Abstract
p73, a member of the p53 family of transcription factors, is upregulated in response to DNA damage, inducing cell cycle arrest and apoptosis. Besides indications that this p73 response is post-transcriptional, little is known about the underlying molecular mechanisms of p73 protein degradation. Ubiquitination and proteasomal-dependent degradation of p53 are regulated by its transcriptional target MDM2. However, unlike p53, p73 binds to, but is not degraded by, MDM2. Here we describe the binding of p73 to Itch, a Hect ubiquitin–protein ligase. Itch selectively binds and ubiquitinates p73 but not p53; this results in the rapid proteasome-dependent degradation of p73. Upon DNA damage Itch itself is downregulated, allowing p73 protein levels to rise and thus interfere with p73 function. In conclusion, we have identified a key mechanism in the control of p73 protein levels both in normal as well as in stress conditions.
Keywords: degradation, E3 ligase, p53, proteasome
Introduction
p73 is a member of the p53 family of transcription factors, and, like p53, has a modular structure (Figure 1A) (Kaghad et al, 1997). p73 shares a high degree of sequence homology with p53 and can bind to p53-responsive elements, activating the transcription of p53 target genes, such as those inducing cell cycle arrest and promoting apoptosis (De Laurenzi et al, 1998, 2000; Catani et al, 2002).
Figure 1.
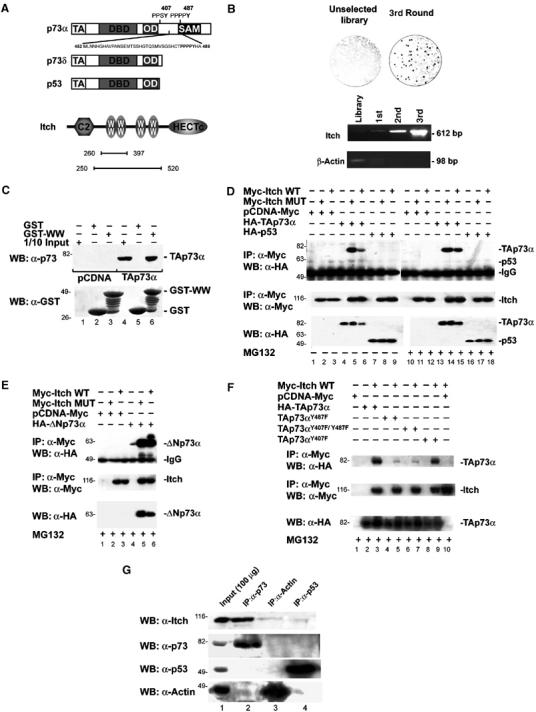
p73 binds to Itch. (A) Schematic representation of the modular structure of the p73α, p73δ, p53, and Itch proteins. The main structural domains are indicated: transactivation domain (TA), DNA-binding domain (DBD), oligomerization domain (OD), sterile alpha motif (SAM), amino-terminal C2 domain (C2), WW domains (WW), and carboxyl-terminal Hect domain (HECTc). The p73 region from Met452 to Ala489, containing the PPxY motif, used as bait in the phage library panning experiment, is also shown. Bars indicate the regions of Itch expressed by the clones selected in the screening. (B) T7 plaques from the unselected library and from the enriched phage pool, transferred to a cellulose membrane and probed with GST-PY are shown. PCR demonstrating the enrichment of clones containing the Itch WW domains is shown in the lower panel. (C) GST pull-down. Hek293 cells were transfected with HA-TAp73α (TAp73α) or empty vector (pCDNA), and lysates were incubated with GST alone, or a GST fusion protein containing the four WW domains of Itch (GST-WW). The retained proteins were detected with anti-p73 antibody (upper panel). The same blot was reprobed with anti-GST polyclonal serum (lower panel). Co-IP of overexpressed proteins: HA-TAp73α or HA-TAp53 (D) or HA-ΔNp73α (E) or TAp73αY487F, TAp73αY407F, and TAp73αY407F/Y487F (F) were transiently transfected in Hek293 cells with either Myc-Itch or Myc-Itch MUT expression vectors. Cells were treated with or without MG132 before lysis. Cell extracts were IP with anti-Myc antibody. The immune complexes were subjected to western blot analysis with anti-HA antibody (upper panels). The same blots were re-probed with anti-Myc antibody (middle panels). Aliquots of total cell extracts from unprocessed cells (25 μg/lane) were directly subjected to immunoblot analysis with anti-HA antibody (lower panels). (G) Co-IP of endogenous p73 and Itch proteins. Cells were IP with antibodies against either p73 (mix of clones C17 and C20, Santa Cruz) or p53 (mix of clones D01 and 1801, Santa Cruz) and western blot was performed with antibody against: Itch, p73, p53, and Actin. As a control, IP was performed also with an anti-Actin antibody.
Unlike p53, however, p73 is expressed as different isoforms (Kaghad et al, 1997; Ueda et al, 1999). Most of the variation generated by alternative splicing occurs at the 3′ end, in a part of the sequence that does not have a counterpart in p53. At least six different p73 proteins (α–η) are generated (De Laurenzi et al, 1998, 1999; Ueda et al, 1999). In addition, the p73 gene exploits an alternative promoter and an extra exon (exon 3′) to generate N-terminally truncated isoforms (ΔNp73). These variants lack the transactivation domain and act as ‘dominant negatives', blocking the function of either p53 or p73 full-length proteins (Yang et al, 2000; Grob et al, 2001; Sayan et al, 2004). The relative levels of TA and ΔN isoforms determine cell fate, resulting in either growth arrest and death or uncontrolled proliferation. TAp73 steady-state protein levels are upregulated in response to DNA damage in a fashion distinct from p53 (Agami et al, 1999; Gong et al, 1999; Yuan et al, 1999), while ΔNp73 is rapidly degraded (Maisse et al, 2004). These observations suggest an important differential role for these isomers in carcinogenesis (Melino et al, 2002, 2003; Stiewe and Putzer, 2002; Zaika et al, 2002).
Despite its importance, however, very little is known of the molecular mechanisms underlying the regulation of p73 protein steady-state levels. MDM2, the E3 ubiquitin (Ub) ligase that regulates the degradation of the cognate protein p53 via a proteasomal-dependent pathway, binds to p73 but does not promote its degradation (Balint et al, 1999; Dobbelstein et al, 1999; Lohrum and Vousden, 1999; Ongkeko et al, 1999; Zeng et al, 1999).
In order to define the degradation pathway of p73, we searched a human cDNA library displayed on bacteriophage capsids for p73-specific binding partners. In order to identify mechanisms distinct from those of p53, we used a p73 C-terminal fragment as a bait, since this region is not present in p53 and contains a PPxY (PY) sequence that has been characterized as a binding motif for a class of WW domains (Figure 1A) (Sudol, 1996).
We found that p73 binds Itch, a human Ub–protein ligase (E3). The Itch mouse homologue gene is absent in the non-agouti-lethal 18H (Itchy) mice, which display profound immune defects (Perry et al, 1998; Fang et al, 2002). Itch belongs to the Nedd4-like E3 family, and is characterized by a modular organization that includes: an N-terminal protein kinase C-related C2 domain, multiple WW domains, and a C-terminal HECT (homologous to the E6-associated protein carboxyl-terminus) Ub–protein ligase domain (Harvey and Kumar, 1999) (Figure 1A). Ub–protein ligases are involved in the multistep process that leads to ubiquitination of protein substrates. In this pathway, E3 catalyzes the final transfer of Ub to a specific substrate, thus governing the specificity of substrate recognition (Hicke, 2001; Kloetzel, 2001; Weissman, 2001).
Here we show that Itch binds and ubiquitinates TAp73 and ΔNp73, but not p53, and determines p73 proteasome-dependent degradation. We also show that Itch expression is downregulated upon DNA damage, thereby allowing stabilization of the TA and ΔN p73 proteins. Our data also show that a second as yet unidentified mechanism is responsible for selective ΔNp73 degradation in response to DNA damage. Therefore, we have identified a relevant mechanism in the control of p73 levels both in normal as well as in stress conditions.
Results
Identification of Itch as a novel p73 interacting partner
To identify new p73-binding proteins, not shared by p53, we fused a fragment of TAp73α (Met 452–Ala 489) to GST protein (Figure 1A) and used it as bait in a ‘phage display' screening (Cesareni et al, 1999; Castagnoli et al, 2001). This fragment contains a region that is not homologous to p53 and contains a protein-protein binding motif, known as a PY motif (Strano et al, 2001) (Figure 1A) characterized by the consensus sequence PPxY. This motif binds to a 40 amino-acid long structural domain known as WW domain, organized to form a three-stranded, antiparallel β sheet, containing two tryptophan (W) residues, spaced 20–22 amino acids apart (Sudol, 1996).
After three rounds of affinity selection, performed on a human brain cDNA library displayed by a T7 phage vector, we analyzed the resulting phage population by a plaque assay. The affinity-selected phage pool contained a high percentage (25%) of positive clones that bound the bait (Figure 1B). By comparing the frequency of positive plaques before and after selection, we estimated an enrichment of at least 60-fold (Figure 1B). Several clones contained overlapping protein fragments (all containing the WW domains) encoded by the Itch gene (Figure 1A).
The enrichment of clones displaying Itch WW domains was further confirmed by performing PCR reactions with specific oligonucleotides flanking the WW domains (from Pro 317 to Pro 520) (Figure 1B). In contrast, clones containing the β-actin gene were rapidly lost during the selection process (Figure 1B).
p73, but not p53, associates with Itch
In order to verify that Itch associates with p73, we performed an in vitro pull-down assay. Human embryonal kidney (Hek)293 cells were transfected with either an empty vector or with a vector encoding HA-tagged TAp73α. Cell lysates were mixed separately with a sepharose resin containing either GST or the WW region of Itch fused to GST (GST-WW). TAp73α was efficiently retained by GST-WW, while no significant binding to GST alone was detected (Figure 1C).
The interaction was also confirmed by co-immunoprecipitation (co-IP) of overexpressed TAp73α and Itch. As shown in Figure 1D, immunoprecipitation (IP) of Myc-tagged Itch resulted in co-IP of TAp73α. Addition of proteasome inhibitor MG132 resulted in stronger interaction. As expected, since p53 does not contain the PY motif (Figure 1A), it did not bind to Itch and could not be precipitated with Itch, regardless of the presence of the proteasome inhibitor (Figure 1D). Similarly, TAp73δ, that also lacks the PY motif (Figure 1A), could not be co-IP with Itch (data not shown). The N-terminally truncated form ΔNp73α also bound Itch (Figure 1E). To confirm that the interaction requires the PY motif of p73, we generated mutants of both the PY motifs found in p73 (Figure 1A). Figure 1F shows that mutants containing the Y487F substitution lost the ability to bind Itch, while the single mutant TAp73αY407F did not.
In order to confirm that this interaction also occurs in cells at physiological concentrations, we performed co-IP of endogenous proteins (Figure 1G). Again IP of endogenous p73 co-precipitated Itch, while IP of p53 did not.
These data clearly demonstrate that endogenous p73α isoforms can bind to the WW domains of Itch through their PY motif and that the interaction is selective for p73 and not shared by p53.
p73 is ubiquitinated by Itch
We next investigated whether p73 can serve as a substrate for the Ub–protein ligase activity of Itch. We used a recombinant Itch (GST-Itch) (Qiu et al, 2000), in a reconstituted in vitro ubiquitination system containing Ub, wheat E1, human E2 (UbcH7), ATP, and in vitro synthesized radiolabelled [35S]TAp73α protein as substrate. In the presence of purified GST-Itch, the TAp73α protein was ubiquitinated, as shown by the appearance of discrete higher molecular weight TAp73α species (Figure 2A, lane 1). To demonstrate that the appearance of ubiquitinated forms of TAp73α requires an intact Itch Hect domain, we used a previously described inactive mutant of Itch (GST-Itch MUT) (C830A) (Winberg et al, 2000). As shown in Figure 2A, this mutant, that retains the ability to bind TAp73α (Figure 1D), lost the ability to promote TAp73α ubiquitination. The inability of Itch to ubiquitinate p73δ and p53 suggests that the PY motifs are required, since these two proteins lack these motifs (Figures 1A and 2A, lanes 4 and 7). These in vitro data also show that no other factor was required for this reaction to occur.
Figure 2.
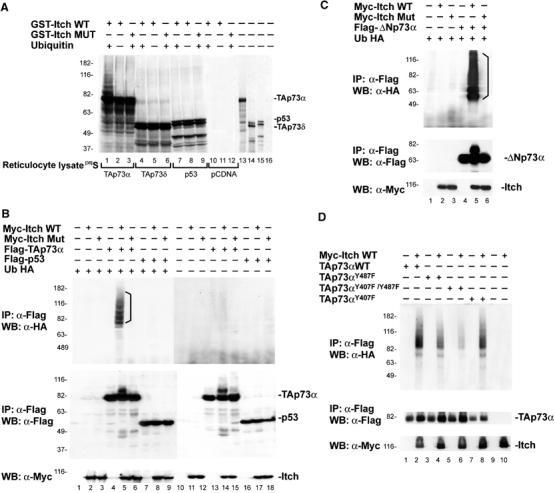
Itch ubiquitinates p73. (A) TAp73α and TAp73δ and p53 proteins were in vitro translated in the presence of [35S]Met and incubated with purified Itch (GST-Itch) or its catalytically inactive mutant (GST-Itch MUT) in the presence of ATP, Ub, and bacterially expressed E1 and E2 (UbcH7). Lanes 10–12 show in vitro translation with the empty vector (pCDNA). Lanes 13–16 show an aliquot (1/10) of the in vitro translated proteins used in the ubiquitination reaction. To demonstrate that p73 ubiquitination could also occurs in a more physiological system, Hek293 cells were transiently co-transfected with expression plasmids for Ub-HA, Flag-TAp73α or Flag-p53 (B), Flag-ΔNp73α (C), or Flag-TAp73αY487F, Flag-TAp73αY407F, and Flag-TAp73αY407F/Y487F (D) and Myc-Itch or Myc-Itch MUT. At 48 h after transfection, cells were treated with MG132 and then collected. Lysates were subjected to IP using an anti-Flag antibody. Immune complexes were revealed with anti-HA antibody (upper panels). No Ub-HA conjugates are present when Ub-HA is omitted from the reaction (B—lanes 10–18). The p73 and p53 protein expression levels are demonstrated by probing the same membranes with anti-Flag antibodies (middle panels), and those of Itch by probing the same membrane with anti-Myc antibodies (lower panels).
We next examined if Itch can catalyze p73 ubiquitination in cells. Extracts of Hek293 cells transfected with plasmids expressing, HA-tagged Ub (Ub-HA), Myc-tagged Itch (Myc-Itch), and Flag-tagged TAp73α or p53 (Flag-TAp73α and Flag-p53), were subjected to IP with anti-Flag antibodies and detected with anti-HA and anti-Flag western blots. As shown in Figure 2B (lane 5), Ub-HA TAp73α conjugates were detected upon co-transfection only with wild-type (WT) Itch (Myc-Itch WT), and not with the catalytically inactive mutant of Itch (Myc-Itch MUT) (Figure 2B, lane 6). Similarly, ΔNp73α was ubiquitinated by Itch (Figure 2C), showing that the N-terminal part of the protein is not required for the ubiquitination. In contrast, p53 was not ubiquitinated by Itch (Figure 2B, lanes 7–9). Although ubiquitination of TAp73αY487F mutant was reduced, a greater reduction was seen with the double mutant TAp73αY407F/Y487F (Figure 2D). Ubiquitination of the TAp73αY407F mutant was similar to that of WT p73. The right panel (lanes 10–18) in Figure 2B demonstrates the specificity of the reaction since no higher molecular weight bands were observed in the absence of Ub-HA. These data clearly show that p73 but not p53 is ubiquitinated by Itch, suggesting that this protein plays an important role in the regulation of p73 steady-state protein levels.
Itch regulates the stability of p73 in cells
Since ubiquitination of proteins is usually associated with their turnover (Weissman, 2001), we investigated if Itch can regulate p73 protein abundance. We measured TAp73α levels in whole-cell extracts in the presence or absence of Itch. Representative data from several independent experiments demonstrate that co-expression of Myc-Itch and TAp73α in cells results in a striking decrease of TAp73α levels (Figure 3A), indicating that the Itch-dependent ubiquitination targets p73 for degradation. Consistently, the catalytic mutant of Itch was not able to alter the concentration of p73 (Figure 3A, lane 3) and the TAp73αY487F (Figure 3B) mutant levels were not affected by Itch overexpression.
Figure 3.
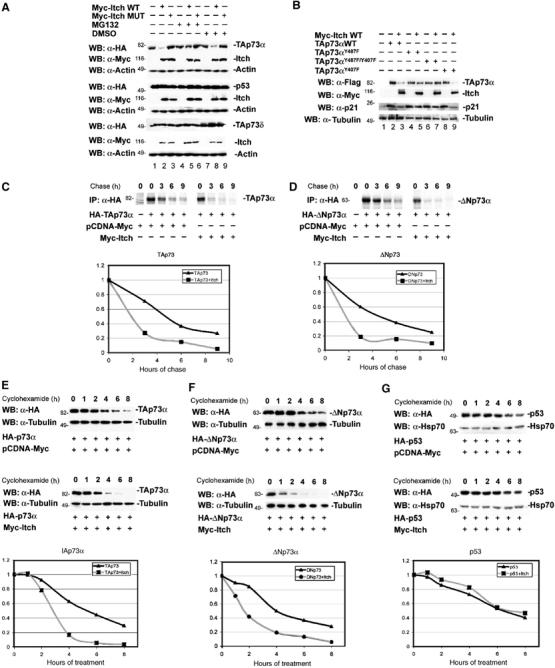
Effect of Itch expression on the steady-state levels of p73. (A) Hek293 cells were transfected with either HA-TAp73α and p53 or HA-TAp73δ (A) or Flag-TAp73αY487F, Flag-TAp73αY407F, and Flag-TAp73αY407F/Y487F (B), together with either Myc-Itch or Myc-Itch MUT. At 48 h after transfection, cells were treated or not with MG132. Equal amounts of total protein cell lysates were subjected to western blotting analysis using anti-HA antibody or anti-Flag antibody (upper rows) to detect the steady-state levels of p73s and p53 proteins. The same blots were re-probed with anti-Myc antibody in order to detect the expression levels of Itch (middle rows), and with anti-Actin antibody to show equal loading (lower rows). 35S pulse chase: H1299 cells were transfected with HA-TAp73α (C) or HA-ΔNp73α (D), together with Myc-Itch or pCDNA-Myc. At 48 h post-transfection, cells were labelled with 250 μCi/ml of Redivue PRO-MIX (L-[35S] in vitro cell-labelling mix). Unlabelled Met and Cys (1 mg/ml) were added and cells were collected at the indicated time points. IP was performed with anti-HA (Y-11) polyclonal antibody. Immunoprecipitates were washed and run on SDS–PAGE and detected by autoradiography. For cycloheximide-blocking experiments, Hek293 cells were transfected with either HA-TAp73α (E), HA-ΔNp73α (F), or HA-p53 (G), together with either Myc-Itch or pCDNA-Myc. At 24 h after transfection, cells were treated with cycloheximide and collected at different time points. Equal amounts of total protein cell lysates were subjected to western blotting analysis using anti-HA antibody. To demonstrate equal loading, the same blots were stripped and re-probed with anti-β-Tubulin or anti-Hsp-70 antibodies for p73s and p53 blots, respectively. (C) Western blots were subjected to densitometric analysis and results were normalized based on β-Tubulin or Hsp-70 expression levels, respectively, and reported in graphical form (lower panels).
As polyubiquitination generally targets proteins for proteasomal degradation (Weissman, 2001), we determined the effect of MG132 on the steady-state levels of TAp73α. As shown in Figure 3A (lanes 4–6), addition of MG132 to cells blocked the Itch-mediated TAp73α degradation and resulted in the accumulation of the ubiquitinated forms of TAp73α (data not shown). This is consistent with previous reports showing that proteasome inhibition leads to the stabilization of endogenous p73 protein (Balint et al, 1999). Again, TAp73δ and p53 levels were not affected by Itch (Figure 3A). We further confirmed these results by measuring p73 half-life in the presence or absence of Itch using two different methods. Both pulse chase using [35S]Met and Cys (Figure 3C and D), and cycloheximide blockade (Figure 3E and F), showed a marked decrease of TAp73 and ΔNp73 half-lives in the presence of Itch. Under similar experimental conditions, no change in p53 half-life was observed (Figure 3G).
To further confirm the importance of Itch in controlling p73 steady-state levels, we reduced endogenous Itch levels by siRNA. To this end, we used Tet-On-inducible Saos-2 cells expressing p73 (Melino et al, 2004). Figure 4A shows that, in this inducible cell line, reduction in Itch expression by siRNA resulted in more rapid induction and higher levels of p73 protein. Moreover, after withdrawal of induction, p73 levels decline more slowly when Itch expression is reduced (Figure 4B). Similarly, the steady-state levels of endogenous TAp73 and ΔNp73 isoforms increased in Saos-2 cells when Itch was downregulated (Figure 4C). This confirms that basal Itch levels are important in controlling basal p73 levels. In agreement, endogenous levels of ΔNp73 (which is the only isoform detectable in these cells) were increased in mouse embryo fibroblasts (MEFs) derived from non-agouti-lethal 18H Itch-deficient mice (MEF Itch−/−) (Figure 4D). Re-introduction of WT Itch into MEFs Itch−/− resulted in reduction of endogenous ΔNp73 levels in these cells (Figure 4E).
Figure 4.
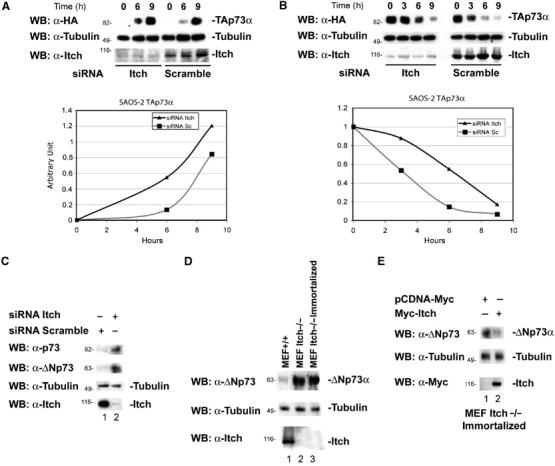
Effects of Itch downregulation on p73 protein levels. (A) Saos-2-TAp73α inducible cells were transfected with siRNA oligonucleotides targetting the Itch sequence or with a scrambled oligonucleotide. After 48 h, cells were induced to express TAp73α for the indicated time points with doxycycline (inducer). p73 levels increase more rapidly and reach higher levels when Itch is downregulated. The lower panel shows endogenous Itch levels. Graphs show densitometric analysis of the p73 western blots normalized for β-Tubulin. (B) Saos-2-TAp73α-inducible cells were transfected with oligos targetting the Itch sequence, or with a scrambled oligo. Cells were induced to express TAp73α for 14 h with doxycycline, the inducer was removed and cells collected at the indicated time points. p73 levels decay more rapidly in cells transfected with the scrambled oligo compared to those in which Itch is downregulated. The lower panel shows endogenous Itch levels. Graphs show densitometric analysis of the p73 western blots normalized for β-Tubulin. (C) Saos-2 cells were transfected with oligos targeting the Itch sequence or with a scrambled oligo and collected 48 h later. Itch downregulation (lower panel) results in an increase of TA and ΔN p73a levels (upper panels). (D) Western blot for endogenous p73 of WT MEFs (MEF+/+), non-agouti-lethal 18H Itch-deficient MEFs (MEF Itch−/−) and a spontaneously immortalized clone of these MEFs (MEF Itch−/− Immortalized). ΔNp73 levels (the only form detectable in these cells) are higher in MEFs Itch−/−. (E) Immortalized MEFs Itch−/− were transfected with Myc-Itch WT and collected 48 h later. Re-introduction of Itch results in ΔNp73 downregulation.
Not all Itch family members had the same effect on p73. Nedd4 bound (Figure 5A) both TAp73 and ΔNp73, but was not capable of catalyzing the ubiquitination of these proteins (Figure 5C), and therefore did not affect their degradation (Figure 5B). Miyazaki et al (2003) demonstrated that NEDL2, another Nedd4 family member, binds, ubiquitinates and stabilizes p73. Thus, different Nedd4-like E3 ligases, although all capable of binding the p73 PY motifs, exert different effects on these proteins.
Figure 5.
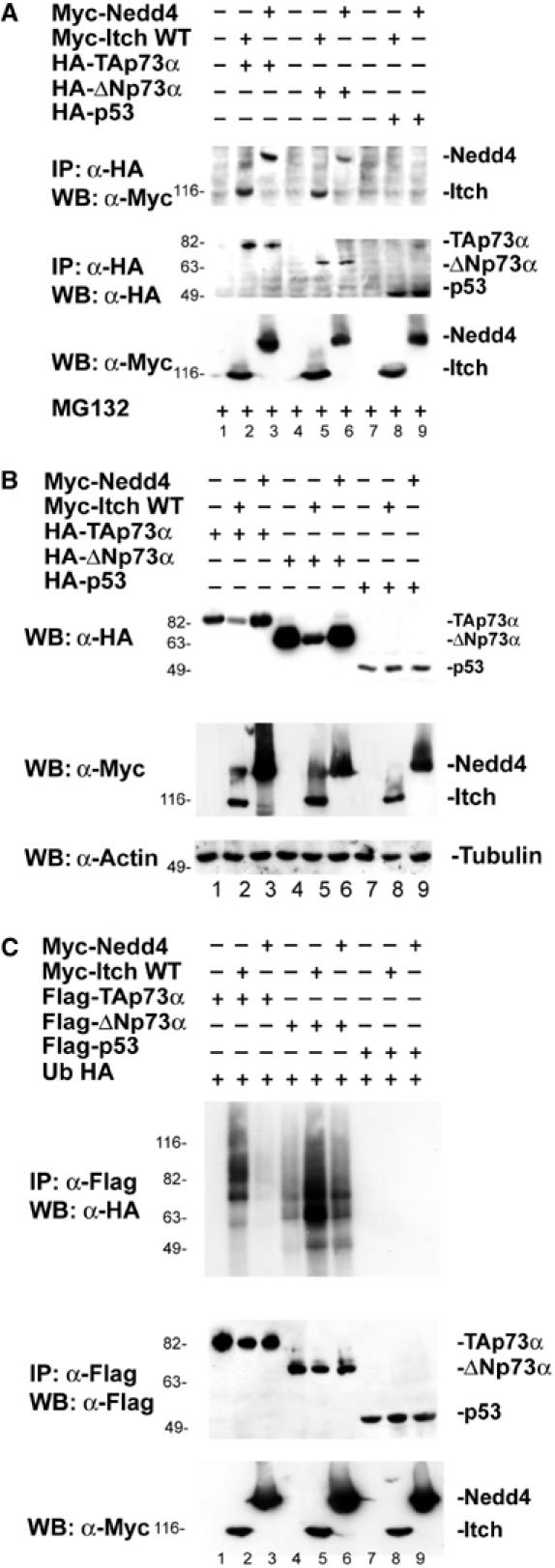
Nedd4 binds p73 but fails to ubiquitinate it. (A) Hek293 cells were transfected with HA-TAp73α, HA-ΔNp73α, HA-p53 or an empty vector, together with Myc-Itch or Myc-Nedd4 in the presence of MG132. Cells were subjected to IP using anti-HA antibody and analyzed by western blot, using an antibody against Myc. Itch and Nedd4 co-IP with p73. (B) Hek293 cells were transfected with HA-TAp73α, HA-ΔNp73α or HA-p53, together with Myc-Itch or Myc-Nedd4. Cell extracts were analyzed by western blot, using an antibody against HA. Expression of Itch but not of Nedd4 results in TA and ΔNp73 downregulation. (C) Cells transfected with the indicated plasmids together with a plasmid expressing Ub-HA were analyzed by western blot using an antibody against HA. Higher molecular bands characteristic of ubiquitination appear when Itch but not Nedd4 is overexpressed.
Itch decreases p73-dependent transcriptional activity
To evaluate if the interaction between p73 and Itch influences its transcriptional activity, we co-transfected H1299 cells with TAp73α and Myc-Itch or Myc-Itch MUT and assessed TAp73α transcription activity by luciferase reporter assay using different p53/p73-responsive promoters (Bax, p21, and MDM2). As shown in Figure 6A, C and E, consistent with a reduction in TAp73 protein levels, co-transfection of Myc-Itch reduced the transcriptional activity of TAp73α on all the promoters tested in a dose-dependent manner. As expected, the mutated form of Itch had no effect on the transcriptional activity of p73 (Figure 6B, D and F). The reduction of the promoter activity was paralleled by a reduction in endogenous levels of p73 target proteins such as p21 (Figure 6G).
Figure 6.
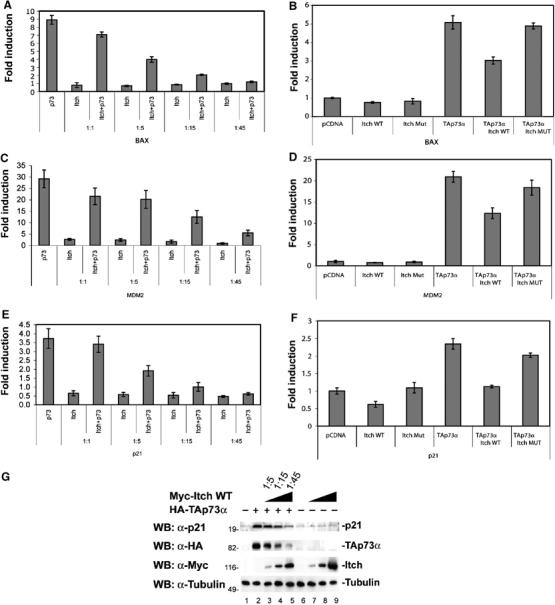
Itch expression reduces the transcriptional activity of p73. H1299 cells were transfected with the indicated combinations of plasmids (at the different indicated ratios) encoding TAp73α, together with Myc-Itch WT (Itch wt) or Myc-Itch MUT (Itch Mut), or empty control vector (pCDNA), together with a Bax- (A, B) or MDM2- (C, D) or p21- (E, F) luciferase reporter plasmid and Renilla luciferase reporter plasmid. Cell extracts were prepared 36 h later and luciferase activity was determined. Results are represented as fold induction of luciferase activity as compared with the control cells. Histograms show the mean of three independent experiments; bars indicate standard deviation. (G) H1299 cells were transfected with HA-TAp73α, together with Myc-Itch at the indicated ratios. Equal amounts of cell extracts were subjected to western blot with anti p21 antibody (upper panel), anti-HA (middle panel), and anti-Myc (lower panel).
Itch is downregulated in response to DNA damage
Since TAp73 protein levels increase in response to DNA damage (Agami et al, 1999; Gong et al, 1999; Yuan et al, 1999), we investigated whether Itch expression is also modulated after DNA damage. In Saos-2 cells, following treatment with doxorubicin, to induce DNA damage, endogenous Itch protein levels were downregulated in a time- (Figure 7A) and dose- (not shown) dependent manner. The reduction of Itch levels was paralleled by an increase in endogenous TAp73 levels (Figure 7A). Reduction of Itch also paralleled an increase in apoptosis (Figure 7B). This response of Itch to doxorubicin treatment is not cell type specific: Figure 7C shows a similar experiment using HeLa, H1299, and Hek293 cell lines treated with doxorubicin. These results suggest that Itch could be an important regulator of TAp73 levels and that upon DNA damage Itch is downregulated, allowing TAp73 levels to rise. The relative contribution of this mechanism in determining apoptosis of damaged cells remains to be quantified. This pathway is p53 independent since the effect on cell cycle and apoptosis was also observed in cell lines lacking p53, such as Saos-2.
Figure 7.
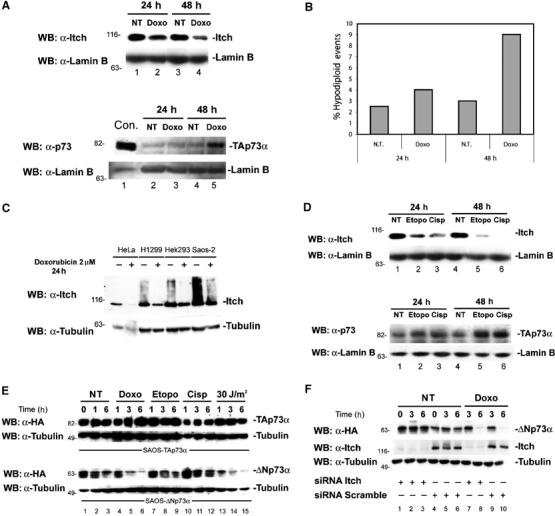
Itch is downregulated by DNA-damaging agents. Saos-2 cells were treated with 2 μM doxorubicin (Doxo) for 24 or 48 h, then collected by trypsinization. Cells were lysed for western blotting (A) and half of the cells were ethanol fixed for apoptosis analysis (B). Western blots were performed with either anti-Itch (upper panel) or anti-p73 (lower panel) antibodies. Blots were stripped and re-probed with anti-Lamin B antibody to show equal loading. Apoptosis was evaluated by flow-cytometric analysis of PI-stained cells. Nontreated controls (NT) are shown. H1299 cells transfected with HA-TAp73α (Con) are loaded as a control for p73 western blot. (C) Western blot using anti-Itch antibody on Hela, H1299, Hek293, and Saos-2 cells treated for 24 h with 2 μM doxorubicin. Blots were stripped and re-probed with anti-β-Tubulin antibody to show equal loading. (D) Western blot using anti-Itch antibody on Saos-2 cells treated for 24 and 48 h with either 15 μM etoposide (Etopo) or 5 μM cisplatin (Cisp). NT cells are also shown. Blots were stripped and re-probed with anti p73 antibody and Lamin B antibody to show equal loading. (E) Saos-2-TAp73α (upper panel) or Saos-2-ΔNp73α (lower panel) were induced with doxycycline for 14 h. Doxycycline was then removed and cells were treated with the indicated drugs and analyzed at the indicated time points by western blot using anti-HA antibody. Blots were re-probed with anti-β-Tubulin to show equal loading. (F) Saos-2-ΔNp73α were transfected with oligos for siRNA against the Itch sequence or with a scrambled sequence, and then induced with doxycycline for 14 h. Doxycycline was then removed and cells were treated with doxorubicin and analyzed at the indicated time points by western blot using anti-HA antibody. Blots were re-probed with anti-β-Tubulin to show equal loading.
The DNA damage-dependent decrease in Itch levels is not specific for doxorubicin treatment, but also occurs when cells are treated with other DNA-damaging agents. As shown in Figure 7D, treatment with cisplatin and etoposide also resulted in a dramatic reduction in Itch protein levels.
Since Itch also affects ΔNp73 levels, an increase in this isoform would also be expected in response to DNA damage. Figure 7E, however, shows that, as we have previously published (Maisse et al, 2004), ΔNp73α unlike TA is rapidly degraded upon DNA damage, regardless of the downregulation of Itch. In order to confirm that ΔNp73 degradation is independent of Itch, we reduced Itch levels in the Saos-2-ΔNp73α-inducible cell line, and then induced ΔNp73 expression by the addition of doxycycline. The inducer was removed and DNA damage was then induced with doxorubicin. As shown in Figure 7F, reduction of Itch levels results in increased stabilization of ΔNp73α, but does not prevent its degradation in response to doxorubicin treatment.
Discussion
p73 activity depends on its steady-state protein levels, and a variety of evidence suggests that post-transcriptional regulation rather than transcriptional control plays a major role in p73 response to DNA damage. Upon DNA damage, p73 becomes activated and stabilized (Catani et al, 2002), and this results in cell cycle arrest and apoptosis (Agami et al, 1999; Gong et al, 1999; Yuan et al, 1999). p73 is rarely mutated in cancers, although altered levels of one or more isoforms of p73 have been found in tumors (Ikawa et al, 1999; Tschan et al, 2000; Casciano et al, 2002; Melino et al, 2002; Putzer et al, 2003; Romani et al, 2003). Little is known about the pathways leading to p73 degradation, despite the likely importance of its consequences in cancer development and therapy. Endogenous p73 steady-state levels increase in the presence of proteasome inhibitors, suggesting a role for this pathway in p73 degradation (Balint et al, 1999; Ongkeko et al, 1999; Zeng et al, 1999). While the homology with p53 suggests that Ub–protein ligase MDM2 could be the required E3 ligase, MDM2 binding to p73 does not lead to p73 ubiquitination and degradation, but instead leads to p73 stabilization (Balint et al, 1999; Dobbelstein et al, 1999; Lohrum and Vousden, 1999; Ongkeko et al, 1999; Zeng et al, 1999).
We have recently reported that, upon activation of p38 MAP kinase, p73 is recruited into the PML-NB and acetylated by p300. Acetylation protects TAp73 from ubiquitination, resulting in its stabilization (Bernassola et al, 2004). NEDL2, a HECT-type E3 ligase, has also been reported to bind and ubiquitinate p73, resulting in its stabilization rather than degradation (Miyazaki et al, 2003). Regulation of p73 stability, and particularly that of its different isoforms, is therefore complex and far from fully understood.
Here, we demonstrate that Itch selectively binds and ubiquitinates p73α. In contrast, it has no such effect on p53. Our results also show a different regulation among the various C-terminal isoforms of p73, since only the α and β isoforms that contain the PY motif are regulated by Itch, while the δ and γ isoforms that lack this domain escape this regulation.
We found that Itch is rapidly downregulated in response to DNA damage, allowing p73 levels to increase. Therefore, this response to DNA damage, selective for p73 and independent of p53, may be important in p53-negative tumors. We have recently shown that, unlike TAp73, the ΔNp73 isoforms are rapidly degraded in response to DNA damage (Maisse et al, 2004). Our results show that ΔNp73 isoforms are also Itch substrates and Itch downregulation results in increased ΔNp73 levels. The downregulation of Itch in response to DNA damage should therefore result in a parallel increase of TAp73 and ΔNp73 forms. This, however, is not the case, and while TAp73 levels actually rise, ΔNp73 is rapidly degraded (Maisse et al, 2004). In our model, Itch seems to be responsible for keeping both TAp73 and ΔNp73 levels low under normal condition (Figure 8). Upon DNA damage, Itch reduction allows stabilization of both TAp73 and ΔNp73. Therefore, a second pathway should specifically target ΔNp73 for degradation. Recently, Toh et al (2004) describe the ability of c-Jun to regulate the stability of p73. Even though the underlying molecular mechanism was not elucidated, here c-Jun affects stability of TAp73 and not ΔNp73, indicating a differential effect on the two major p73 isoforms.
Figure 8.
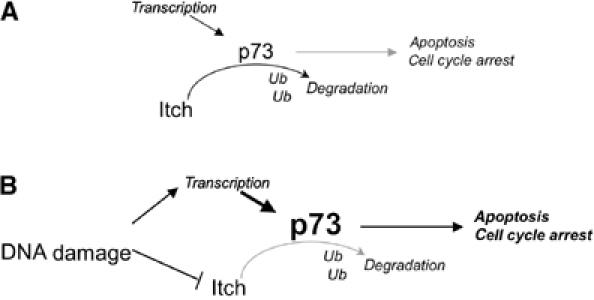
Itch controls p73 levels in resting conditions and in response to DNA damage. (A) Schematic representation of the functional interaction between p73 and Itch. Under nonstressed conditions, basal levels of both TAp73 and ΔNp73 are kept low; in this situation, Itch binds to p73 and promotes its ubiquitination and proteasome-dependent degradation. (B) In response to DNA damage, Itch is rapidly degraded, reducing p73 turn over. TAp73 levels increase while ΔNp73 remain low due to the activation of a DNA damage-dependent ΔN-specific degradation pathway. The final outcome is the induction of cell cycle arrest and apoptosis by p73.
In conclusion, we provide a mechanism by which p73 levels are normally kept low in cells, in accordance with the low basal levels found in many tissues and cell lines, and are increased in response to DNA damage (Figure 8).
Materials and methods
Plasmids
Myc-Itch and Myc-Itch MUT plasmids (C830A) were provided by Dr T Pawson (Winberg et al, 2000). Itch GST fusion proteins were generated by subcloning PCR fragments into the BamHI and SalI sites of pGEX-6P1 (Amersham Pharmacia Biotech). The following fragments were cloned: GST-Itch, from Thr 277 to Glu 903 (lacking the N-terminal C2 domain), and GST-WW, from Pro 317 to Pro 520 (spanning only the four WW domains). HA-p73α and HA-p73δ were described previously (De Laurenzi et al, 1998). GST–p73 fusion protein encompassing only the PY motif (GST-PY, from Met 452 to Ala 489) was obtained by subcloning into BamHI and NotI sites of the pGEX-6P1. The poly-HA-Ub construct (HA-Ub) was kindly provided by Dr D Bohmann (Treier et al, 1994). pET23a-Ubch7 (E2) bacterial expression vectors directing the synthesis of the E2 enzyme (Ubch7) and the E1 Ub-activating enzyme Uba1 were a gift of Dr PM Howley (Kumar et al, 1997). The reporter plasmid containing the luciferase cDNA under control of the Bax and p21 promoter were provided by Dr Levrero (Vossio et al, 2002); that under the control of the MDM2 promoter was provided by Dr Blandino (Strano et al, 2001). Nedd4-myc construct was a gift from Dr D Rotin. All p73 mutants were generated by site-directed mutagenesis using the Quickchange kit (Stratagene), according to the manufacturer's instructions.
Cell culture and transfection
Hek cells (Hek293), HeLa cells, and MEFs were grown in Dulbecco's modified Eagle medium (DMEM) (GibcoBRL); the human lung carcinoma cells H1299 and the human osteosarcoma cells Saos-2 were cultured in RPMI (GibcoBRL). MEF Itch−/− cells were prepared from Itch−/− mice (Y-C Liu) or they were generous gifts from Neil Copeland, Lynda Matesic, Nancy Jenkins. All media were supplemented with 10% (v/v) fetal bovine serum (FBS) (GibcoBRL). Saos-2-TAp73α were a kind gift of K Vousden (Nakano et al, 2000). Saos-2-ΔNp73α were generated as described before (Maisse et al, 2004). For the TET-On cell lines, media was supplemented with 10% (v/v) tetracycline-free FBS (Clonetech). Genes were induced by adding 0.5 μg/ml doxycycline. All cell lines were grown at 37°C in a humidified atmosphere of 5% (v/v) CO2 in air. Transient transfection was performed with lipofectamine 2000 reagent according to the manufacturer's instructions.
Apoptosis was analyzed by flow cytometric evaluation of DNA fragmentation.
Western blot and antibodies
Proteins were separated on SDS–PAGE and blotted onto nitrocellulose membranes. Filters were blocked with TBST 5% nonfat dry milk and incubated with primary antibodies for 2 h at room temperature (RT). Filters were incubated for 1 h at RT using the appropriate horseradish peroxidase-conjugated secondary antibody (rabbit and mouse BioRad; goat, Santa Cruz). Detection was performed with the enhanced chemiluminescence Supersignal West Pico (Pierce).
Anti-p73 is a monoclonal antibody (ER15) (Neomarkers). Endogenous Itch was detected with a mouse monoclonal antibody (BD Bioscience), Actin (sc-1615) polyclonal goat antibody (Santa Cruz), Lamin B (sc-6217) goat polyclonal antibody (Santa Cruz), Hsp70 (sc-1060) polyclonal goat antibody (Santa Cruz), β-Tubulin (sc-9104) polyclonal rabbit antibody (Santa Cruz), p21 (sc-756) polyclonal rabbit antibody (Santa Cruz), p53 DO-1, and Pab1801 monoclonal mouse antibodies (Santa Cruz). c-Myc-tagged constructs were detected or immunoprecipitated with the sc-40 monoclonal mouse antibody (Santa Cruz), HA-tagged with the sc-805 polyclonal rabbit antibody (Santa Cruz), and the Flag-tagged constructs with the M2 monoclonal mouse antibody (Sigma).
Affinity selection
An aliquot of a T7 phage display library of human brain cDNA (Novagen) was incubated overnight at 4°C, with 50 μg of GST-PY p73α fusion protein immobilized on glutathione–sepharose beads (Amersham Biosciences), in the presence of 1% bovine serum albumin. Unbound phages were removed by washing five times with 1 ml of 0.1% Tween 20 in PBS, and then once in 1 ml PBS, and resuspended in 50 μl of PBS. The bound phages were recovered by incubating at 37°C with 2 ml of Escherichia coli BLT5615 (Novagen) induced with 1 mM IPTG for 30 min before phage addition. After cell lysis, the phage lysate was clarified by centrifugation, and used in a new selection round. After three selection cycles, the resulting phages were analyzed by a plaque assay (Zucconi et al, 2001), using soluble purified GST-PY at a concentration of 5 μg/ml. Positive plaques were isolated from plates. In all, 1 μl of the suspension was amplified by PCR using primers to the regions flanking the insert, and fragments were sequenced. To estimate the enrichment of Itch, 1 μl of clarified phage lysate corresponding to each of the different selection rounds was amplified by PCR (20 amplification cycles).
GST fusion proteins and pull-down assays
GST fusion proteins were expressed in E. coli BL21 (DE3) and purified on glutathione–sepharose beads (Amersham Biosciences) following standard procedures. For the pull-down assay, Hek293 cells transfected with HA-TAp73α or empty vector were lysed in 50 mM Tris–HCl, pH 8, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 10% glycerol, 100 mM NaF, and 1% Triton X-100 containing protease cocktails (Sigma), and centrifuged to precipitate cellular debris. In all, 1.5 mg of total cellular proteins were first precleared with glutathione–sepharose beads and then incubated for 2 h at 4°C with immobilized GST fusion proteins (25 μg). Unbound proteins were removed by washing four times with 0.1% Tween 20 in PBS, and the precipitates were resolved by SDS–PAGE. The immunoblots were probed with the indicated antibodies.
Immunoprecipitation
Following a previously published procedure (Gottifredi et al, 1999), Hek293 cells were transiently transfected with 8 μg of total DNA of the indicated mammalian expression plasmids and harvested 48 h after transfection. Cells were then lysed as described above. Following preclearing for 1 h at 4°C, we performed IP by incubating 1.5 mg of whole-cell extracts with the indicated antibodies, rocking at 4°C for 1 h. The immunocomplexes were collected by incubating with protein G agarose (KPL), washed with Net-gel buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.25% gelatin, and 0.1% NP40). The beads were then resuspended in 5 × Laemmli buffer, and subjected to western blot with the indicated primary antibodies.
Ubiquitination assays
In vitro assays were performed as described previously (Hamilton et al, 2001), using in vitro translated radiolabelled p73 and p53 proteins. In vitro transcription/translation of proteins was performed using the rabbit reticulocyte lysate system TNT kit (Promega), in the presence of [35S]Met (Amersham Biosciences), according to the manufacturer's protocol. The ubiquitination reaction mixture contained: 2 μl of E. coli BL21 bacterial extracts overexpressing wheat E1 and 2 μl of a human E2 (UbcH7), 5 μg of purified E3 enzyme (either GST-Itch or GST-Itch MUT), 25 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM dithiothreitol, 2.5 mM ATP, 4 mM MgCl2, and 10 μg of bovine Ub. After incubation for 90 min at 37°C, the reactions were terminated by adding 5 × Laemmli buffer, resolved by SDS–PAGE, followed by autoradiography.
For in vivo experiments, Hek293 cells were transiently transfected with mammalian expression plasmids for HA-Ub, with the indicated combination of plasmids. At 48 h after transfection, cells were harvested, the insoluble fraction was removed by a high-speed spin, and 1 mg of total cellular proteins of the clarified supernatant was subjected to IP using anti-Flag antibodies (Sigma). Ub conjugates were detected by western immunoblot analysis using an anti-HA antibodies (Santa Cruz).
Measurement of p73 half-life
Decay of p73 protein levels in the presence of cycloheximide. Cycloheximide (20 μg/ml) was added to Hek293 cells 24 h after transfection, with a total of 3 μg of the indicated plasmids in a 1:5 ratio of p73/Itch and p53/Itch. Protein levels were determined by collecting cells at the indicated time points and performing immunoblotting as described above. The relative amount of p73 protein was evaluated by densitometry and normalized on β-Tubulin.
35S pulse chase. H1299 cells were transfected with a total of 3 μg of the indicated plasmids in a 1:2 ratio of p73/Itch. At 48 h post-transfection, cells were starved for 30 min in DMEM with dialyzed serum and then labelled with 250 μCi/ml of Redivue PRO-MIX (L-[35S] in vitro cell labelling mix) for 60 min. Unlabelled Met and Cys were added and cells were collected in RIPA buffer (200 mM Tris, pH 8, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP40 and 0.2 mM EDTA) at the indicated times. IPs were performed with 150 μg of total protein lysate and 4 μl of anti-HA (Y-11) polyclonal antibody (Santa Cruz). Immunoprecipitates were washed six times in RIPA buffer and six times in NET gel, and run on SDS–PAGE and detected by autoradiography.
Steady-state protein level analysis
The concentrations of TAp73α, TAp73δ, and p53 were monitored by western blotting. At 48 h after co-transfection with HA-TAp73, HA-TAp73δ, and HA-p53 and either Myc-Itch or Myc-Itch MUT, Hek293 cells were treated for 40 min with or without proteasome inhibitor MG132, at a final concentration of 50 μM in DMSO. In all, 25 μg of cell lysates were subjected to western blotting. p73 proteins were detected by an anti-HA antibody. The same blots were re-probed with anti-Myc antibody to detect Itch and with anti-Actin antibody to show equal loading.
Promoter reporter
H1299 cells were transfected with the indicated combinations of plasmids encoding p73α (10 ng/well of 96-well plate), Myc-Itch or Myc-Itch MUT, or empty control vectors, together with the indicated luciferase reporter plasmid (60 ng/well) and Renilla luciferase reporter (1.2 ng/well). The total amount of transfected DNA in each well was kept constant by the addition of different amounts of empty vector. The luciferase activity was quantified using a commercially available kit (Dual-Glo, Promega) according to the manufacturer's instructions.
Itch siRNA
Saos-2 cells were electroporated with 20 μl of a 20 μM solution of 21-nucleotide RNA (Qiagen) using a BIO-RAD electroporation apparatus. A mix of two different oligos was used to downregulate Itch. The Itch target sequences were AAGTGCTTCTCAGAATGATGA and AACCACAACACACGAATTACA, and the scrambled sequence was AATTCTCCGAACGTGTCACGT. Cells were collected after 48 h and cell lysates were subjected to western blot for Itch detection, as described above.
Acknowledgments
This work was supported by the following grants: AIRC, MIUR (Progetto genomica Funzionale), and an FIRB grant (RBNE01KXC9) to GC; EU grants (QLK-CT-2002-01956), progetto Genomica Funzionale COMETA, COMETA CNR/MIUR Legge 449/97-DM 20/10/2000, FIRB 2001, MIUR 2002, Istituto Naz Tumori 8114, AIRC, and Ministero Sanita', Telethon to GM, Telethon (E1224), MIUR, and Ministero Sanita' to VDL, and NIH grant AI52735 to DRG. We thank T Pawson, D Bohmann, PM Howley, and Dr D Rotin for strains and reagents. We also thank Dr P Salomoni for his help with siRNA experiments. We are particularly grateful to RA Knight for helpful suggestions. We thank Neil Copeland for the Itch−/− cells.
References
- Agami R, Blandino G, Oren M, Shaul Y (1999) Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature 399: 809–813 [DOI] [PubMed] [Google Scholar]
- Balint E, Bates S, Vousden KH (1999) Mdm2 binds p73 alpha without targeting degradation. Oncogene 18: 3923–3929 [DOI] [PubMed] [Google Scholar]
- Bernassola F, Salomoni P, Oberst A, Di Como CJ, Pagano M, Melino G, Pandolfi PP (2004) Ubiquitin-dependent degradation of p73 is inhibited by PML. J Exp Med 199: 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casciano I, Mazzocco K, Boni L, Pagnan G, Banelli B, Allemanni G, Ponzoni M, Tonini GP, Romani M (2002) Expression of DeltaNp73 is a molecular marker for adverse outcome in neuroblastoma patients. Cell Death Differ 9: 246–251 [DOI] [PubMed] [Google Scholar]
- Castagnoli L, Zucconi A, Quondam M, Rossi M, Vaccaro P, Panni S, Paoluzi S, Santonico E, Dente L, Cesareni G (2001) Alternative bacteriophage display systems. Comb Chem High Throughput Screen 4: 121–133 [DOI] [PubMed] [Google Scholar]
- Catani MV, Costanzo A, Savini I, Levrero M, de Laurenzi V, Wang JY, Melino G, Avigliano L (2002) Ascorbate up-regulates MLH1 (Mut L homologue-1) and p73: implications for the cellular response to DNA damage. Biochem J 364: 441–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesareni G, Castagnoli L, Cestra G (1999) Phage displayed peptide libraries. Comb Chem High Throughput Screen 2: 1–17 [PubMed] [Google Scholar]
- De Laurenzi V, Catani MV, Terrinoni A, Corazzari M, Melino G, Costanzo A, Levrero M, Knight RA (1999) Additional complexity in p73: induction by mitogens in lymphoid cells and identification of two new splicing variants epsilon and zeta. Cell Death Differ 6: 389–390 [DOI] [PubMed] [Google Scholar]
- De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G (1998) Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med 188: 1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurenzi V, Raschella G, Barcaroli D, Annicchiarico-Petruzzelli M, Ranalli M, Catani MV, Tanno B, Costanzo A, Levrero M, Melino G (2000) Induction of neuronal differentiation by p73, in a neuroblastoma cell line. Journal Biological Chemistry 275: 15226–15231 [DOI] [PubMed] [Google Scholar]
- Dobbelstein M, Wienzek S, Konig C, Roth J (1999) Inactivation of the p53-homologue p73 by the mdm2-oncoprotein. Oncogene 18: 2101–2106 [DOI] [PubMed] [Google Scholar]
- Fang D, Elly C, Gao B, Fang N, Altman Y, Joazeiro C, Hunter T, Copeland N, Jenkins N, Liu YC (2002) Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat Immunol 3: 281–287 [DOI] [PubMed] [Google Scholar]
- Gong JC, Costanzo A, Yang HQ, Melino G, Kaelin WG Jr, Levrero M, Wang JY (1999) The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399: 806–809 [DOI] [PubMed] [Google Scholar]
- Gottifredi V, Pelicci G, Munarriz E, Maione R, Pelicci PG, Amati P (1999) Polyomavirus large T antigen induces alterations in cytoplasmic signalling pathways involving Shc activation. J Virol 73: 1427–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob TJ, Novak U, Maisse C, Barcaroli D, Luthi AU, Pirnia F, Hugli B, Graber HU, De Laurenzi V, Fey MF, Melino G, Tobler A (2001) Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ 8: 1213–1223 [DOI] [PubMed] [Google Scholar]
- Hamilton MH, Tcherepanova I, Huibregtse JM, McDonnell DP (2001) Nuclear import/export of hRPF1/Nedd4 regulates the ubiquitin-dependent degradation of its nuclear substrates. J Biol Chem 276: 26324–26331 [DOI] [PubMed] [Google Scholar]
- Harvey KF, Kumar S (1999) Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol 9: 166–169 [DOI] [PubMed] [Google Scholar]
- Hicke L (2001) Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol 2: 195–201 [DOI] [PubMed] [Google Scholar]
- Ikawa S, Nakagawara A, Ikawa Y (1999) p53 family genes: structural comparison, expression and mutation. Cell Death Differ 6: 1154–1161 [DOI] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D (1997) Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90: 809–819 [DOI] [PubMed] [Google Scholar]
- Kloetzel PM (2001) Antigen processing by the proteasome. Nat Rev Mol Cell Biol 2: 179–187 [DOI] [PubMed] [Google Scholar]
- Kumar S, Kao WH, Howley PM (1997) Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. J Biol Chem 272: 13548–13554 [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Vousden KH (1999) Regulation and activation of p53 and its family members. Cell Death Differ 6: 1162–1168 [DOI] [PubMed] [Google Scholar]
- Maisse C, Munarriz E, Barcaroli D, Melino G, De Laurenzi V (2004) DNA damage induces the rapid and selective degradation of the DeltaNp73 isoform, allowing apoptosis to occur. Cell Death Differ 11: 685–687 [DOI] [PubMed] [Google Scholar]
- Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M, Knight RA, Green DR, Thompson C, Vousden KH (2004) p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem 279: 8076–8083 [DOI] [PubMed] [Google Scholar]
- Melino G, De Laurenzi V, Vousden KH (2002) p73: Friend or foe in tumorigenesis. Nat Rev Cancer 2: 605–615 [DOI] [PubMed] [Google Scholar]
- Melino G, Lu X, Gasco M, Crook T, Knight RA (2003) Functional regulation of p73 and p63: development and cancer. Trends Biochem Sci 28: 663–670 [DOI] [PubMed] [Google Scholar]
- Miyazaki K, Ozaki T, Kato C, Hanamoto T, Fujita T, Irino S, Watanabe K, Nakagawa T, Nakagawara A (2003) A novel HECT-type E3 ubiquitin ligase, NEDL2, stabilizes p73 and enhances its transcriptional activity. Biochem Biophys Res Commun 308: 106–113 [DOI] [PubMed] [Google Scholar]
- Nakano K, Balint E, Ashcroft M, Vousden KH (2000) A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene 19: 4283–4289 [DOI] [PubMed] [Google Scholar]
- Ongkeko WM, Wang XQ, Siu WY, Lau AW, Yamashita K, Harris AL, Cox LS, Poon RY (1999) MDM2 and MDMX bind and stabilize the p53-related protein p73. Curr Biol 9: 829–832 [DOI] [PubMed] [Google Scholar]
- Perry WL, Hustad CM, Swing DA, O'Sullivan TN, Jenkins NA, Copeland NG (1998) The itchy locus encodes a novel ubiquitin pro-tein ligase that is disrupted in a18H mice. Nat Genet 18: 143–146 [DOI] [PubMed] [Google Scholar]
- Putzer BM, Tuve S, Tannapfel A, Stiewe T (2003) Increased DeltaN-p73 expression in tumors by upregulation of the E2F1-regulated, TA-promoter-derived DeltaN′-p73 transcript. Cell Death Differ 10: 612–614 [DOI] [PubMed] [Google Scholar]
- Qiu L, Joazeiro C, Fang N, Wang HY, Elly C, Altman Y, Fang D, Hunter T, Liu YC (2000) Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J Biol Chem 275: 35734–35737 [DOI] [PubMed] [Google Scholar]
- Romani M, Tonini GP, Banelli B, Allemanni G, Mazzocco K, Scaruffi P, Boni L, Ponzoni M, Pagnan G, Raffaghello L, Ferrini S, Croce M, Casciano I (2003) Biological and clinical role of p73 in neuroblastoma. Cancer Lett 197: 111–117 [DOI] [PubMed] [Google Scholar]
- Sayan AE, Rossi M, Melino G, Knight RA (2004) p73: in silico evidence for a putative third promoter region. Biochem Biophys Res Commun 313: 765–770 [DOI] [PubMed] [Google Scholar]
- Stiewe T, Putzer BM (2002) Role of p73 in malignancy: tumor suppressor or oncogene? Cell Death Differ 9: 237–245 [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G (2001) Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem 276: 15164–15173 [DOI] [PubMed] [Google Scholar]
- Sudol M (1996) Structure and function of the WW domain. Prog Biophys Mol Biol 65: 113–132 [DOI] [PubMed] [Google Scholar]
- Toh WH, Siddique MM, Boominathan L, Lin KW, Sabapathy K (2004) c-Jun regulates the stability and activity of the p53 homologue, p73. Advance online publication 9 August 2004; DOI:10.1074/JBC.M407672200 [DOI] [PubMed]
- Treier M, Staszewski LM, Bohmann D (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78: 787–798 [DOI] [PubMed] [Google Scholar]
- Tschan MP, Grob TJ, Peters UR, De Laurenzi V, Huegli B, Kreuzer KA, Schmidt CA, Melino G, Fey MF, Tobler A, Cajot J-F (2000) Enhanced p73 expression during differentiation and complex p73 isoforms in myeloid leukemia. Biochemical Biophysical Research Communications 277: 62–65 [DOI] [PubMed] [Google Scholar]
- Ueda Y, Hijikata M, Takagi S, Chib T, Shimotohno K (1999) New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene 18: 4993–4998 [DOI] [PubMed] [Google Scholar]
- Vossio S, Palescandolo E, Pediconi N, Moretti F, Balsano C, Levrero M, Costanzo A (2002) DN-p73 is activated after DNA damage in a p53-dependent manner to regulate p53-induced cell cycle arrest. Oncogene 21: 3796–3803 [DOI] [PubMed] [Google Scholar]
- Weissman AM (2001) Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2: 169–178 [DOI] [PubMed] [Google Scholar]
- Winberg G, Matskova L, Chen F, Plant P, Rotin D, Gish G, Ingham R, Ernberg I, Pawson T (2000) Latent membrane protein 2A of Epstein-Barr virus binds WW domain E3 protein-ubiquitin ligases that ubiquitinate B-cell tyrosine kinases. Mol Cell Biol 20: 8526–8535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D (2000) p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404: 99–103 [DOI] [PubMed] [Google Scholar]
- Yuan ZM, Shioya H, Ishiko T, Sun X, Gu J, Huang YY, Lu H, Kharbanda S, Weichselbaum R, Kufe D (1999) p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature 399: 814–817 [DOI] [PubMed] [Google Scholar]
- Zaika AI, Slade N, Erster SH, Sansome C, Joseph TW, Pearl M, Chalas E, Moll UM (2002) DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med 196: 765–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Chen L, Jost CA, Maya R, Keller D, Wang X, Kaelin WG Jr, Oren M, Chen J, Lu H (1999) MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol 19: 3257–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucconi A, Dente L, Santonico E, Castagnoli L, Cesareni G (2001) Selection of ligands by panning of domain libraries displayed on phage lambda reveals new potential partners of synaptojanin 1. J Mol Biol 307: 1329–1339 [DOI] [PubMed] [Google Scholar]


