Abstract
The mRNA export adaptor Yra1p/REF contributes to nascent mRNP assembly and recruitment of the export receptor Mex67p. yra1 mutants exhibit mRNA export defects and a decrease in LacZ reporter and certain endogenous transcripts. The loss of Mlp1p/Mlp2p, two TPR-like proteins attached to nuclear pores, rescues LacZ mRNA levels and increases their appearance in the cytoplasm, without restoring bulk poly(A)+ RNA export. Chromatin immunoprecipitation, FISH and pulse-chase experiments indicate that Mlps downregulate LacZ mRNA synthesis in a yra1 mutant strain. Microarray analyses reveal that Mlp2p also reduces a subset of cellular transcripts in the yra1 mutant. Finally, we show that Yra1p genetically interacts with the shuttling mRNA-binding protein Nab2p and that loss of Mlps rescues the growth defect of yra1 and nab2 but not other mRNA export mutants. We propose that Nab2p and Yra1p are required for proper mRNP docking to the Mlp platform. Defects in Yra1p prevent mRNPs from crossing the Mlp gate and this block negatively feeds back on the transcription of a subset of genes, suggesting that Mlps link mRNA transcription and export.
Keywords: Mlp proteins, mRNP biogenesis and export, Nab2p, transcription, Yra1p
Introduction
Nascent mRNAs rapidly assemble into RNA protein complexes and undergo a series of processing steps, which result in export-competent messenger RNA ribonucleoprotein complexes (mRNPs). Two essential mRNA export factors Yra1p/REF and Sub2p, a DEHX box ATPase/RNA helicase, are recruited during transcription. The binding of Sub2p to mRNA is facilitated by Hpr1p, a component of the THO complex implicated in transcription elongation (Chavez et al, 2000; Lei et al, 2001; Strasser et al, 2002; Zenklusen et al, 2002). Yra1p and Sub2p copurify with THO in a complex called TREX, establishing a functional link between transcription and mRNA export (Strasser et al, 2002). Yra1p and other adaptor proteins such as Npl3p (Gilbert and Guthrie, 2004) facilitate the recruitment of the export receptor Mex67p to the mRNP. Mex67p then promotes mRNP export through the nuclear pore complex (NPC) via direct interactions with FG-nucleoporins (reviewed in Stutz and Izaurralde, 2003).
Recent data indicate that Rrp6p, an exonuclease specific for the nuclear exosome, participates in the degradation of nuclear pre-mRNAs, unadenylated and 3′-unprocessed mRNAs, as well as mRNPs produced in yeast TREX mutants (Bousquet-Antonelli et al, 2000; Burkard and Butler, 2000; Hilleren et al, 2001; Libri et al, 2002; Torchet et al, 2002; Zenklusen et al, 2002). The enhanced susceptibility to the nuclear exosome observed in TREX mutants has been attributed to defects in early mRNP assembly and recruitment of maturation or export factors. Mutations in THO components, Sub2p or Yra1p result in reduced mRNA levels and sequestration of newly synthesized transcripts within nuclear foci at or close to the site of transcription (Jensen et al, 2001; Zenklusen et al, 2002; Thomsen et al, 2003). The deletion of Rrp6p in these strains releases transcripts from the dots and restores mRNA levels, suggesting that the exosome retains aberrant mRNPs and favours their destruction (Hilleren et al, 2001; Libri et al, 2002; Zenklusen et al, 2002).
Although defects in early mRNP assembly and export appear to affect mRNA stability, independent studies suggested that the low LacZ mRNA levels detected in THO, sub2 or yra1 mutants resulted from transcriptional defects (Jimeno et al, 2002; Huertas and Aguilera, 2003). These two proposals are not mutually exclusive and inefficient mRNP assembly may trigger both a slow-down of mRNA transcription and increased susceptibility to degradation (Vinciguerra and Stutz, 2004).
The yeast myosin-like proteins Mlp1p and Mlp2p are large nonessential proteins proposed to form intranuclear filamentous structures anchored at the NPC nuclear basket. Various studies have indicated that these two related proteins have distinct but clearly overlapping functions (Strambio-de-Castillia et al, 1999; Galy et al, 2000; Kosova et al, 2000; Feuerbach et al, 2002; Hediger et al, 2002). Mlp1p and Mlp2p are homologous to hTPR, and related proteins have been identified in Xenopus, Drosophila and Schizosaccharomyces pombe (Cordes et al, 1997; Zimowska et al, 1997; Strambio-de-Castillia et al, 1999). Yeast Mlp proteins and hTPR contain a long N-terminal domain predicted to form coiled-coils, which mediates homodimerization and includes the NPC-binding site (Bangs et al, 1998; Cordes et al, 1998; Strambio-de-Castillia et al, 1999). Several interactions have been described between yeast Mlp proteins and mRNP components. The globular C-terminal region of Mlp1p directly interacts with Nab2p and Npl3p, two shuttling hnRNP proteins required for mRNA export (Green et al, 2003). Mlp1p also copurified with Sub2p, and Mlp2p was found in association with Mex67p (Kosova et al, 2000; Strasser et al, 2002). Despite these interactions, no poly(A)+ RNA export defect was detected in strains lacking Mlp1p, Mlp2p or both (Strambio-de-Castillia et al, 1999; Kosova et al, 2000), indicating that these proteins fulfil no essential role in mRNA export. Interestingly, Mlp1p was recently proposed to contribute to a new step of mRNP surveillance by mediating nuclear retention of unprocessed pre-mRNAs (Galy et al, 2004).
This study provides evidence that both Mlp1p and Mlp2p also retain intronless mRNP complexes produced in a yra1 mutant background, that is, mRNPs with early assembly defects. More specifically, the loss of Mlp1p or Mlp2p increases the levels of transcripts reduced in yra1 and substantially rescues the growth defect of both yra1 and nab2 mutants, consistent with a role of Yra1p and Nab2p in proper mRNP docking to Mlp proteins. The data also show that mRNP complexes loaded with mutant Yra1p are sequestered on the Mlp platform and unable to proceed along the export pathway. This block negatively impacts on the expression of a subset of genes, potentially located at the nuclear periphery. The data suggest that Mlp proteins may establish a link between mRNA synthesis and export through the nuclear pores.
Results
Loss of Mlp proteins rescues the temperature-sensitive phenotype of GFP-yra1-8
The yra1-8 mutant protein, containing two amino-acid substitutions (D10K and E11K), confers a weak growth defect on its own, but acquires a strong and tight temperature-sensitive (ts) phenotype when fused to GFP. At the nonpermissive temperature, the GFP-yra1-8 mutant exhibits nuclear retention of poly(A)+ RNA and accumulation of newly made heat-shock transcripts within nuclear foci. In addition, some transcripts are poorly expressed in this background and sensitive to degradation by the nuclear exosome (Zenklusen et al, 2002). Cells expressing wild-type (wt) GFP-Yra1p have no growth phenotype (data not shown).
To extend the functional analysis of Yra1p, we searched for high-copy suppressors of GFP-yra1-8 and identified a genomic clone expressing the first 1000 amino acids of Mlp2p (data not shown). Further characterization of the relationship between Yra1p and Mlp proteins revealed that the deletion of MLP2 or MLP1 also substantially rescued the ts phenotype of GFP-yra1-8, whereas the combined deletion of MLP1 and MLP2 had no beneficial effect (Figure 1A; see Discussion). Overexpression of the N-terminus may interfere with the formation of functional Mlp2p homodimers (Kosova et al, 2000) and thus phenocopy an MLP2 deletion. Because the GFP-yra1-8 growth defect was more efficiently rescued by loss of Mlp1p or Mlp2p than by overexpression of the N-terminal domain of Mlp2p (data not shown), all the following experiments were carried out in the context of MLP gene disruptions. Further genetic analyses showed that the deletion of MLP2 not only suppressed the GFP-yra1-8 phenotype, but was even able to bypass the unviable phenotype of a Δyra1 knockout strain (Figure 1B). These observations taken together indicate that loss of Mlp2p, and to a lesser extent of Mlp1p, alleviates the requirement for intact Yra1p, suggesting a functional interaction between Yra1p and Mlp proteins.
Figure 1.
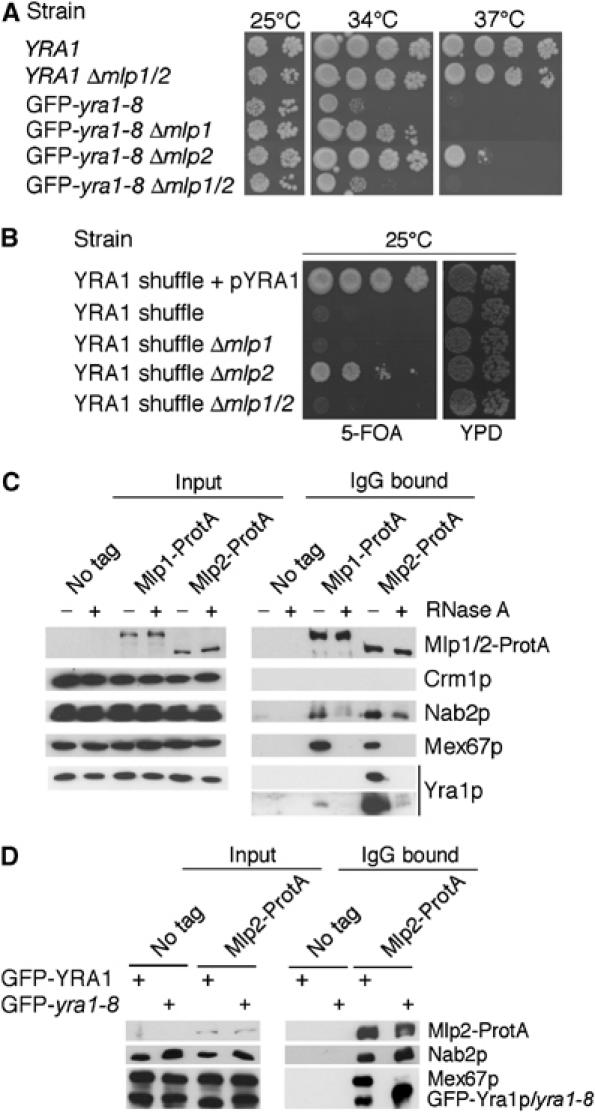
(A) Deletion of MLP2 or MLP1 rescues the ts phenotype of GFP-yra1-8. In all, 10-fold dilutions of wt YRA1 or GFP-yra1-8 strains as such, or in combination with Δmlp1, Δmlp2 or Δmlp1Δmlp2, were spotted on YEPD plates and incubated at 25 (two last dilutions only), 34 or 37°C for 3 days. (B) Deletion of MLP2 bypasses the requirement for Yra1p. The YRA1 shuffle strain (yra1::HIS3, pURA3-YRA1) as such, or combined with Δmlp1, Δmlp2 or Δmlp1/2, was spotted on a control plate (YEPD; two last dilutions only) or on medium containing 5-FOA to select against pURA3-YRA1. The YRA1 shuffle strain transformed with a wt YRA1 plasmid (pFS1877) served as positive control. (C) Mlp proteins physically interact with Yra1p and Mex67p in an RNA-dependent manner. Extracts prepared from a nontagged strain (FSY1026) or from strains expressing Mlp1-ProtA (FSY1567) or Mlp2-ProtA (FSY1351) fusions were treated (+) or not treated (−) with RNase A and purified on Pan Mouse IgG Dynabeads. Total extracts (input, left) or affinity-purified (IgG, right) extracts were analysed by Western blotting with antibodies against ProtA or the indicated proteins. Two different exposures are shown for the immunoprecipitation of Yra1p, expressed from the YRA1 cDNA. (D) The GFP-yra1-8 mutation affects mRNP composition and interaction with Mlp2p. Extracts prepared from nontagged or Mlp2-ProtA-tagged strains expressing wt GFP-Yra1p or GFP-yra1-8 and shifted to 37°C for 2.5 h were purified on Pan Mouse IgG Dynabeads. Total or affinity-purified extracts were analysed by Western blotting as above. GFP-yra1-8 migrates slightly more slowly than GFP-Yra1p.
Mlp proteins physically interact with mRNP components
To define whether Yra1p and additional mRNP components interact with Mlps, proteins copurifying with ProteinA-tagged Mlp1p or Mlp2p were examined by Western blotting (Figure 1C). As these interactions may be mediated by RNA, they were examined in untreated extracts or extracts subjected to a preliminary RNase A treatment. No association was detected between Mlp proteins and the export receptor Crm1p. In contrast, Yra1p, Nab2p and Mex67p, as well as Npl3p, Sub2p and Cbp80p (data not shown), were selected on IgG beads in the presence of Mlp1-ProtA and Mlp2-ProtA, but not with untagged extracts. These observations indicate that Mlp proteins specifically interact with these mRNP components. Yra1p interacts more strongly with Mlp2p than Mlp1p, suggesting an overlapping but distinct functional relationship of Yra1p with either protein, potentially related to the ability of Δmlp2, but not Δmlp1, to bypass Δyra1. The selection of Yra1p and Mex67p on IgG beads was sensitive to RNase A, indicating that interaction of these export factors with Mlp proteins is indirect and mediated by the mRNP. In contrast, interaction of Nab2p with Mlp1p or Mlp2p was only partially sensitive to RNase A, consistent with the proposition that Nab2p can directly interact with Mlp1p (Green et al, 2003) or Mlp2p (A Corbett, personal communication).
The GFP-yra1-8 mutation alters interaction with Mlp2p and affects mRNP composition
To determine whether the GFP-yra1-8 mutant protein is still capable of interacting with Mlp2p and whether the binding of Mex67p or Nab2p with Mlp2p is altered in the GFP-yra1-8 background, proteins copurifying with Mlp2-ProtA were compared in strains expressing wt GFP-Yra1p or mutant GFP-yra1-8 (Figure 1D). Significantly higher amounts of GFP-yra1-8 copurified with Mlp2-ProtA compared to wt GFP-Yra1p. The levels of Nab2p bound to Mlp2-ProtA were modestly increased in GFP-yra1-8, but also slightly more abundant in total extracts (see input samples and Figure 6D). Importantly, the GFP-yra1-8 mutation strongly inhibited the association of Mex67p with Mlp2-ProtA. These observations suggest that mRNPs loaded with GFP-yra1-8 are retained on Mlp2p and unable to recruit Mex67p.
Figure 6.
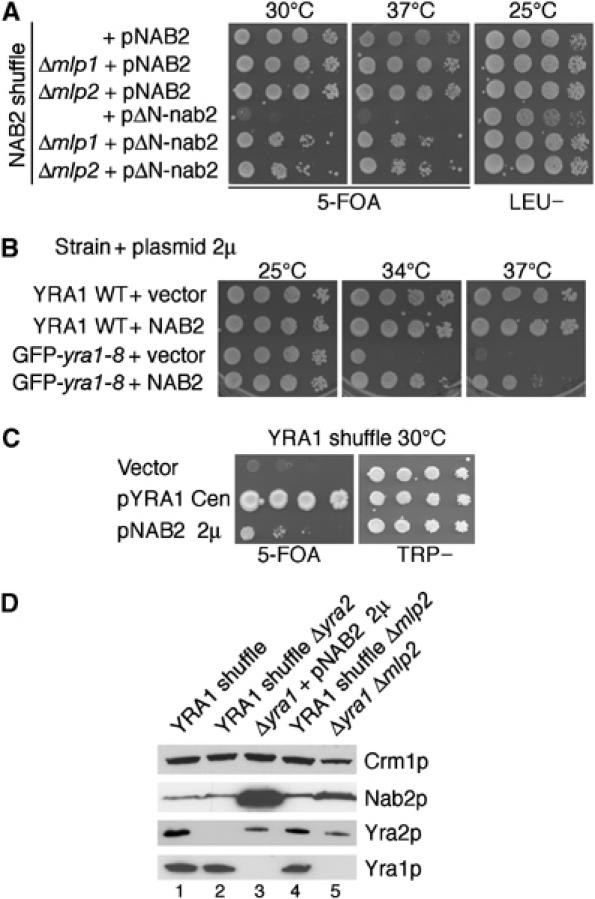
Nab2p genetically interacts with Mlp proteins and Yra1p. (A) Deletion of MLP1 or MLP2 rescues the ts phenotype of ΔN-nab2. The NAB2 shuffle strain (nab2::HIS3, pURA3-NAB2) as such, or deleted for MLP1 or MLP2, was transformed with wt NAB2 (pAC717) or ΔN-nab2 (pAC1152) plasmids. Transformants were spotted as 10-fold dilutions on 5-FOA to select against the wt pURA3-NAB2 plasmid or on a LEU− selective plate to control for cell number, and incubated for 5 days at 25, 30 or 37°C as indicated. (B) Overexpression of Nab2p partially rescues the GFP-yra1-8 ts phenotype. Wt YRA1 and GFP-yra1-8 mutant strains were transformed with empty vector or a high-copy plasmid expressing wt NAB2 (pFS2764). Transformants were spotted on selective plates and incubated for 3 days at 25, 34 or 37°C. (C) Overexpression of Nab2p bypasses the requirement for Yra1p. The YRA1 shuffle strain (yra1::HIS3, pURA3-YRA1) was transformed with an empty vector, a wt YRA1 plasmid (pFS1878) or a high-copy plasmid expressing NAB2 (pFS2764). Transformants were spotted as 10-fold serial dilutions on 5-FOA or on a selective plate as above. Plates were incubated for 5 days at 30°C. (D) The loss of Mlp2p or overexpression of Nab2p does not bypass the requirement for Yra1p by inducing Yra2p overexpression. The indicated strains were grown at 25°C in YEPD and proteins from equal amounts of cells were extracted and analysed by Western blotting with antibodies against the indicated proteins.
Deletion of MLP1 and MLP2 in GFP-yra1-8 partially rescues LacZ mRNA levels and β-galactosidase activity
The rescue of the GFP-yra1-8 ts phenotype in the absence of Mlp2p or Mlp1p suggested that the loss of these proteins may reduce the poly(A)+ RNA export block observed in this mutant strain. However, the analysis of poly(A)+ RNA distribution in GFP-yra1-8 combined with Δmlp1, Δmlp2 or Δmlp1Δmlp2 showed no significant decrease in nuclear poly(A)+ RNA signal relative to GFP-yra1-8 alone (Supplementary Figure 1), suggesting that the better growth of GFP-yra1-8 in the absence of Mlp2p or Mlp1p is not due to a detectable increase in bulk poly(A)+ RNA export.
To examine the effect of these mutations on a specific transcript, wt or GFP-yra1-8 mutant strains as such or in combination with Δmlp1, Δmlp2 or Δmlp1Δmlp2 were transformed with pLGSD5, a high-copy plasmid carrying a galactose-inducible and intronless LacZ gene (Legrain and Rosbash, 1989). The levels of LacZ transcripts and β-galactosidase activity were measured after inducing the cells for 2.5 h at 37°C (Figure 2A). LacZ transcripts were barely detectable in GFP-yra1-8 (0.6% of wt, lanes 1 and 5, light grey bars). These very low levels of LacZ transcripts probably result from the combination of a defect in Yra1p and the absence of a bona fide transcription terminator on the pLGSD5 reporter (Long et al, 1995, and see below). Deletion of Mlp1p or Mlp2p had marginal effects in the wt strain, but restored pLGSD5-encoded LacZ mRNA levels up to 50–60% of wt in GFP-yra1-8 (Figure 2A, lanes 1–3 and 5–7). Thus, Mlp proteins significantly affect LacZ mRNA synthesis and/or stability in the mutant, but not in the wt background. Mlp1p and Mlp2p are probably involved in the same process, as the double deletion did not lead to higher LacZ mRNA levels than the single ones (lanes 6–8).
Figure 2.
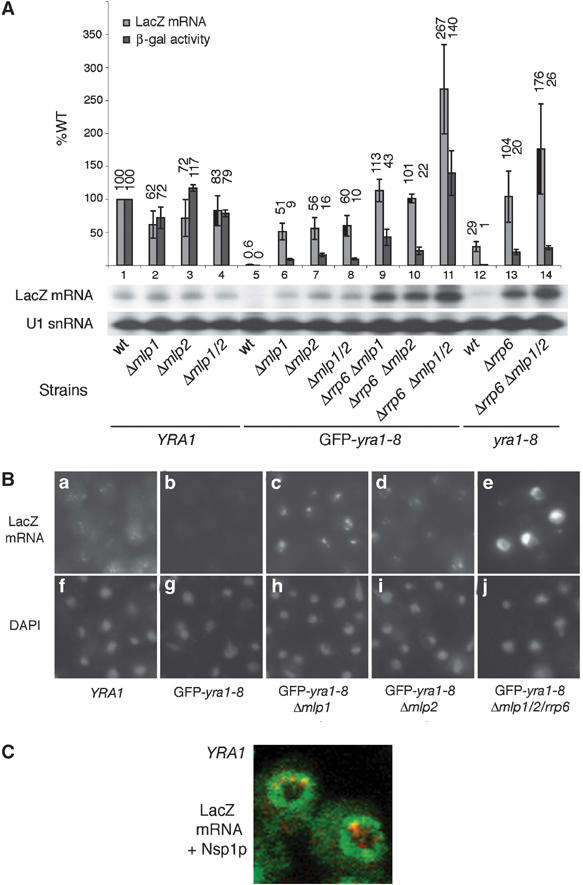
(A) Loss of Mlp1p and Mlp2p restores LacZ mRNA levels in GFP-yra1-8. Strains expressing wt Yra1p (FSY1485) or mutant GFP-yra1-8 (FSY1486) or yra1-8 (FSY1786) proteins as such, or in combination with the indicated Δmlp1, Δmlp2 or Δrrp6 simple, double or triple disruptions, were transformed with the reporter plasmid pLGSD5 (Legrain and Rosbash, 1989) encoding LacZ from a galactose-inducible promoter. Transformants were pregrown in selective medium containing 2% glycerol/2% lactate/0.05% glucose, and LacZ expression was induced by the addition of 3% galactose/1% raffinose for 2.5 h at 37°C. LacZ mRNA levels (light grey bars), defined by primer extension analysis of total RNA, were quantified and normalized to the U1 snRNA internal control. The same strains were analysed for β-galactosidase (β-gal) activity (dark grey bars). LacZ mRNA levels and β-galactosidase enzymatic activities were expressed as a percentage of wt and compared on the same histogram. Error bars have been derived from three independent experiments. (B) In situ localization of LacZ transcripts. The indicated strains transformed with pLGSD5 were induced with galactose as above and processed for in situ hybridization with Cy3-conjugated oligonucleotide probes specific for LacZ mRNA (panels a–e). DAPI staining indicates the position of the nucleus (panels f–j). RNA distribution was examined in fixed cells using a Zeiss axioplan fluorescent microscope with × 100 objective. Exposure time was of 4 s in panels a–d and only 1 s in panel e. (C) LacZ transcripts are located at the nuclear periphery. wt cells transformed with pLGSD5 were induced and prepared as above. Fixed cells were hybridized with Cy3-conjugated LacZ probes and subsequently immunostained with anti-Nsp1p monoclonal and FITC-labelled secondary antibodies. Samples were analysed and images taken with a Zeiss LSM510 confocal microscope.
LacZ mRNAs are a substrate for the nuclear exosome when Yra1p is defective (Zenklusen et al, 2002). To investigate whether Mlps participate in the exosome-dependent degradation, we examined LacZ mRNA levels in strains lacking both Rrp6p and Mlps. Δrrp6 is lethal in combination with GFP-yra1-8, but viable when Mlp1p and/or Mlp2p are already missing from the mutant strain (Supplementary Figure 2). The loss of Rrp6p in addition to Mlp1p and Mlp2p in GFP-yra1-8 had a clear additive effect on LacZ mRNA levels, which reached up to 270% of wt (lane 11). To corroborate these observations, we used the milder yra1-8 mutant, which is viable in the absence of Rrp6p and allows to analyse the effect of Δrrp6 alone (Zenklusen et al, 2002). Loss of Rrp6p in yra1-8 restored LacZ transcripts to wt levels (compare lanes 1, 12 and 13), and loss of Mlp1p and Mlp2p in addition to Rrp6p yielded even higher LacZ mRNA amounts (up to 176% of wt; compare lanes 13 and 14). These cumulative effects suggest that Mlp proteins do not contribute to Rrp6p activity, but rather that Rrp6p and Mlp proteins affect LacZ mRNA levels via distinct mechanisms.
β-Galactosidase enzymatic activities were measured in all these strains to evaluate what fraction of LacZ transcripts was exported. Like LacZ mRNA levels, β-galactosidase activity was not significantly altered by loss of Mlp1p and/or Mlp2p in wt cells, confirming that Mlp proteins have no major effect on mRNA amounts and export under normal conditions (compare lanes 1–4; dark grey bars). β-Galactosidase activity was undetectable or extremely low in the GFP-yra1-8 or yra1-8 mutants respectively (lanes 5 and 12), consistent with the low mRNA levels. Importantly, loss of Mlp1p, Mlp2p or Rrp6p, which greatly increased the LacZ mRNA levels in these strains, led to marginal increases in enzymatic activity (lanes 6–8 and 13). Thus, only a small fraction of these transcripts was able to reach the cytoplasm, in agreement with the persistence of the poly(A)+ RNA export block in these cells (Supplementary Figure 1). The higher β-galactosidase activity in GFP-yra1-8 Δmlp1Δmlp2Δrrp6 (140% of wt, lane 11) may result from the extremely high LacZ mRNA levels present in this strain (270% of wt). One possibility is that loss of Mlp proteins increases LacZ mRNA access to the cytoplasm.
Localization of LacZ transcripts by in situ hybridization
To assess the cellular localization of the LacZ transcripts more directly, pLGSD5-transformed strains were induced with galactose at 37°C and examined by in situ hybridization with LacZ-specific probes (Figure 2B). In a wt strain, a weak and diffuse signal was detected in the cytoplasm but LacZ transcripts also accumulated as small nuclear foci, probably corresponding to nascent transcripts encoded by the multicopy plasmid (panel a). This distribution was specific for LacZ mRNA, as no signal was detected in noninduced cells (data not shown). Simultaneous detection of LacZ transcripts and the NPC protein Nsp1p indicated that LacZ nuclear foci were predominantly located at the nuclear periphery (Figure 2C). Consistent with the primer extension analyses, no LacZ transcripts were detected in GFP-yra1-8 (Figure 2B, panel b), but the signal was restored in the absence of Mlp1p or Mlp2p (panels c and d). In these strains, LacZ transcripts were detected primarily in the nucleus where they accumulated within a few foci. The cytoplasmic signal appeared weaker in these cells compared to wt, but slightly stronger than in GFP-yra1-8, in agreement with the appearance of a small fraction of LacZ transcripts in the cytoplasm. Finally, the LacZ signal was extremely strong and mainly nuclear in GFP-yra1-8Δmlp1Δmlp2Δrrp6, in agreement with the high mRNA levels detected in this strain (Figure 2A and B, panel e). The cytoplasmic signal appears weak in these cells, as exposure time was four times less in panel e to avoid overexposure of the nuclear signal.
The same strains were examined for the distribution of endogenous SSA4 transcripts induced at 42°C (Supplementary Figure 3). In GFP-yra1-8, SSA4 mRNAs accumulated within a single nuclear focus, which persisted and became even slightly more intense in the absence of Mlp1p, Mlp2p or both (Supplementary Figure 3A). In contrast, the nuclear dot disappeared when RRP6 was deleted in these strains (Supplementary Figure 3B), indicating that the retention of heat-shock transcripts within foci depends on Rrp6p and not on Mlp proteins. These observations provide additional evidence for independent roles of Rrp6p and Mlp proteins in mRNP biogenesis.
LacZ transcripts are not more stable in GFP-yra1-8 Δmlp2 than in GFP-yra1-8
To investigate how the absence of Mlp2p or Mlp1p may increase LacZ mRNA levels, mRNA turnover rates were compared in wt or GFP-yra1-8 strains in the presence or absence of Mlp2p. pLGSD5-encoded LacZ transcripts were not suitable for mRNA decay studies, since they were quasi-undetectable in GFP-yra1-8 (Figure 2A, lane 5). Thus, wt YRA1, YRA1Δmlp2, GFP-yra1-8 and GFP-yra1-8Δmlp2 strains were transformed with pGAL1-LacZ, a centromeric reporter construct containing a bona fide terminator (Chavez et al, 2000) and producing higher levels of LacZ transcripts in GFP-yra1-8 than pLGSD5 (Figure 3). After 2.5 h of galactose induction at 37°C, LacZ transcription was shut-off by shifting the cells to medium containing glucose, and mRNA decay was followed over time. In wt YRA1 and YRA1Δmlp2, LacZ transcripts accumulated to similar levels and decayed with a comparable half-life of 4.5 min (lanes 1–8). This half-life likely reflects cytoplasmic mRNA turnover, as LacZ mRNAs are efficiently assembled and exported in these two strains. In GFP-yra1-8, LacZ mRNAs accumulated to only 28% of wt and decayed more slowly, with a half-life of 13 min (lanes 9–12). Importantly, LacZ mRNA levels in GFP-yra1-8Δmlp2 accumulated to 68% of wt but decayed with a half-life of 6 min (lanes 13–16), an intermediate value between that obtained in wt and GFP-yra1-8. Notably, LacZ transcripts accumulate to higher steady-state levels in GFP-yra1-8Δmlp2 despite a shorter half-life, suggesting that loss of Mlp2p in GFP-yra1-8 does not stabilize LacZ transcripts, but results in increased mRNA synthesis. The data therefore support the view that Mlp2p downregulates mRNA synthesis in the GFP-yra1-8 mutant background. The drop in LacZ mRNA half-life, from 13 min in GFP-yra1-8 to 6 min in GFP-yra1-8Δmlp2, may reflect an increase in cytoplasmic LacZ mRNA levels, or changes in mRNP quality (Jensen et al, 2004).
Figure 3.
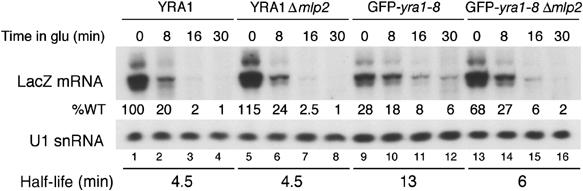
LacZ mRNA decay rates differ in GFP-yra1-8 and GFP-yra1-8Δmlp2. Exponential cultures of wt YRA1, YRA1Δmlp2, GFP-yra1-8 and GFP-yra1-8Δmlp2 strains transformed with the pGAL1-LacZ reporter construct were induced in medium containing 3% galactose/1% raffinose for 2.5 h at 37°C, transferred to preheated medium containing 2% glucose and collected after various times in glucose at 37°C. LacZ transcripts were quantified by primer extension and their levels expressed as a percentage of wt at time 0. LacZ mRNA half-lives were calculated as described (Materials and methods).
The LacZ mRNA decay analyses as well as the two next experiments were performed in GFP-yra1-8Δmlp2 but not in GFP-yra1-8Δmlp1. Based on the similar phenotypes induced by the absence of Mlp1p or Mlp2p in GFP-yra1-8 (Figures 1 and 2), we speculate that loss of Mlp1p may have effects overlapping those induced by loss of Mlp2p in these other assays as well.
Mlp2p downregulates LacZ mRNA synthesis in GFP-yra1-8
To evaluate the effect of Mlp2p on GAL1-LacZ mRNA transcription more directly, we used chromatin immunoprecipitation (ChIP) experiments to compare the association of the TATA-box-binding protein (TBP) with the GAL1 promoter in the wt YRA1, YRA1Δmlp2, GFP-yra1-8 and GFP-yra1-8Δmlp2 strains transformed with pGAL1-LacZ. Real-time PCR quantification of pGAL1-LacZ plasmid confirmed identical copy numbers of this centromeric reporter construct in the four strains (data not shown). Four independent ChIP experiments showed that the amount of TBP bound to the GAL1 promoter was more than two-fold lower in GFP-yra1-8 compared to wt (Figure 4). Importantly, loss of Mlp2p in GFP-yra1-8 increased TBP binding by at least two-fold, restoring nearly wt levels of TBP association. In contrast, loss of Mlp2p in wt YRA1 slightly reduced TBP binding (80% of wt). Additional ChIP analyses revealed comparable relative changes in the association of RNA polymerase II (Pol II) over the LacZ coding region in these four strains (data not shown). The two- to three-fold increase in TBP or RNA Pol II binding over the GAL1-LacZ transcription unit parallels the two- to three-fold increase in LacZ mRNA levels in GFP-yra1-8Δmlp2 relative to GFP-yra1-8 (Figure 3). Western blot analyses confirmed that the total levels of TBP were identical in all four strains (data not shown). These observations taken together show that Mlp2p downregulates LacZ gene transcription initiation and elongation in GFP-yra1-8 but not in the wt background.
Figure 4.
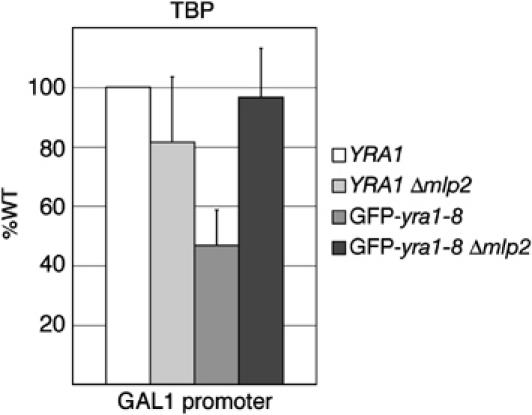
ChIP analysis of TBP binding to pGAL1-LacZ promoter. The indicated strains transformed with the pGAL1-LacZ reporter were induced with galactose for 2.5 h at 37°C prior to crosslinking with formaldehyde. Crosslinked and sonicated extracts were immunoprecipitated with antibodies against TBP, and copurifying DNA was amplified by real-time PCR with primers specific for the GAL1 promoter. The relative abundance of the GAL1 promoter segment in each immunoprecipitate was expressed as a fold increase with respect to a nontranscribed intergenic region value set to 1. TBP binding in the different strains was then expressed as a percentage of binding in wt YRA1. In wt YRA1, a 20-fold (±5) increase of TBP was measured at the GAL1 promoter in the presence of galactose. Values were derived from four independent experiments.
Mlp2p affects the levels of cellular transcripts
To assess the effect of Mlp2p on gene expression in GFP-yra1-8 at a global level, DNA microarrays (Affymetrix) were used to compare mRNA levels in wt YRA1 to those in YRA1Δmlp2, GFP-yra1-8 and GFP-yra1-8Δmlp2 after a 2.5 h shift at 37°C. An initial comparison identified a set of 194 transcripts, which decreased by two-fold or more (median log ratio equal to or less than −1) in GFP-yra1-8 compared to wt YRA1 (Supplementary data and Supplementary Table I; Miamexpress accession number E-MEXP-130). Comparison of the mean intensity values revealed that most (85%) of these 194 transcripts increased in GFP-yra1-8Δmlp2 versus GFP-yra1-8, of which 58 augmented by 1.5-fold or more (dark green genes in Supplementary Table I). Thus, Mlp2p significantly downregulates at least 30% of the cellular transcripts most strongly affected in GFP-yra1-8 (Figure 5A). In contrast, the intensity values of the same set of 194 transcripts remained unchanged or became slightly lower in YRA1Δmlp2 versus wt YRA1 (Figure 5B). The 58 Mlp2p-sensitive genes in GFP-yra1-8 include low and highly expressed genes, located all over the genome and encoding proteins with various functions (Supplementary Table I and data not shown). Only three of these genes are essential and encode RNase P RNA (RPR1), or proteins involved in fatty acid biosynthesis (FAS2 and ACC1). It is unclear whether improper expression of these genes and/or a combination of the nonessential genes is responsible for the phenotypes observed in GFP-yra1-8.
Figure 5.
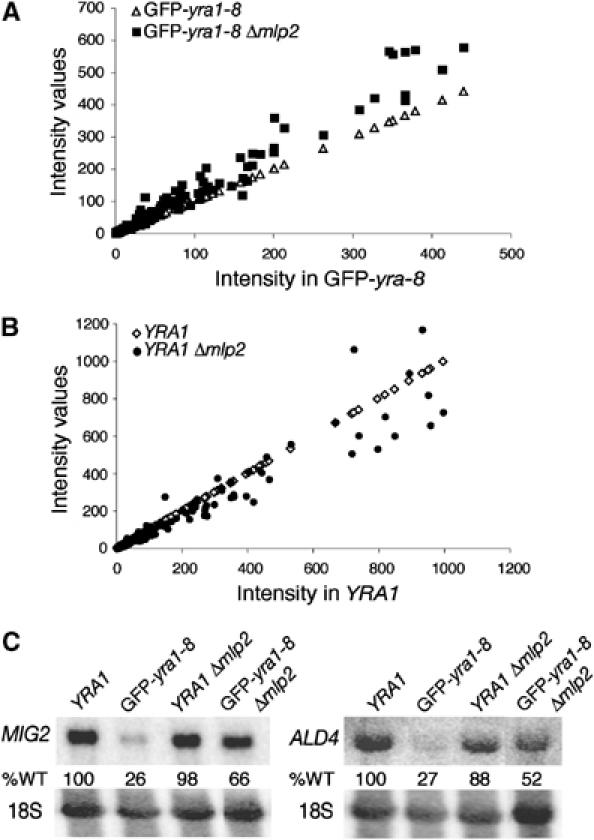
Mlp2p downregulates a subset of cellular transcripts in GFP-yra1-8 but not in wt YRA1. (A) The DNA microarray intensity values of the 194 transcripts decreased in GFP-yra1-8 (open triangles), and their values in GFP-yra1-8Δmlp2 (closed squares) were plotted as a function of the intensity of these 194 transcripts in GFP-yra1-8. (B) The intensity values of the same 194 selected transcripts in wt YRA1 (open diamonds) and YRA1Δmlp2 (closed circles) were plotted as a function of transcript intensity in wt YRA1. (C) Mlp2p downregulates MIG2 and ALD4 transcripts in GFP-yra1-8 but not in wt YRA1. Exponentially growing cultures of the indicated strains were shifted for 2.5 h to 37°C. Total RNA was extracted and examined by Northern blot analysis with MIG2- and ALD4-specific probes. The same blots were rehybridized with a probe specific for 18S RNA to correct for unequal loading.
The microarray data were confirmed by Northern blotting of two highly expressed transcripts, MIG2 and ALD4, strongly affected in GFP-yra1-8 and rescued upon loss of Mlp2p (Figure 5C). MIG2 RNA levels increased from 26% of wt in GFP-yra1-8 to 66% in GFP-yra1-8Δmlp2 (25 and 76% by microarray; MIG2 is gene no. 6 in Supplementary Table I). Similarly, ALD4 transcripts rose from 27% in GFP-yra1-8 to 52% in GFP-yra1-8Δmlp2 (49 and 75% by microarray; ALD4 is gene no. 52 in Supplementary Table I). In contrast, MIG2 and ALD4 mRNA levels in YRA1Δmlp2 were 98 and 88% of wt (88 and 96% by microarray), confirming the marginal effect of Mlp2p on these genes in a wt background. These observations support the view that Mlp2p downregulates a subset of transcripts in response to a defect in Yra1p.
Genetic interactions functionally link Mlp proteins and Yra1p to Nab2p
To address the functional relationship between Mlp proteins and mRNA export factors distinct from Yra1p, MLP1 or MLP2 was deleted in several other ts mutants affecting early or late steps of mRNA biogenesis and export, and the growth phenotypes of single and double mutants examined (Table I). Loss of Mlp1p or Mlp2p had no significant effect on growth in most mRNA export mutant backgrounds, with the exception of ΔN-nab2. The ΔN-nab2 mutant lacks the 100 amino-terminal amino acids proposed to interact with the mRNA export machinery (Marfatia et al, 2003). The ts phenotype of ΔN-nab2 was efficiently rescued in the absence of Mlp1p or Mlp2p (Figure 6A). The deletion of MLP1 or MLP2 even weakly bypassed the lethal Δnab2 phenotype (data not shown). The better growth of the GFP-yra1-8 and ΔN-nab2 mutants in the absence of Mlp1p or Mlp2p suggests that loss of these perinuclear proteins alleviates the requirement for fully functional Yra1p and Nab2p. These two mRNA export factors may therefore play a crucial role in docking the mRNP to the Mlp platform, in agreement with the proposed direct interaction between Nab2p and Mlp1p (Green et al, 2003).
Table 1.
Growth phenotypes of mRNA export mutants combined with Δmlp1 or Δmlp2 at 34°C
| Strains |
wt |
Δmlp1 |
Δmlp2 |
|---|---|---|---|
| wt | ++++ | ++++ | ++++ |
| GFP-yra1-8 | − | ++ | +++ |
| ΔN-nab2 | − | +++ | +++ |
| sub2-201 | − | − | − |
| Δhpr1 | + | ++ | ++ |
| Δcbp20 | +++ | +++ | +++ |
| Δcbp80 | +++ | +++ | +++ |
| rna14-1 | + | + | + |
| rna15-1 | − | − | − |
| pap1-1 | − | − | − |
| npl3-1 | − | − | − |
| Δrrp6 | + | + | ++ |
| mex67-5 | − | − | +/− |
| rat8-2 | + | − | − |
|
rat7-1 |
+/− |
ND |
+/− |
| The indicated strains were grown for 3 days at 34°C on YPD as 10-fold dilution series. The growth of wt was arbitrarily set to ‘++++'. Each‘+' roughly corresponds to a 10-fold difference in growth at 34°C. See Supplementary data for strain descriptions and references. | |||
| ND: not determined. | |||
Interestingly, overexpression of Nab2p efficiently suppresses the GFP-yra1-8 mutant and even bypasses the lethal Δyra1 phenotype (Figure 6B and C). These genetic observations suggest either that Nab2p can substitute for Yra1p or that Yra1p contributes to Nab2p function. Our earlier data have shown that the lethal phenotype of Δyra1 can also be suppressed by overexpression of the nonessential Yra1p-related protein Yra2p (Zenklusen et al, 2001). To confirm that the bypass of Δyra1, in Δmlp2 or when Nab2p is in excess, is not due to overexpression of endogenous Yra2p, the amounts of this protein were examined in the relevant strains (Figure 6D). The levels of Yra2p are comparable in the YRA1 shuffle and YRA1 shuffle Δmlp2 strains (lanes 1 and 4), and remain largely unchanged in Δyra1Δmlp2 or in Δyra1 overexpressing Nab2p (lanes 5 and 3). Thus, the growth of the strains deleted for YRA1 is not due to elevated amounts of Yra2p. In contrast, the levels of Nab2p are increased in Δyra1Δmlp2, consistent with the ability of Nab2p to compensate for the absence of Yra1p (Figure 6C). These data taken together support the view that Yra1p, Nab2p, Mlp2p, and to some extent Mlp1p, participate in a common step along the mRNP biogenesis pathway.
Discussion
In this study, we found that the loss of Mlp2p, and to a lesser extent Mlp1p, substantially rescues the ts phenotype of the GFP-yra1-8 mutant without detectably restoring poly(A)+ RNA export (Figure 1 and Supplementary Figure 1). LacZ mRNAs, which are poorly expressed in GFP-yra1-8, increase in the absence of Mlp1p and/or Mlp2p. Analysis of β-gal enzymatic activity in these strains however indicates that only a small fraction of these transcripts reaches the cytoplasm for translation (Figure 2A). Thus, although the loss of Mlp proteins in GFP-yra1-8 does not significantly rescue poly(A)+ RNA export, it nevertheless reduces the retention of incorrectly assembled mRNP complexes. Deletion of MLP1 and/or MLP2 increases LacZ transcripts in the mutant but not in the wt background, suggesting that Mlp proteins lower LacZ mRNA levels in response to a mutation in Yra1p (Figure 2). Similarly, genome-wide analyses indicate that Mlp2p negatively affects the levels of a fraction of endogenous transcripts in GFP-yra1-8 but not in a wt strain (Figure 5). These observations suggest that the loss of Mlp2p, and to a lesser extent of Mlp1p, may rescue the GFP-yra1-8 ts phenotype by restoring the levels of limiting cellular transcripts, a fraction of which is able to reach the cytoplasm. The data further support the view that Mlp2p lowers the expression levels of a subset of transcripts in GFP-yra1-8 by reducing mRNA synthesis rather than stability (see below).
Mlp proteins retain malformed mRNP complexes within the nucleus
Mlp1p was proposed to participate in mRNP surveillance by retaining unspliced pre-mRNAs via interaction with early splicing factors associated with unprocessed transcripts (Galy et al, 2004). Our data indicate that the GFP-yra1-8 mutant protein more strongly interacts with Mlp2p than its wt counterpart, suggesting that mutant mRNP complexes get trapped on Mlp2p. The better growth of GFP-yra1-8 in the absence of Mlp2p is likely due to increased mobility of malformed mRNP complexes. Loss of Mlps however did not result in a significant flow-off of poly(A)+ RNA into the cytoplasm (Supplementary Figure 1), probably because Mex67p is very poorly recruited to mRNPs in the presence of GFP-yra1-8 (Figure 1D). Thus, only properly packaged mRNPs may be able to interact productively with the Mlp platform and subsequently access the central channel of the NPC. One prediction of this model is that mRNP components, whose main function is to mediate the interaction with Mlp proteins, would become dispensable in the absence of this perinuclear gate. In this view, the ability of Δmlp2 to bypass the lethal phenotype of Δyra1 is consistent with a role of Yra1p in mRNP docking to Mlp2p. Δmlp2 rescued GFP-yra1-8 more efficiently than Δmlp1. These distinct phenotypes could reflect preferential interaction of either Mlp protein with specific subsets or types of cellular transcripts. In contrast, loss of both Mlp1p and Mlp2p had no beneficial effect on GFP-yra1-8 growth. Possibly, the Δmlp1Δmlp2 double deletion more severely affects NPC structure and nuclear envelope integrity (Hediger et al, 2002), and totally prevents mRNPs poorly loaded with Mex67p to reach the cytoplasm.
Besides GFP-yra1-8, the ΔN-nab2 strain was the only mutant among a number of conditional mRNA export mutants, whose growth phenotype was efficiently rescued in the absence of Mlp1p or Mlp2p (Figure 6A and Table I). These genetic data suggest a key role for Nab2p in mRNP docking to the Mlp platform. Consistently, Nab2p directly interacts with the C-terminal domain of Mlp1p (Green et al, 2003) and Mlp2p (A Corbett, personal communication). Interestingly Nab2p has also been proposed to participate in polyA tail length regulation (Hector et al, 2002), a process important for mRNA export. Thus Nab2p could mediate mRNP docking by signalling proper 3′ end formation. Furthermore, the ability of Nab2p overexpression to suppress defects in Yra1p (Figure 6B and C) is consistent with a function of both proteins in a common step of mRNA export. One possibility could be that Yra1p facilitates the binding of Nab2p to Mlp proteins or to some other export-related factor.
Mlp proteins reduce mRNA transcription in response to inefficient mRNP assembly
Although Mlp proteins have no described exonuclease activity, they could contribute to mRNA degradation by facilitating the interaction of the mRNP with the nuclear exosome. Our data however indicate that Mlp proteins act independently of Rrp6p and are more likely to downregulate LacZ gene transcription rather than to reduce mRNA stability in GFP-yra1-8. First, the combined deletions of RRP6 and MLP1/MLP2 in a yra1 mutant strain have additive effects on LacZ mRNA levels, indicating that Mlp proteins and Rrp6p probably act at different steps of mRNA biogenesis (Figure 2A). Second, whereas Δrrp6 is synthetically lethal with GFP-yra1-8 (Zenklusen et al, 2002), the loss of Mlp proteins rescues the mutant growth defect (Figure 1A), further indicating that Rrp6p and Mlp proteins have distinct functional relationships to Yra1p. Finally, the sequestration of newly made SSA4 transcripts within nuclear foci in GFP-yra1-8 disappeared upon loss of Rrp6p, but persisted in the absence of Mlp proteins alone (Supplementary Figure 3). Similarly, LacZ transcripts accumulated in a few intense nuclear foci in GFP-yra1-8 lacking Mlp1p or Mlp2p (Figure 2B). These results indicate that Rrp6p, but not Mlp proteins, is implicated in the retention of mutant mRNPs within nuclear foci.
The strong increase and accumulation of LacZ signal within nuclear dots in GFP-yra1-8Δmlp1 and GFP-yra1-8Δmlp2 is compatible with higher LacZ gene transcription in the absence of these perinuclear proteins (Figure 2B). An effect of Mlp2p on LacZ transcription was also inferred from the analysis of LacZ mRNA decay in GFP-yra1-8 versus GFP-yra1-8Δmlp2 (Figure 3). Finally, ChIP experiments showed that the association of TBP and RNA Pol II with the GAL1-LacZ gene was reduced in GFP-yra1-8 relative to wt, but increased by at least two-fold in GFP-yra1-8Δmlp2 (Figure 4 and data not shown), confirming that Mlp2p downregulates LacZ gene transcription initiation and elongation in response to a defect in Yra1p.
Mlp proteins may link transcription and mRNA export
How could Mlp proteins affect mRNA transcription? mRNP complexes produced in GFP-yra1-8 more strongly interact with Mlp proteins, presumably resulting in the stalling of mRNP particles along the mRNA export pathway. This block may negatively feed back on the transcription machinery via some yet undefined reverse flow of information (Figure 7). Interestingly, Mlp proteins are present only on sections of the nuclear envelope adjacent to chromatin (Galy et al, 2004), suggesting that they may contact nascent transcripts, thereby linking mRNA synthesis and export. The frequent localization of galactose-induced LacZ transcripts at the nuclear periphery (Figure 2C) suggests that transcription of the reporter construct may indeed occur in the vicinity of NPCs. Recent studies have established functional links between the NPC and gene expression. One provided evidence that transcriptional activation of the GAL genes results in their association with nuclear pore proteins, including Mlps (Casolari et al, 2004). Another identified physical interactions between components of the transcription apparatus and the pore-associated mRNA export machinery (Rodriguez-Navarro et al, 2004). Earlier experiments also revealed that physical tethering of genomic loci to the NPC can alter their transcriptional activity (Ishii et al, 2002). Finally, proteomic studies indicated association of Mlp1p with various transcription components (Gavin et al, 2002). Therefore, the effect of Mlp proteins on gene expression may reflect physical interactions with growing mRNPs and/or components of the transcription machinery.
Figure 7.
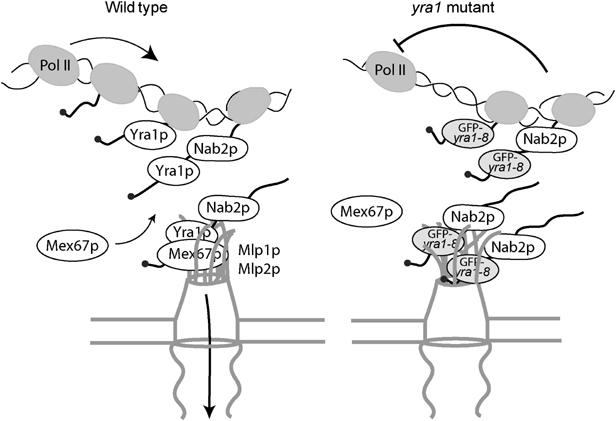
Model. Mlp proteins establish a link between mRNA synthesis and export. In wt cells, the mRNA-binding proteins Yra1p and Nab2p are essential for mRNP docking to the Mlp export gate at the nuclear periphery. mRNP complexes produced in the GFP-yra1-8 mutant strain are retained by the Mlp selective filter and mRNP stalling negatively feeds back on mRNA synthesis. Loss of Mlp1p or Mlp2p alleviates the negative effect on mRNA synthesis and allows a fraction of transcripts to reach the cytoplasm. Whether Mlp proteins physically interact with nascent transcripts has to be determined.
Our genome-wide analyses show that a subset of transcripts decreased in GFP-yra1-8 are significantly upregulated by the loss of Mlp2p. This raises the question of how Mlp2p discriminates between the same mutant Yra1p loaded on different mRNAs. The finding that only a minor subset of transcripts are affected, including those of the LacZ reporter gene apparently transcribed in the proximity of the nuclear envelope, might indicate that Mlp proteins are involved in the regulation of a small group of genes located at the nuclear periphery. It will be interesting to define whether the LacZ reporter or the cellular genes whose expression levels are influenced by Mlp proteins in GFP-yra1-8 preferentially localize at the nuclear envelope. Future experiments will focus on the functional interactions between Yra1p, Nab2p and Mlp proteins and determine whether these perinuclear proteins become associated with nascent transcripts.
Materials and methods
Plasmids and yeast strains
Plasmids and yeast strains used in this study are described in Supplementary data section and summarized in Supplementary Tables II and III.
Yeast extracts, affinity purifications of ProtA fusions and Western blotting
Mlp1-ProtA, Mlp2-ProtA and nontagged Yra1 shuffle strains were grown in YEPD at 30°C as 800 ml cultures to OD600=1 (1.5 × 107 cells/ml). Cells were collected by centrifugation and washed once with cold 1 × PBS. Pellets were resuspended as 130 OD600/ml in lysis buffer (20 mM Hepes–KOH pH 7.5, 100 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.25% Triton X-100 and complete protease inhibitor mix (Roche)) and lysed with 4 ml glass beads by vortexing 10 times for 1 min in the cold. The soluble fraction was recovered after centrifugation for 45 min at 10 000 g. For RNase A treatment, lysates were split and incubated at 25°C for 30 min with or without 150 μg/ml of RNase A. Lysates were then incubated with 50 μl of Dynabeads Pan Mouse IgG (Dynal biotech) for 20 h at 4°C. Beads were washed four times with 4 ml cold lysis buffer prior to boiling in protein sample buffer. Immunoprecipitates were examined by Western blotting as described (Supplementary data). Mlp2-ProtA and nontagged Yra1 shuffle strains shuffled with GFP-YRA1 (YRA1 cDNA) or GFP-yra1-8 constructs were grown in YEPD at 25°C as 400 ml cultures to 0.75 × 107 cells/ml. These 400 ml were shifted to 37°C for 2.5 h. The subsequent lysis and immunoprecipitation steps were performed as described above, except that Triton X-100 was used at 0.5%.
RNA extractions, primer extensions and Northern blotting
RNA extractions, Northern blotting and primer extensions were performed as described (Zenklusen et al, 2002). The following primers were used for primer extension: OFS506 (5′CACCAGTGAGACGGGC3′) for pLGSD5-encoded LacZ, OFS790 (5′GGGTTTTCCCAGTCACGACG) for pGAL1-LacZ-encoded LacZ and OFS645 (5′GGACTTCTTGATCTCCTCTG3′) for U1 snRNA.
To define LacZ mRNA half-lives, counts remaining after the shift to glucose were expressed as ln(%WT cpm) and plotted as a function of time. mRNA half-lives were obtained by using the relationship T1/2=ln 2/k, where k is the slope of the best-fit line.
β-Galactosidase assays
Cells transformed with the LacZ reporter construct pLGSD5 were grown in selective medium containing 2% lactate/2% glycerol/0.05% glucose to 3 × 106 cells/ml. Cells were shifted to 37°C and induced for 2.5 h by the addition of 1/10 volume 30% galactose/10% raffinose. A 1 ml portion of cells was collected by centrifugation and analysed for β-galactosidase activity.
In situ hybridizations and immunostaining
Fixed cells were incubated with a 3′Cy3-labelled oligo-dT55 probe essentially as described (Zenklusen et al, 2002). SSA4 and LacZ-specific in situ hybridization with a mixture of two Cy3-labelled oligonucleotide primers was performed as described (Thomsen et al, 2003; Galy et al, 2004). In double-labelling experiments, NPCs were visualized by incubating in situ-hybridized cells with anti-Nsp1p monoclonal (EnCor Biotech; dilution 1:500) and FITC-conjugated secondary antibodies as described (Thomsen et al, 2003).
Chromatin immunoprecipitation analysis
Cells transformed with the LacZ reporter construct pGAL1-LacZ were grown at 25°C in selective medium containing 2% lactate/2% glycerol/0.05% glucose to 3 × 106 cells/ml. Cells were shifted to 37°C and LacZ transcription was induced for 2.5 h by the addition of 1/10 volume 30% galactose/1% raffinose. ChIPs were performed as described previously (Zenklusen et al, 2002), except that cells were incubated with 1.2% formaldehyde for only 10 min. Crosslinked and sonicated extracts were precleared for 1 h and immunoprecipitated overnight with antibodies specific for TBP (a gift of Martine Collart) in the presence of Protein A–Sepharose beads. Immunoprecipitated DNA was analysed by quantitative PCR. Each ChIP was repeated four times with independent extracts and each PCR was run in duplicate. To calculate the fold increase in signal on the GAL1 promoter, the absolute values obtained by quantitative PCR were normalized to the values obtained with a nontranscribed intergenic region (as described in Supplementary data).
Supplementary Material
Supplementary Material
Supplementary Tables
Acknowledgments
We thank A Aguilera, C Cole, A Corbett, E Izaurralde, A Jacquier, TH Jensen, D Libri, I Mattaj, L Minvielle-Sebastia, U Nehrbass and P Silver for strains or plasmids, M Collart, A Corbett, TH Jensen and C Dargemont for antibodies and TH Jensen and U Nehrbass for providing respectively SSA4 and LacZ modified oligonucleotide probes. We are grateful to T Laroche for support with confocal microscopy and to S Gasser, TH Jensen, D Libri and K Strub for reading the manuscript. This work was supported by grants (3130-048850, 3100-61378 and 3100A0-102235) from the Swiss National Science Foundation and from the Leenaards Foundation to FS.
References
- Bangs P, Burke B, Powers C, Craig R, Purohit A, Doxsey S (1998) Functional analysis of Tpr: identification of nuclear pore complex association and nuclear localization domains and a role in mRNA export. J Cell Biol 143: 1801–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Burkard KT, Butler JS (2000) A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol 20: 604–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA (2004) Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439 [DOI] [PubMed] [Google Scholar]
- Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A (2000) A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Hase ME, Muller L (1998) Molecular segments of protein Tpr that confer nuclear targeting and association with the nuclear pore complex. Exp Cell Res 245: 43–56 [DOI] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW (1997) Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol 136: 515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerbach F, Galy V, Trelles-Sticken E, Fromont-Racine M, Jacquier A, Gilson E, Olivo-Marin JC, Scherthan H, Nehrbass U (2002) Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat Cell Biol 4: 214–221 [DOI] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U (2004) Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U (2000) Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403: 108–112 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Gilbert W, Guthrie C (2004) The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell 13: 201–212 [DOI] [PubMed] [Google Scholar]
- Green DM, Johnson CP, Hagan H, Corbett AH (2003) The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc Natl Acad Sci USA 100: 1010–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, Wilson SM, Minvielle-Sebastia L, Swanson MS (2002) Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBOJ 21: 1800–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F, Dubrana K, Gasser SM (2002) Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J Struct Biol 140: 79–91 [DOI] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH (2001) Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK (2002) Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109: 551–562 [DOI] [PubMed] [Google Scholar]
- Jensen TH, Boulay J, Rosbash M, Libri D (2001) The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr Biol 11: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Jensen TH, Boulay J, Olesen JR, Colin J, Weyler M, Libri D (2004) Modulation of transcription affects mRNP quality. Mol Cell 16: 235–244 [DOI] [PubMed] [Google Scholar]
- Jimeno S, Rondon AG, Luna R, Aguilera A (2002) The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosova B, Pante N, Rollenhagen C, Podtelejnikov A, Mann M, Aebi U, Hurt E (2000) Mlp2p, a component of nuclear pore attached intranuclear filaments, associates with nic96p. J Biol Chem 275: 343–350 [DOI] [PubMed] [Google Scholar]
- Legrain P, Rosbash M (1989) Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57: 573–583 [DOI] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA (2001) Messenger RNAs are recruited for nuclear export during transcription. Genes Dev 15: 1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH (2002) Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol Cell Biol 22: 8254–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Elliott DJ, Stutz F, Rosbash M, Singer RH (1995) Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA 1: 1071–1078 [PMC free article] [PubMed] [Google Scholar]
- Marfatia KA, Crafton EB, Green DM, Corbett AH (2003) Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J Biol Chem 278: 6731–6740 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Perez-Ortin JE, Reed R, Hurt E (2004) Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86 [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, Rout MP (1999) Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol 144: 839–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Stutz F, Izaurralde E (2003) The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol 13: 319–327 [DOI] [PubMed] [Google Scholar]
- Thomsen R, Libri D, Boulay J, Rosbash M, Jensen TH (2003) Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9: 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D (2002) Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol Cell 9: 1285–1296 [DOI] [PubMed] [Google Scholar]
- Vinciguerra P, Stutz F (2004) mRNA export: an assembly line from genes to nuclear pores. Curr Opin Cell Biol 16: 285–292 [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Strahm Y, Stutz F (2001) The yeast hnRNP-like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol 21: 4219–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Wyss JC, Stutz F (2002) Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 22: 8241–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimowska G, Aris JP, Paddy MR (1997) A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci 110 (Part 8): 927–944 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Tables


