Abstract
olig genes encode a previously unrecognized group of vertebrate-specific basic helix–loop–helix transcription factors. As shown in mice, chickens, and zebrafish, two members of this group, olig1 and olig2, are involved in the differentiation of motoneurons and oligodendrocytes, but nothing is known about the role of the third member, olig3. Here, we show that olig3 plays an essential role in the establishment of the neural crest–lateral neural plate boundary. In zebrafish embryos, morpholino-induced olig3 inactivation dramatically increases the number of neural crest cells, but lateral neural plate fates (interneurons and astrocytes) are missing. Zebrafish swirl mutants that have impaired bone morphogenetic protein signaling and lack neural crest cells display an expanded olig3 expression domain. Moreover, olig3 is up-regulated in mindbomb mutants lacking the neural crest because of an impaired notch signaling, and olig3 repression in such mutants rescues the neural crest. In addition, olig3 regulates ngn1 and deltaA expression in interneuron precursors. Our results indicate that olig3 has an essential proneural activity in the dorsal spinal cord and cooperates with the Delta/Notch regulatory loop to establish the boundary between the neural crest and the lateral neural plate. Thus, a proper regulation of the olig gene family is essential for the formation of three cell types (oligodendrocytes, astrocytes, and neural crest) that are unique to vertebrates.
Keywords: interneurons, neural crest, olig3, zebrafish, glia
The spinal cord of vertebrates is originated by remodeling of the neural plate. Its general plan comprises motoneurons located ventrally and interneurons in a more dorsal position. In zebrafish, Rohon–Beard (RB) cells (primary sensory neurons) are generated at the outer border of the neural plate (within the neural crest domain) and migrate in the dorsal spinal cord, whereas cells of the dorsal root ganglia (also originated in the neural crest) migrate ventrally without entering the spinal cord. Two main signaling pathways are thought to establish the dorsoventral patterning of the embryonic neural tissue: bone morphogenetic protein (BMP) and Hedgehog (Hh). Hh signaling regionalizes the ventral neural tube and restricts the expression of some genes to dorsal regions (1). In parallel, BMP signaling determines a gradient of positional information throughout the entire neural plate that defines the establishment of dorsal and intermediate neuronal cell types of the spinal cord (2). The current view is that BMP and Hh morphogenetic activities generate zones of competence within which other factors will subsequently establish different cell fates. In this regard, components of the Delta/Notch signaling pathway are involved in the generation of different neuronal and glial sub-types with a mechanism called lateral specification. Components of the Delta/Notch genetic loops are often members of the basic helix–loop–helix (bHLH) family of transcription factors. The Neurogenin (Ngn) subfamily of bHLH is critical to establish the neurogenic program and maintain the Delta/Notch regulatory feedback that creates differences within the nervous system (3–5). Recently, a previously unrecognized vertebrate-specific group of bHLH has been shown to participate in neural fate decisions mediated by Delta/Notch signaling: the known members of this family are olig1, olig2, and olig3 (6–9). The discovery that, in mice, chickens, and zebrafish, olig2 is necessary for the differentiation of motoneurons and oligodendrocytes from a ventral population of multipotent neural precursors raised the crucial issue of whether other neuroepithelial domains in the ventral or dorsal spinal cord can switch from neuron to glial cell production: for example, from neurons to astrocytes (6–8). In mice, the expression domains of olig1 and olig2 are partially overlapped and distinct from that of olig3 (9). Because the function of olig3 has not been determined yet, we have isolated and analyzed the zebrafish olig3 gene. Our results show that (i) olig3 is expressed in lateral domains of the neural plate and (ii) its proneural activity is necessary for the development of neural populations derived from the dorsal neuroepithelium, such as interneurons and astrocytes. Moreover, olig3 activity is required to establish the boundary between the neural plate and the neural crest, indicating that the three oligs collectively participate in the formation of astrocytes, oligodendrocytes, and the neural crest.
Materials and Methods
Cloning of Zebrafish olig3 Gene. The olig3 sequence was initially retrieved from zebrafish genomic DNA by using combinations of degenerate primer pairs targeting the bHLH domain: bHLH-F1, AAGAAGGCNAAYGAYCGNGA; bHLH-R1, CADATGTARTTRTGNGCGAA; bHLH-F2, CGNGARCGNAAYCGNATGCA; and bHLH-R2, GTNAGNGCCCADATGTARTT. (n = A, G, C, and T; Y = C and T; R = A and G; and D = A, G, and T.)
The full-length coding sequence was determined and amplified from a zebrafish shield-stage cDNA library (GeneFinder library 567, RZPD) by using the following specific primers: for olig3HLH-F, GCACGACCTCA ACCAGACTA, and for olig3HLH-R, ATGTAGTTTCTGGCGAGCAG, combined with vector oligos. For the antisense riboprobe, the olig3 partial CDS/3′ UTR was cloned into pCRII-TOPO cut with KpnI and transcribed with T7 RNA polymerase.
Embryo Manipulations. The following morpholinos (MOs) (Gene Tools, Philomath, OR) were used: olig3AUGMO, TCTGAATCCATCTTTGAAAGTCTTC (spanning the olig3 AUG region); olig3UTRMO, GAAACAATCCGCTTGAAGTGAGGCG (targeting the olig3 5′ UTR); olig3mismMO, GAAAgAATaCGaTTGAAGTcAGcCG (mismatched MO; mutations are lowercase); and olig2MO, TCCATGGCGTTCAGTGCGCTCTCAG (spanning the olig2 AUG region).
olig3AUGMO and olig3UTRMO acted synergistically, and, when used independently at similar concentrations, they elicited the same phenotype. Unless indicated, the experiments were performed comparing the activity of olig3UTRMO (olig3MO) with that of olig3mismMO. The phenotype of embryos injected with the olig3mismMO was normal in shape and pigmentation.
Capped olig3, Xnotch-ICD (3), β-catenin, dnTCF (10), and GFP messengers were synthesized by linearization of the expression vectors and transcription with SP6 RNA polymerase (mMessage mMachine in vitro transcription kit, Ambion, Austin, TX).
Notch signaling was inactivated in vivo by N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) treatment (11).
Embryo Staining and Whole-Mount in Situ Hybridization. Whole-mount in situ hybridization was performed according to Thisse et al. (12). Whenever possible, MO- or mRNA-injected embryos were stained in the same tube with control embryos (which had the tip of the tail cut). RB cells have been revealed at the two-somite stage in ngn1:GFP-transgenic embryos (13) by using a GFP antisense probe.
Results
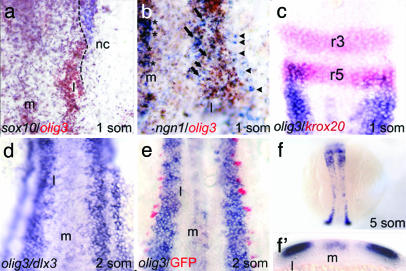
olig3 Is Expressed in the Dorsal Neural Plate. By PCR-based screening, we retrieved olig3 cDNA from libraries of zebrafish at different developmental stages, including a shield-stage cDNA pool. By in situ hybridization, olig3 transcripts are first detected at the shield stage in a stripe of the deep cell layer (not shown). This early and wide expression is peculiar to olig3 and is not observed for the other members of the olig gene family (8, 9). At the tail bud stage (Fig. 6a), olig3-positive cells are arranged in two bilateral domains located in the prospective lateral neural plate and along the midline. Double-staining experiments performed at the one-somite stage with sox10 and ngn1 show that olig3 is expressed medially to neural crest markers (Fig. 1a) and that the lateral (l) olig3 expression domain comprises the region where ngn1-positive interneurons differentiate (Fig. 1b). At the same stage, the anterior border of the olig3 domain is delimited by krox20 expression in rhombomere-5 (r5) (Fig. 1c). At the two-somite stage, olig3 is expressed at the medial side of dlx3-expressing cells, directly adjacent to the prospective RB cells (Fig. 1 d and e). It is noteworthy that, at the two-somite stage, the earliest and anterior RB-cell precursors have already migrated in the neural plate, whereas the more posterior RB-cell precursors (originated later) are still in the neural crest domain (Fig. 1e). Because in ngn1:GFP transgenics the number of RB/GFP-expressing cells is normal and not increased as expected by a shift of the ngn1 expression domain (13), in this specific case, it is unlikely that segregation of fates is achieved by changes in gene expression domain (without movement of cells).
Fig. 6.
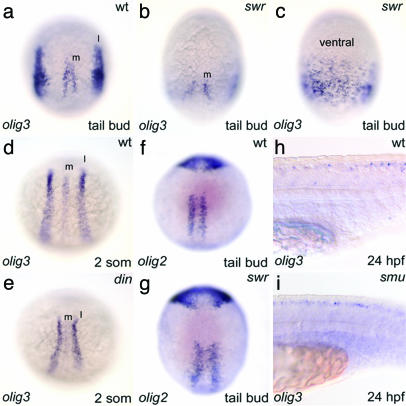
BMP-dependent restriction of olig3 expression domains. (a) Normal embryo. (b) swr embryo showing a widened space between the two medial domains (m). (c) Ventral view of a swr embryo showing that the lateral olig3 domains are broadened and merge on the ventral side. (d) Normal embryo. (e) din embryo with reduced lateral (l) domain. (f and g) Normal (f) and swr (g) embryos show a similar olig2 pattern. (h and i) Normal expression of olig3 in the dorsal spinal cord of the smu embryo. Embryos in a–g are in dorsal view with anterior to the top, except in c (ventral view). Embryos in h and i are in lateral view, anterior to the left.
Fig. 1.
olig3 expression pattern. (a) High magnification of the right side of the rhomboencephalon region showing that olig3 (brown) is expressed medially to the neural crest marker sox10 (blue); the dashed line represents the boundary of expression of the two markers. (b) High magnification of the right side of the trunk region shows that the lateral (l) olig3 (brown) expression domain comprises the region where ngn1-positive interneurons (arrows) differentiate. RB cells (arrowheads) are in a more lateral position and are outside the olig3 expression domain. (c) The anterior border of the olig3 domain (blue) is delimited by krox20 (red) in rhombomere-5 (r5). (d) The olig3 domain has elongated rostrocaudally, and a two-cell-wide gap separates olig3 and dlx3. (e) ngn1:GFP transgenics showing that RB-cell precursors (red) and olig3-expressing cells are in close contact. (f) At the five-somite stage, the lateral domain has further elongated. (f′) Transverse section at the level of the posterior trunk of the embryo in f. The embryos are in dorsal view, anterior to the top.
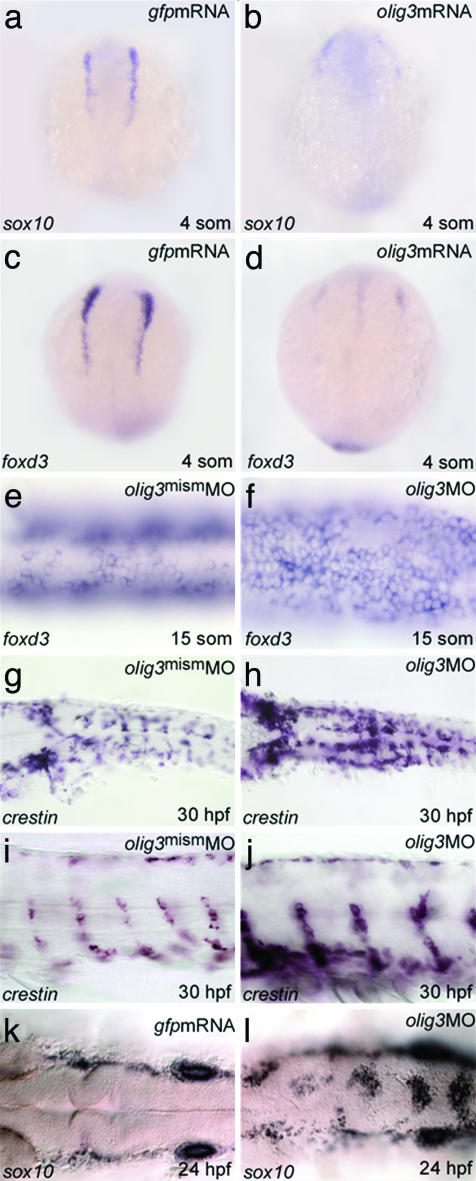
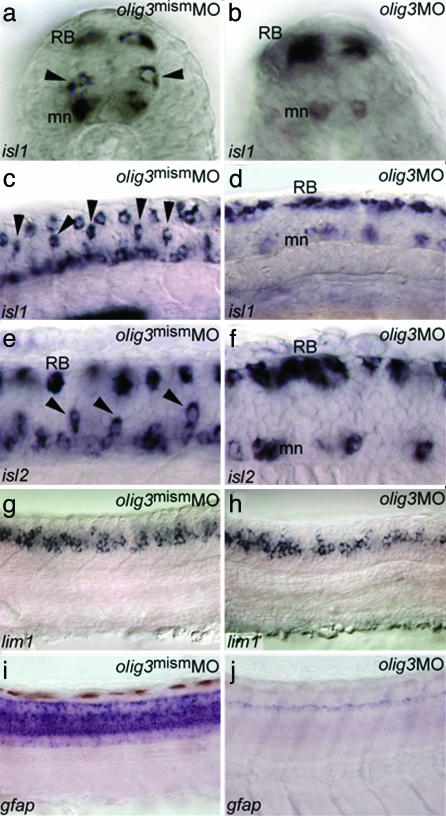
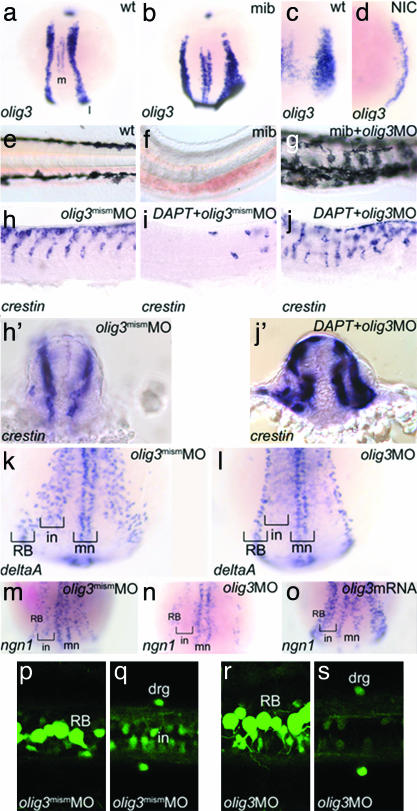
olig3 Is Necessary for the Differentiation of the Dorsal Spinal Cord. The early and strong expression and the lack of information about olig3 function prompted us to address its role in development by means of loss and gain of function studies. The targeted inactivation of olig3 was achieved by using antisense MO oligo-nucleotides (14). The MO-injected embryos (morphants) display a general retardation of development, a curved tail, a scarce motility, and, strikingly, an increased pigmentation at 36 hours postfertilization (hpf) (not shown). Injection of a mismatched MO (Figs. 2 and 3) as well as rescue experiments in which the olig3UTRMO was injected with olig3 mRNA demonstrate the specificity of the MO used in these analyses (line 6 of Table 1). To investigate whether the increased pigmentation in olig3 morphants reflects an increment of neural crest cells, we have analyzed the expression pattern of neural crest-expressed markers. It can be observed (Fig. 2 e–l) that foxd3/fkd6, crestin, and sox10 (15–17) are expressed at considerably higher levels in olig3 morphants compared with siblings injected with a mismatched olig3MO. Moreover, in the morphants, these markers are expressed ectopically and along the dorsal midline (Fig. 2 e–l). Conversely, misexpression of olig3 messengers is associated with decreased or absent pigmentation (lines 4 and 5 of Table 1) and reduced expression of early neural crest markers (Fig. 2 a–d). These results show that olig3 is able to block the specification of trunk neural crest precursors. We next analyzed the neural populations deriving from the lateral neural plate and neural crest: astrocytes, sensory neurons, and different classes of interneurons (D1–D3) (18, 19). Analysis of olig3 morphants with isl1 and isl2 indicates that D2 interneurons are missing; lim1 reveals that D3 interneurons are fairly normal or slightly decreased (Fig. 3 a–h). In addition, the RB cells, deriving from the neural crest, are increased in number in olig3 morphants compared with control embryos (Figs. 3 b, d, and f and 4r). As shown in Fig. 3j, olig3 inactivation affects the expression of the glial fibrillar acidic protein gene, which is normally expressed in astrocytes (Fig. 3i and ref. 20). In summary, olig3 morphants lack neurons and glial cells derived from the neural plate domain between D3 interneurons and the neural crest. Because levels of cell death and proliferation are comparable in olig3 morphants and controls, apoptosis and control of cell cycle cannot explain the lack of astrocytes or interneurons in morphants (not shown).
Fig. 2.
olig3 controls specification of the neural crest. (a–d) The levels and the expression domains of sox10 and foxd3 are decreased in four-somite embryos injected with olig3 mRNA (b and d) (phenotype in >90% of injected embryos, n = 30). (e and f) The expression of foxd3 is increased in 15-somite embryos treated with olig3MO (f) (morphant phenotype > 90%, n = 20). Similarly, the expression of crestin is increased in 30-hpf embryos treated with olig3MO (h and f) (morphant phenotype > 90%, n = 40). Similar results are obtained in 24-hpf embryos hybridized with sox10 (k and l) (morphant phenotype > 90%, n = 30). Embryos are in dorsal (a–h, k, and l) or lateral (i and j) view, anterior to the top (a–d) or to the left (e–l).
Fig. 3.
olig3 knockdown affects differentiation of lateral neural plate cells. (a–f) isl1/2-expressing interneurons (a and b; black arrowheads in a) (18, 19) are absent in 20-somite (b and d) and 24-hpf (f) embryos injected with olig3MO. Conversely, RB cells (RB) are increased in number in olig3 morphants (b, d, and f) (morphant isl1-phenotype > 90%, n = 40; morphant isl2-phenotype > 90%, n = 30). mn, motoneurons. (g and h) lim1 is expressed at 24 hpf in D3 interneurons of the dorsal spinal cord (g). D3 interneurons are slightly affected in embryos injected with olig3MO (h) (morphant phenotype > 90%, n = 15). (i and j) The glial fibrillar acidic protein gene (gfap) is expressed at 3 days postfertilization in astrocytes of the dorsal spinal cord (i). gfap expression is absent in embryos injected with olig3MO (j) (morphant phenotype > 90%, n = 20). Embryos are in transverse sections (a and b) or in lateral view with anterior to the left (c–j).
Table 1. Results of microinjection experiments.
| Construct injected | Amount injected | No. surviving 1 day after fertilization/no. injected (%) | Pigmentation 2 days after fertilization |
|---|---|---|---|
| olig3mismMO | 10 ng | 92/97 (95) | Normal |
| olig3UTRMO | 10 ng | 100/159 (63) | High |
| olig3 mRNA | 40 pg | 0/101 (0) | — |
| olig3 mRNA + olig3UTRMO | 40 pg + 10 ng | 26/46 (57) | Low (60%) or absent (40%) |
| olig3 mRNA | 20 pg | 64/84 (76) | Low (70%) or normal (30%) |
| olig3 mRNA + olig3UTR MO | 20 pg + 10 ng | 54/66 (82) | Normal |
Fig. 4.
olig3 is controlled by Notch and regulates ngn1 and deltaA expression in the lateral neural plate. (a–d) Notch signaling controls olig3 expression. (a) olig3 expression at the two-somite stage showing the medial (m) and lateral (l) domains. (b) mib embryo at the two-somite stage showing increased and broadened expression of olig3. (c and d) Tail bud-stage embryo injected with Notch-ICD (NIC) showing dramatic reduction of olig3 expression (d) (shown phenotype > 90%, n = 20). (e–g) olig3MO rescues pigmentation in mib mutants. (e) Trunk pigmentation at 30 hpf. (f) mib embryo lacks melanocytes. (g) Rescue of the pigmentation in a mib mutant injected with olig3MO (morphant phenotype 9/10). (h–j, h′, and j′) olig3MO rescues the neural crest in DAPT-treated embryos. (h) crestin expression in a normal embryo at 30 hpf. (h′) Transverse section of the embryo in h. (i) A DAPT-treated embryo lacks most of the neural crest cells. (j) Injection of olig3MO in an embryo treated with DAPT rescues neural crest cells (morphant phenotype 9/10). (j′) Transverse section of the embryo in j. (k and l) olig3 controls the expression of deltaA in interneurons. (k) deltaA expression pattern in a four-somite-stage control embryo injected with olig3mismMO. (l) The stripe of deltaA-positive cells corresponding to interneurons (in) is missing in embryos injected with olig3MO (morphant phenotype > 90%, n = 40). (m–o) In situ hybridizations at two-somite stage showing that olig3 activates ngn1 expression in the lateral neural plate. (m) Expression of ngn1 in motoneurons (mn), interneurons (in), and RB cells (RB) in control embryos injected with olig3mismMO. (n) In an olig3MO-injected embryo, ngn1 is expressed only in the medialmost (mn) and lateralmost (RB) neuronal populations (morphant phenotype > 90%, n = 20). (o) Injection of olig3 mRNA up-regulates ngn1 expression (shown phenotype > 90%, n = 20). (p–s) RB and dorsal root ganglion (drg) cells are present in ngn1:GFP-transgenic embryos (13) injected with olig3MO (r and s), as in controls (p and q). Interneurons (in) are reduced or absent (q and s). Embryos are shown in dorsal view with anterior to the top (a–d and k–o), lateral view with anterior to the left (e–j), and dorsal view with anterior to the left (p–s).
olig3 Is a Proneural Gene. Notch signaling, by repressing the expression of its ligands (Delta and Jagged), couples cell-autonomous and non-cell-autonomous regulatory loops of gene expression that lead to boundary formation or cell-lineage divergence (21). Because Notch signaling is necessary for neural crest specification (4, 22), we analyzed olig3 expression in mindbomb (mib) embryos that bear a mutation in a ubiquitin ligase gene necessary for efficient Delta activation of Notch (23). mib embryos are characterized by excess RB cells and interneurons at the expense of pigment cells and other trunk neural crest derivatives (4, 24, 25). In mib mutants, expression of olig3 is increased (Fig. 4 a and b), suggesting that it is down-regulated by Notch signaling. Further evidence that Notch controls olig3 expression comes from the misexpression of Notch-ICD, a dominant form of Notch intracellular that activates Notch signaling constitutively (3). Embryos injected with Notch-ICD mRNA and analyzed at gastrula stage displayed reduced olig3 expression compared with controls (Fig. 4 c and d). We reasoned that, if olig3 is under negative control of Notch signaling and its activity represses neural crest fates, its inactivation in mib mutants should restore neural crest development. Consistent with this hypothesis, we found that olig3 inactivation, induced with MO, restores pigmentation in mib mutant embryos (Fig. 4 e–g). Moreover, we tested the rescue of neural crest cells in embryos in which the intracellular release of Notch was blocked with DAPT, a γ-secretase inhibitor (11). Zebrafish embryos treated with DAPT display a phenotype that strictly resembles that of mib mutants (11), and their expression of crestin is dramatically reduced (Fig. 4i). However, when injected with olig3MO, DAPT-treated embryos show complete rescue of crestin expression, although the migratory path of crest cells is altered according to the disorganization of somites (Fig. 4 j and j′). These experiments demonstrate that functional repression of olig3, downstream of inactive Notch signaling, restores neural crest formation. Misexpression of proneural genes up-regulates delta genes in frog and mouse embryos (3, 26). We therefore tested whether olig3, in a manner similar to proneural genes, also regulates the levels of delta gene expression. Indeed, in olig3 morphants analyzed at the four-somite stage, the deltaA expression domain corresponding to interneurons (4) is missing (Fig. 4 k and l). Moreover, olig3 overexpression causes increased deltaA expression (not shown). Thus, our results indicate that olig3 behaves like a proneural gene and is required in some cell lineages for the expression of Notch ligands.
Some aspects of olig3 function are similar to those described for the ngn1 gene. During early somitogenesis, ngn1 is expressed in three proneural domains in cells fated to differentiate into sensory neurons (RB cells and dorsal root ganglia in the lateral neural plate), interneurons (intermediate neural plate) and motoneurons (medial neural plate). ngn1 is required for the formation of RB cells at the border of neural and nonneural ectoderm and its knockdown partially resumes the formation of pigments cells in mib mutants (4). Moreover, previous studies revealed interactions between ngn and olig2 during the differentiation of motoneurons and oligodendrocytes (7, 27). We found that olig3 morphants lack the ngn1-expressing interneuron precursors, whereas ngn1 is normally expressed in the lateralmost and medialmost domains that correspond to the neural progenitors of the neural crest and motoneurons, respectively (Fig. 4 m, n, and p–s). In addition, olig3 overexpression leads to increased ngn1 levels (Fig. 4o). Conversely, ngn1 mutants (28) have a normal expression pattern of olig3 (not shown). All of these observations suggest that the intermediate ngn1 domain, fated to interneuron precursors, depends on the expression of olig3.
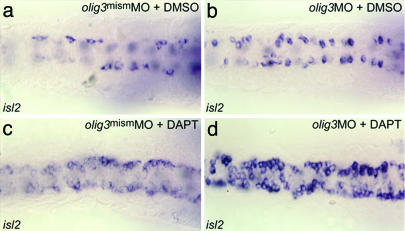
We then tested whether Notch could regulate mediolateral specification through olig3 expression or whether olig3 acts like ngn1 in promoting RB-cell fates at the expense of the neural crest. To test these alternatives, we treated zebrafish embryos with DAPT and olig3MO, supposing that if olig3 is acting like ngn1, then the olig3MO would decrease the number of RB cells in DAPT-treated animals. Results show that DAPT treatment and olig3MO act synergistically (Fig. 5 b–d), confirming the role of olig3 in Notch-mediated mediolateral specification of the neural tube.
Fig. 5.
olig3 mediates mediolateral specification. isl2 staining of RB cells is shown. RB cells originating in the neural crest domain are increased in number in olig3MO-injected embryos (morphant isl2 phenotype > 90%, n = 20) (b) as well as in DAPT-treated embryos (morphant isl2 phenotype > 90%, n = 40) (c) compared with controls (a). DAPT treatment and olig3MO act synergistically (d) (morphant isl2 phenotype > 90%, n = 20). Embryos are shown in dorsal view with anterior to the left.
Regulation of olig3 During Gastrulation. The expression of olig3 in the lateral domain of the neuroectoderm at the shield stage suggests that early genetic events restrict its expression ventrally. Because Wnt signaling is required for polarization of the early vertebrate embryo and the specification of the dorsal neuroectoderm (29, 30), olig3 might be implicated in the regulation of neural plate and neural crest fates under the control of the blastula Wnt gradient. To test this hypothesis, we analyzed the olig3 expression pattern in embryos in which the Wnt signaling is increased or impaired. In embryos in which Wnt signal is increased either by lithium treatment (31, 32) or by injection of β-catenin (10), the expression domain of olig3 is expanded in the prospective ventral side (not shown). Because Wnt signaling, by activating chordin, also represses the dorsal activity of the BMP gradient (33), it could be either directly or indirectly (by means of BMP) involved in regulating the olig3 expression domain. To test this hypothesis, we analyzed the olig3 expression pattern in embryos dorsalized with lithium treatment and injected with a chordin antisense MO (14). The results show that the olig3 domain is not expanded as would be expected in lithium-treated embryos; rather, it is restricted dorsally. This result indicates that BMP, downstream of Wnt signaling, is directly involved in regulating olig3 expression (not shown). To further test this hypothesis, we analyzed the olig3 expression pattern in swirl/bmp2b (swr) and dino/chordin (din) mutants in which the BMP signal is impaired and increased, respectively (34–36). In swr embryos that lack the neural crest (37, 38), the lateral domain of olig3 is expanded and surrounds the embryo completely (Fig. 6 a–c). Conversely, in din mutants, the lateral olig3 domain is reduced (Fig. 6 d and e), and overexpression of bmp2b suppresses olig3 expression (not shown; n = 10). Thus, BMP signaling limits the olig3 domain and, by relieving olig3 repression of neural crest-specific genes (such as foxd3, sox10, and crestin), allows an ectodermal subset of competent cells to acquire neural crest fates.
The olig2 gene is expressed in the ventral neural plate in regions that do not overlap with olig3 domains (8, 9), and its expression is controlled by Shh (8). As shown in Fig. 6 (f and g), the olig2 expression pattern is fairly normal in swr mutants. Conversely, olig3 expression in the dorsal spinal cord is unaffected in both zebrafish smu mutants and cyclopamine-treated embryos, in which the shh pathway is impaired (39) (Fig. 6 h and i and not shown). Thus, Hedgehog signaling is required to establish olig2 expression and ventral neuroectoderm competence (6, 8), whereas Bmp signaling establishes the olig3 expression domain and dorsal neuroectoderm competence.
Discussion
olig3 is a gene expressed during early gastrulation in the prospective dorsal spinal cord at the proximal boundary with the prospective neural crest. The shape and formation of the outer (lateral) boundaries of the neural crest can be revealed and studied with a large number of markers (i.e., dlx genes). However, to our knowledge, olig3 is the only example of a gene revealing and precisely defining the inner (medial) boundary of the neural crest. As expected for a gene regulating dorsal neural tube differentiation, its expression is under the control of BMP, rather than Shh. Because olig3 is expressed in the presumptive ectoderm at a specific position along the anteroposterior and mediolateral axes, it is possible that the early BMP/Wnt signaling affects its expression indirectly; however, this hypothesis is unlikely, because olig3 is already expressed at epiboly. We propose a genetic model describing the role of olig3 in cell fate decisions taken at the neural crest–lateral neural plate boundary. According to this model, BMP and Notch signals restrict olig3 transcription. In addition, swirl/bmp2b is essential to establish neural crest and lateral neural plate zones of competence, as postulated by Nguyen et al. (37). This zone of competence is likely to be generated at the cleavage stage by a Wnt-mediated dorsal repression of Bmp (33).
In the prospective lateral neural plate, Chordin, secreted by the organizer, binds to BMP and releases the transcription of olig3, which (i) represses the expression of neural crest-specific genes, (ii) non-cell-autonomously activates Notch in prospective neural crest cells, and (iii) promotes some lateral neural plate fates, such as that of astrocytes and deltaA- and ngn1-positive interneurons. The initial mediolateral unbalance of BMP signaling creates an asymmetry in olig3 expression that, by means of the Delta/Notch genetic loop, establishes the boundary between the neural crest and the lateral neural plate. This model predicts that, in mib mutants, the lack of Notch signaling allows general olig3 expression, which, in turn, represses neural crest-specific genes, as seen in misexpressed olig3 embryos. Conversely, when olig3 is knocked down, the expression of neural crest-specific genes is up-regulated in wild-type embryos or rescued in mib mutants. This effect is more dramatic, compared with ngn1 knockdown (4), suggesting that ngn1 might be involved in specific cell fate subdecisions downstream of the olig3 activity. The significant increase of RB cells in olig3MO-injected embryos implies that not only the neural crest, but also the RB-cell specification is negatively regulated by olig3. The dramatic increase of RB and neural-crest cells, due to the MO-mediated block of olig3 translation, is associated with reduction of dorsal neural cell types, such as interneurons and astrocytes. Notably, these effects are not accompanied by changes in cell death or proliferation, implying that dorsal neural populations lacking olig3 activity are fated to neural crest and RB cells. Overall, our finding would suggest that olig3 promotes the astrocyte/interneuron domain at the expense of the dorsal domain that corresponds to RB/crest cells. Thus, olig3 appears to act like olig2, which promotes the motoneuron/oligodendrocyte domain at the expense of the dorsal astrocyte/interneuron domain. Notably, the specific function of olig2 and olig3 in neuronal development reveals a spatial model of neuron/glia differentiation, with motoneurons/oligodendrocytes and interneurons/astrocytes originating in the ventral (olig2+) and dorsal (olig3+) neuroepithelium, respectively. In conclusion, Olig proteins form a vertebrate-specific group of bHLH transcription factors controlling the development of neural crest cells, astrocytes, and oligodendrocytes, three neuroectodermal-derived cell types occurring only in vertebrates. A better knowledge of the genetic pathways that control activation and repression of olig genes will help to dissect the mechanisms leading to either differentiation or malignant transformation of these cell lineages.
Acknowledgments
We thank Dirk Meyer for assistance in some crucial experiments, Stefan Pauls and Wolfgang Driever for help and assistance, Bruce Appel (Vanderbilt University, Nashville) and Stefano Piccolo (Universita' di Padua) for critical reading of the manuscript and for providing plasmids, and Margaret Kirk for her kind revision. Zebrafish strains were kindly provided as follows: ngn1:GFP, Uwe Strahle (Forschungszentrum Karlsruhe, Karlsruhe, Germany); mib, Ajay Chitnis (National Institutes of Health/National Institute of Child Health and Human Development); swr and din, Steve Wilson (University College London, London); and smu, Zoltan Varga (University of Oregon, Eugene). This work was supported by the European Commissions as part of the ZF-MODELS Integrated Project in the Sixth Framework Program (Contract no. LSHG-CT-2003-503496).
Author contributions: A.F., N.T., G.D., and E.Z. performed research; A.F., N.T., G.D., E.Z., M.B., and F.A. analyzed data; N.T. and F.A. designed research; and N.T., M.B., and F.A. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: bHLH, basic helix–loop–helix; BMP, bone morphogenetic protein; RB, Rohon–Beard; Ngn, Neurogenin; MO, morpholino; olig3MO, olig3UTRMO; DAPT, N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester; hpf, hours postfertilization; mib, mindbomb; swr, swirl/bmp2b; din, dino/chordin.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AJ488292 (olig2) and AJ488293 (olig3)].
References
- 1.Ericson, J., Muhr, J., Jessell, T. M. & Edlund, T. (1995) Int. J. Dev. Biol. 39, 809–816. [PubMed] [Google Scholar]
- 2.Lee, K. J. & Jessell, T. M. (1999) Annu. Rev. Neurosci. 22, 261–294. [DOI] [PubMed] [Google Scholar]
- 3.Chitnis, A. & Kintner, C. (1996) Development (Cambridge, U.K.) 122, 2295–2301. [DOI] [PubMed] [Google Scholar]
- 4.Cornell, R. A. & Eisen, J. S. (2002) Development (Cambridge, U.K.) 129, 2639–2648. [DOI] [PubMed] [Google Scholar]
- 5.Blader, P., Fischer, N., Gradwohl, G., Guillemont, F. & Strahle, U. (1997) Development (Cambridge, U.K.) 124, 4557–4569. [DOI] [PubMed] [Google Scholar]
- 6.Lu, Q. R., Yuk, D., Alberta, J. A., Zhu, Z., Pawlitzky, I., Chan, J., McMahon, A. P., Stiles, C. D. & Rowitch, D. H. (2000) Neuron 25, 317–329. [DOI] [PubMed] [Google Scholar]
- 7.Zhou, Q. & Anderson, D. J. (2002) Cell 109, 61–73. [DOI] [PubMed] [Google Scholar]
- 8.Park, H. C., Mehta, A., Richardson, J. S. & Appel, B. (2002) Dev. Biol. 248, 356–368. [DOI] [PubMed] [Google Scholar]
- 9.Takebayashi, H., Ohtsuki, T., Uchida, T., Kawamoto, S., Okubo, K., Ikenaka, K., Takeichi, M., Chisaka, O. & Nabeshima, Y. (2002) Mech. Dev. 113, 169–174. [DOI] [PubMed] [Google Scholar]
- 10.Agius, P. E., Piccolo, S. & De Robertis, E. M. (1999) J. Soc. Biol. 193, 347–354. [PMC free article] [PubMed] [Google Scholar]
- 11.Geling, A., Steiner, H., Willem, M., Bally-Cuif, L. & Haass, C. (2002) EMBO Rep. 3, 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thisse, C., Thisse, B., Schilling, T. F. & Postlethwait, J. H. (1993) Development (Cambridge, U.K.) 119, 1203–1215. [DOI] [PubMed] [Google Scholar]
- 13.Blader, P., Plessy, C. & Strahle, U. (2003) Mech. Dev. 120, 211–218. [DOI] [PubMed] [Google Scholar]
- 14.Nasevicius, A. & Ekker, S. C. (2000) Nat. Genet. 26, 216–220. [DOI] [PubMed] [Google Scholar]
- 15.Dutton, K. A., Pauliny, A., Lopes, S. S., Elworthy, S., Carney, T. J., Rauch, J., Geisler, R., Haffter, P. & Kelsh, R. N. (2001) Development (Cambridge, U.K.) 128, 4113–4125. [DOI] [PubMed] [Google Scholar]
- 16.Rubinstein, A. L., Lee, D., Luo, R., Henion, P. D. & Halpern, M. E. (2000) Genesis 26, 86–97. [DOI] [PubMed] [Google Scholar]
- 17.Odenthal, J. & Nusslein-Volhard, C. (1998) Dev. Genes Evol. 208, 245–258. [DOI] [PubMed] [Google Scholar]
- 18.Lee, K. J., Dietrich, P. & Jessell, T. M. (2000) Nature 403, 734–740. [DOI] [PubMed] [Google Scholar]
- 19.Tamme, R., Wells, S., Conran, J. G. & Lardelli, M. (2002) BMC Dev. Biol. 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen, A. L. & Jorgensen, A. L. (2003) Gene 310, 123–132. [DOI] [PubMed] [Google Scholar]
- 21.Haddon, C., Jiang, Y. J., Smithers, L. & Lewis, J. (1998) Development (Cambridge, U.K.) 125, 4637–4644. [DOI] [PubMed] [Google Scholar]
- 22.Glavic, A., Silva, F., Aybar, M. J., Bastidas, F. & Mayor, R. (2004) Development (Cambridge, U.K.) 131, 347–359. [DOI] [PubMed] [Google Scholar]
- 23.Itoh, M., Kim, C. H., Palardy, G., Oda, T., Jiang, Y. J., Maust, D., Yeo, S. Y., Lorick, K., Wright, G. J., Ariza-McNaughton, L., et al. (2003) Dev. Cell 4, 67–82. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, Y. J., Brand, M., Heisenberg, C. P., Beuchle, D., Furutani-Seiki, M., Kelsh, R. N., Warga, R. M., Granato, M., Haffter, P., Hammerschmidt, M., et al. (1996) Development (Cambridge, U.K.) 123, 205–216. [DOI] [PubMed] [Google Scholar]
- 25.Schier, A. F., Neuhauss, S. C., Harvey, M., Malicki, J., Solnica-Krezel, L., Stainier, D. Y., Zwartkruis, F., Abdelilah, S., Stemple, D. L., Rangini, Z., et al. (1996) Development (Cambridge, U.K.) 123, 165–178. [DOI] [PubMed] [Google Scholar]
- 26.Ma, Q., Chen, Z., del Barco Barrantes, I., de la Pompa, J. L. & Anderson, D. J. (1998) Neuron 20, 469–482. [DOI] [PubMed] [Google Scholar]
- 27.Novitch, B. G., Chen, A. I. & Jessell, T. M. (2001) Neuron 31, 773–789. [DOI] [PubMed] [Google Scholar]
- 28.Golling, G., Amsterdam, A., Sun, Z., Antonelli, M., Maldonado, E., Chen, W., Burgess, S., Haldi, M., Artzt, K., Farrington, S., et al. (2002) Nat. Genet. 31, 135–140. [DOI] [PubMed] [Google Scholar]
- 29.Nieto, M. A. (2001) Mech. Dev. 105, 27–35. [DOI] [PubMed] [Google Scholar]
- 30.Muroyama, Y., Fujihara, M., Ikeya, M., Kondoh, H. & Takada, S. (2002) Genes Dev. 16, 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stachel, S. E., Grunwald, D. J. & Myers, P. Z. (1993) Development (Cambridge, U.K.) 117, 1261–1274. [DOI] [PubMed] [Google Scholar]
- 32.Klein, P. S. & Melton, D. A. (1996) Proc. Natl. Acad. Sci. USA 93, 8455–8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oelgeschlager, M., Kuroda, H., Reversade, B. & De Robertis, E. M. (2003) Dev. Cell 4, 219–230. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen, V. H., Schmid, B., Trout, J., Connors, S. A., Ekker, M. & Mullins, M. C. (1998) Dev. Biol. 199, 93–110. [DOI] [PubMed] [Google Scholar]
- 35.Lee, K. H., Marden, J. J., Thompson, M. S., MacLennan, H., Kishimoto, Y., Pratt, S. J., Schulte-Merker, S., Hammerschmidt, M., Johnson, S. L., Postlethwaite, J. H., et al. (1998) Dev. Genet. (Amsterdam) 23, 97–103. [DOI] [PubMed] [Google Scholar]
- 36.Schulte-Merker, S., Lee, K. J., McMahon, A. P. & Hammerschmidt, M. (1997) Nature 387, 862–863. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen, V. H., Trout, J., Connors, S. A., Andermann, P., Weinberg, E. & Mullins, M. C. (2000) Development (Cambridge, U.K.) 127, 1209–1220. [DOI] [PubMed] [Google Scholar]
- 38.Barth, K. A., Kishimoto, Y., Rohr, K. B., Seydler, C., Schulte-Merker, S. & Wilson, S. W. (1999) Development (Cambridge, U.K.) 126, 4977–4987. [DOI] [PubMed] [Google Scholar]
- 39.Varga, Z. M., Amores, A., Lewis, K. E., Yan, Y. L., Postlethwait, J. H., Eisen, J. S. & Westerfield, M. (2001) Development (Cambridge, U.K.) 128, 3497–3509. [DOI] [PubMed] [Google Scholar]