernandes AW, Iorga SR, Greth W, Bancroft T. Epidemiology of systemic sclerosis in a large U.S. managed care population. J Rheumatol 39: 784–786, 2012. doi: 10.3899/jrheum.111106. [DOI] [PubMed] [Google Scholar]
- 18.Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, Sirri V. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J Cell Sci 122: 489–498, 2009. doi: 10.1242/jcs.042382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guarente L. Calorie restriction and sirtuins revisited. Genes Dev 27: 2072–2085, 2013. doi: 10.1101/gad.227439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guarente L, Picard F. Calorie restriction—the SIR2 connection. Cell 120: 473–482, 2005. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 21.Herzog EL, Mathur A, Tager AM, Feghali-Bostwick C, Schneider F, Varga J. Review: interstitial lung disease associated with systemic sclerosis and idiopathic pulmonary fibrosis: how similar and distinct? Arthritis Rheumatol 66: 1967–1978, 2014. doi: 10.1002/art.38702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425: 191–196, 2003. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 23.Huang XZ, Wen D, Zhang M, Xie Q, Ma L, Guan Y, Ren Y, Chen J, Hao CM. Sirt1 activation ameliorates renal fibrosis by inhibiting the TGF-β/Smad3 pathway. J Cell Biochem 115: 996–1005, 2014. doi: 10.1002/jcb.24748. [DOI] [PubMed] [Google Scholar]
- 24.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol 24: 464–471, 2014. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaeberlein M, Burtner CR, Kennedy BK. Recent developments in yeast aging. PLoS Genet 3: e84, 2007. doi: 10.1371/journal.pgen.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol 2: E296, 2004. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13: 2570–2580, 1999. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karim MF, Yoshizawa T, Sato Y, Sawa T, Tomizawa K, Akaike T, Yamagata K. Inhibition of H3K18 deacetylation of Sirt7 by Myb-binding protein 1a (Mybbp1a). Biochem Biophys Res Commun 441: 157–163, 2013. doi: 10.1016/j.bbrc.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 29.Kenyon CJ. The genetics of ageing. Nature 464: 504–512, 2010. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 30.Kiran S, Chatterjee N, Singh S, Kaul SC, Wadhwa R, Ramakrishna G. Intracellular distribution of human SIRT7 and mapping of the nuclear/nucleolar localization signal. FEBS J 280: 3451–3466, 2013. doi: 10.1111/febs.12346. [DOI] [PubMed] [Google Scholar]
- 31.Kiran S, Oddi V, Ramakrishna G. Sirtuin 7 promotes cellular survival following genomic stress by attenuation of DNA damage, SAPK activation and p53 response. Exp Cell Res 331: 123–141, 2015. doi: 10.1016/j.yexcr.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Lee N, Kim DK, Kim ES, Park SJ, Kwon JH, Shin J, Park SM, Moon YH, Wang HJ, Gho YS, Choi KY. Comparative interactomes of SIRT6 and SIRT7: Implication of functional links to aging. Proteomics 14: 1610–1622, 2014. doi: 10.1002/pmic.201400001. [DOI] [PubMed] [Google Scholar]
- 33.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289: 2126–2128, 2000. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 34.Liu JP, Chen R. Stressed SIRT7: facing a crossroad of senescence and immortality. Clin Exp Pharmacol Physiol 42: 567–569, 2015. doi: 10.1111/1440-1681.12423. [DOI] [PubMed] [Google Scholar]
- 35.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell 153: 1194–1217, 2013. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luckhardt TR, Thannickal VJ. Systemic sclerosis-associated fibrosis: an accelerated aging phenotype? Curr Opin Rheumatol 27: 571–576, 2015. doi: 10.1097/BOR.0000000000000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luzina IG, Kopach P, Lockatell V, Kang PH, Nagarsekar A, Burke AP, Hasday JD, Todd NW, Atamas SP. Interleukin-33 potentiates bleomycin-induced lung injury. Am J Respir Cell Mol Biol 49: 999–1008, 2013. doi: 10.1165/rcmb.2013-0093OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luzina IG, Lockatell V, Todd NW, Kopach P, Pentikis HS, Atamas SP. Pharmacological in vivo inhibition of S-nitrosoglutathione reductase attenuates bleomycin-induced inflammation and fibrosis. J Pharmacol Exp Ther 355: 13–22, 2015. doi: 10.1124/jpet.115.224675. [DOI] [PubMed] [Google Scholar]
- 39.Massudi H, Grant R, Braidy N, Guest J, Farnsworth B, Guillemin GJ. Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS One 7: e42357, 2012. doi: 10.1371/journal.pone.0042357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16: 4623–4635, 2005. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM, Chen D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347: 1374–1377, 2015. doi: 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mortuza R, Chen S, Feng B, Sen S, Chakrabarti S. High glucose induced alteration of SIRTs in endothelial cells causes rapid aging in a p300 and FOXO regulated pathway. PLoS One 8: e54514, 2013. doi: 10.1371/journal.pone.0054514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124: 315–329, 2006. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 44.Pérez-Bocanegra C, Solans-Laqué R, Simeón-Aznar CP, Campillo M, Fonollosa-Pla V, Vilardell-Tarrés M. Age-related survival and clinical features in systemic sclerosis patients older or younger than 65 at diagnosis. Rheumatology (Oxford) 49: 1112–1117, 2010. doi: 10.1093/rheumatology/keq046. [DOI] [PubMed] [Google Scholar]
- 45.Pfister JA, Ma C, Morrison BE, D’Mello SR. Opposing effects of sirtuins on neuronal survival: SIRT1-mediated neuroprotection is independent of its deacetylase activity. PLoS One 3: e4090, 2008. doi: 10.1371/journal.pone.0004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pochetuhen K, Luzina IG, Lockatell V, Choi J, Todd NW, Atamas SP. Complex regulation of pulmonary inflammation and fibrosis by CCL18. Am J Pathol 171: 428–437, 2007. doi: 10.2353/ajpath.2007.061167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, Lynch DA, Ryu JH, Swigris JJ, Wells AU, Ancochea J, Bouros D, Carvalho C, Costabel U, Ebina M, Hansell DM, Johkoh T, Kim DS, King TE Jr, Kondoh Y, Myers J, Müller NL, Nicholson AG, Richeldi L, Selman M, Dudden RF, Griss BS, Protzko SL, Schünemann HJ; on behalf of the ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis . An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med 183: 788–824, 2011. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raghu G, Weycker D, Edelsberg J, Bradford WZ, Oster G. Incidence and prevalence of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 174: 810–816, 2006. doi: 10.1164/rccm.200602-163OC. [DOI] [PubMed] [Google Scholar]
- 49.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA 101: 15998–16003, 2004. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rubio-Rivas M, Royo C, Simeón CP, Corbella X, Fonollosa V. Mortality and survival in systemic sclerosis: systematic review and meta-analysis. Semin Arthritis Rheum 44: 208–219, 2014. doi: 10.1016/j.semarthrit.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 51.Ryu D, Jo YS, Lo Sasso G, Stein S, Zhang H, Perino A, Lee JU, Zeviani M, Romand R, Hottiger MO, Schoonjans K, Auwerx J. A SIRT7-dependent acetylation switch of GABPβ1 controls mitochondrial function. Cell Metab 20: 856–869, 2014. doi: 10.1016/j.cmet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 52.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675, 2012. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwartz MI, King TE. Interstitial Lung Disease. Shelton, CT: People's Medical Publishing House-USA, 2010. [Google Scholar]
- 54.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143: 802–812, 2010. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sosulski ML, Gongora R, Feghali-Bostwick C, Lasky JA, Sanchez CG. Sirtuin 3 deregulation promotes pulmonary fibrosis. J Gerontol A Biol Sci Med Sci 72: 595–602, 2017. doi: 10.1093/gerona/glw151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steen VD, Medsger TA. Changes in causes of death in systemic sclerosis, 1972-2002. Ann Rheum Dis 66: 940–944, 2007. doi: 10.1136/ard.2006.066068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem 282: 6823–6832, 2007. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 58.Todd NW, Luzina IG, Atamas SP. Molecular and cellular mechanisms of pulmonary fibrosis. Fibrogenesis Tissue Repair 5: 11, 2012. doi: 10.1186/1755-1536-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics 11: 60–76, 2012. doi: 10.1074/mcp.A111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai YC, Greco TM, Cristea IM. Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Mol Cell Proteomics 13: 73–83, 2014. doi: 10.1074/mcp.M113.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res 102: 703–710, 2008. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 62.Vazquez BN, Thackray JK, Simonet NG, Kane-Goldsmith N, Martinez-Redondo P, Nguyen T, Bunting S, Vaquero A, Tischfield JA, Serrano L. SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J 35: 1488–1503, 2016. doi: 10.15252/embj.201593499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature 477: E1–E2, 2011. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- 64.Wei J, Ghosh AK, Chu H, Fang F, Hinchcliff ME, Wang J, Marangoni RG, Varga J. The histone deacetylase sirtuin 1 is reduced in systemic sclerosis and abrogates fibrotic responses by targeting transforming growth factor β signaling. Arthritis Rheumatol 67: 1323–1334, 2015. doi: 10.1002/art.39061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wronska A, Lawniczak A, Wierzbicki PM, Kmiec Z. Age-related changes in sirtuin 7 expression in calorie-restricted and refed rats. Gerontology 62: 304–310, 2016. doi: 10.1159/000441603. [DOI] [PubMed] [Google Scholar]
- 66.Zerr P, Palumbo-Zerr K, Huang J, Tomcik M, Sumova B, Distler O, Schett G, Distler JH. Sirt1 regulates canonical TGF-β signalling to control fibroblast activation and tissue fibrosis. Ann Rheum Dis 75: 226–233, 2016. doi: 10.1136/annrheumdis-2014-205740. [DOI] [PubMed] [Google Scholar]
- 67.Zhang PY, Li G, Deng ZJ, Liu LY, Chen L, Tang JZ, Wang YQ, Cao ST, Fang YX, Wen F, Xu Y, Chen X, Shi KQ, Li WF, Xie C, Tang KF. Dicer interacts with SIRT7 and regulates H3K18 deacetylation in response to DNA damaging agents. Nucleic Acids Res 44: 3629–3642, 2016. doi: 10.1093/nar/gkv1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng CF, Guan KL. Cytoplasmic localization of the mitogen-activated protein kinase activator MEK. J Biol Chem 269: 19947–19952, 1994. [PubMed] [Google Scholar]