Abstract
In recent years, considerable progress has been made in unraveling the control mechanisms operating on the plant cell cycle and most of the key regulators have now been identified, including cyclin-dependent kinases (CDKs), cyclins, CDK-inhibitory proteins, the WEE kinase and proteins of the retinoblastoma-related protein (RBR)/E2F/DP pathway. The review discusses recent developments in our understanding of the plant cell cycle machinery and highlights the role of the cell cycle in plant development.
Keywords: Arabidopsis, endoreduplication, plant cell cycle, plant cell differentiation, plant development
Introduction
The cell cycle is one of the most comprehensively studied biological processes, particularly given its importance for growth and development and in many human disorders. Research on yeast, worms, flies, frogs, mammals, and recently also plants, has contributed to a kind of universal picture on how the basic cell cycle machinery is regulated. However, much of this picture is based on experiments performed on single cells. It is surprising to note that the role of the cell cycle machinery during development has received relatively little attention. To understand how, in different organisms, the basic cell cycle machinery integrates with development is an important scientific challenge for the coming years. Plants offer exceptional opportunities to significantly contribute to such a challenge.
In contrast to animals, plant development is largely post-embryonic. New organs, such as roots, stems, leaves and flowers, originate from life-long iterative cell divisions followed by cell growth and differentiation. Such cell divisions occur at specialized zones known as meristems. Leaves and flowers are formed at the shoot and floral meristems, respectively, whereas the root meristems continuously add new cells to the growing root. Plant development is also unique because any cell migration is prevented by the rigid cell walls surrounding plant cells. All these aspects make the analysis of the plant cell cycle of fundamental interest.
The plant cell cycle
Cyclin-dependent kinases
Cyclin-dependent kinases (CDKs) govern, as in other eukaryotic organisms, the plant cell cycle. All eukaryotic organisms studied to date possess at least one CDK with the PSTAIRE hallmark. In plants too, a bona fide PSTAIRE CDK, designated CDKA, plays a pivotal role in both the G1/S and G2/M transitions. In agreement with a dual role in the cell cycle, overexpression of a dominant-negative CDKA completely abolishes cell cycle progression and arrests cells in both G1 and G2 (Hemerly et al, 1995; Joubès et al, 2004). No orthologs of the mammalian G1/S-specific CDK4 and CDK6 genes have been found in plant genomes to date. However, besides CDKA, plants contain a second, so-called B-type, class of CDKs that seem to be unique to plants. The PSTAIRE hallmark present in CDKAs is replaced by either PPTALRE or PPTTLRE, reflecting the existence of two subgroups, CDKB1 and CDKB2 (Vandepoele et al, 2002). CDKBs accumulate at the G2- and M-phase and are essential for regulating the G2/M transition (Magyar et al, 1997; Porceddu et al, 2001). Overexpression of a dominant-negative CDKB interferes with cell cycle progression and causes a G2 arrest (Porceddu et al, 2001; Boudolf et al, 2004a).
Cyclins
Plants contain many more cyclins than previously described in other organisms (Vandepoele et al, 2002; Wang et al, 2004). For example, despite its small genome size, Arabidopsis thaliana contains at least 32 cyclins with a putative role in cell cycle progression. The plant cyclin nomenclature is based on the functional similarity with the mammalian counterparts. Arabidopsis gene annotation identified 10 A, 11 B, 10 D and one H cyclins (Vandepoele et al, 2002; Wang et al, 2004). In a broad sense, D cyclins are thought to regulate the G1/S transition and appear to act as integrators of various signals; A cyclins are of importance for the S-to-M phase control; B cyclins generally play a role in the G2/M transition and intra-M phase control; and the H cyclin is part of the CDK-activating kinase. Although the complexity of plant cyclins can be attributed partly to extensive duplications of the Arabidopsis genome (Simillion et al, 2002), the large number of cyclins may reflect the high developmental plasticity of sessile plants to respond to both intrinsic developmental signals and environmental cues. The different cyclins may have a wide range of expression patterns and might confer different substrate specificities. Although much of this is speculation, the expression of plant cyclins, more specifically D cyclins, has been shown to be modulated by plant growth factors, such as cytokinins, brassinosteroids, sucrose and gibberellins (Stals and Inzé, 2001). Figure 1 gives an overview of how developmental signals and environmental cues impinge on the cell cycle machinery. Some cyclins might act as key switches; for example, overexpression of CYCD3;1 is sufficient to compensate for the lack of cytokinins in the culture medium (Riou-Khamlichi et al, 1999). However, the precise role of cyclins in the cell cycle, their stability throughout the cell cycle and their subcellular location are poorly understood.
Figure 1.
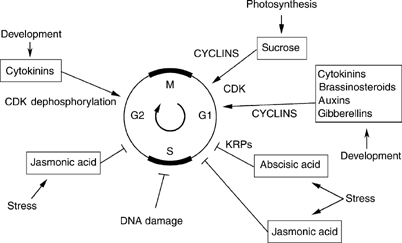
Schematic view of the various developmental and environmental signals that impinge on the plant cell cycle. For details, see text.
Proteolysis
Plant cyclins are, as in other organisms, also subject to extensive regulation by proteolysis. B1 cyclins, a subclass of B cyclins, contain a destruction box and are a substrate for a ubiquitin-dependent protein ligase complex that strongly resembles the anaphase-promoting complex or APC/C (Criqui et al, 2000). The functional significance of this cyclin destruction is best exemplified by the observation that constitutive overexpression of a nondegradable B1 cyclin, lacking the destruction box, causes severe growth retardation and abnormal development, with a higher percentage of cells exhibiting duplicated ploidy levels than the controls (Weingartner et al, 2004). On the other hand, proteolysis of B2 cyclins at prometaphase appears to be proteosome independent (Weingartner et al, 2003). Recently, CYCD3;1, but not CYCD2;1, has been shown to be a highly unstable protein whose proteolysis is mediated by a proteasome-dependent pathway (Planchais et al, 2004). Other cell cycle regulators, such as CDC6 (Castellano et al, 2001) and E2Fc (del Pozo et al, 2002), are destroyed via the ubiquitin/26S proteasome pathway as well. With the exception of the apparent involvement of SKP2 in SCF-targeted proteolysis of E2Fc (del Pozo et al, 2002), it is currently unknown which of the 694 F-box proteins of Arabidopsis are involved in the specific recognition of the cell cycle regulatory proteins.
CDK phosphorylation
Similarly to that in yeasts and animals, the activity of plant CDK/cyclin complexes is regulated by phosphorylation/dephosphorylation and the interaction with regulatory proteins. Yeast CDK/cyclin complexes are subject to an inhibitory phosphorylation of an N-terminal Tyr residue in the CDK partner, whereas in vertebrates CDKs are phosphorylated on both an N-terminal Tyr and Thr residue. This phosphorylation is catalyzed by the WEE1 kinase and is counteracted by the dual-specificity phosphatase CDC25. Plants possess a WEE kinase, which is involved in the inhibitory phosphorylation of CDKAs (Sun et al, 1999; Sorrell et al, 2002; Vandepoele et al, 2002; our unpublished results). Recently, the primitive unicellular algae, Ostreococcus tauri, has been found to contain a bona fide CDC25 (Khadaroo et al, 2004). However, in both Arabidopsis and rice, no genes with high homology to yeast or animal CDC25 genes have been identified (Vandepoele et al, 2002; our unpublished data). Nevertheless, both biochemical and genetic evidence suggest that higher plants too have a phosphatase that can activate CDK/cyclin complexes. Recently, a small protein with dual-specificity CDC25-like phosphatase has been identified in Arabidopsis. This CDC25-like protein consists of a sole catalytic domain and is able to stimulate Arabidopsis CDK activity (Landrieu et al, 2004). The in vivo role of this CDC25-like protein, however, remains to be determined.
CDK inhibitors
Plants also contain CDK/cyclin inhibitory proteins with a somewhat peculiar structure. All known plant CDK inhibitors share a 31-amino-acid domain with p27Kip1, a member of the mammalian Kip/Cip family of CDK inhibitors. This conserved domain is essential for the interaction between CDKs and cyclins and is located in the N-terminus of the Kip/Cip proteins and in the C-terminus in the plant inhibitors (Wang et al, 1997; De Veylder et al, 2001). Based on this similarity, plant CDK inhibitors were designated Kip-Related Proteins (KRPs, De Veylder et al, 2001). KRPs interact with both CDKAs (but not CDKBs) and with D cyclins (Wang et al, 1998; De Veylder et al, 2001). The expression of KRP genes is consistent with a role in regulating the cell cycle during development and in response to environmental signals. For example, auxins trigger the re-entry of quiescent root pericycle cells into the cell cycle and this event is preceded by a very specific downregulation of KRP2 gene expression in the auxin-responsive pericycle cells (Himanen et al, 2002). Furthermore, KRP1 was upregulated by abscisic acid, a plant hormone implicated in stress-induced cell cycle arrest (Wang et al, 1998). No plant homologs to the INK4 family of inhibitors have been found (Vandepoele et al, 2002).
The RB/E2F/DP pathway
It is astonishing to observe that, despite one billion years of evolution separating animals and plants, both types of organisms use the same RB/E2F/DP pathway to control the G1/S transition. Even the canonical DNA sequence (TTTCCCGC) recognized by the E2F transcription factors of animals and plants is identical (Ramirez-Parra and Gutierrez, 2000; de Jager et al, 2001). This identity argues in favor of the hypothesis that the RB/E2F/DP pathway had already evolved in primitive organisms, before the branching between animal and plant taxa. On the other hand, this well-studied pathway does not occur as such in yeasts and has long been thought to be specific for multicellular organisms. However, this idea has been challenged by the discovery of a retinoblastoma-related protein (RBR) in the unicellular green flagellate Chlamydomonas reinhardtii (Umen and Goodenough, 2001) and the recent identification in yeast of Whi5, an inhibitor of G1-specific transcription that is antagonized by Cln/CDK activity (Costanzo et al, 2004; de Bruin et al, 2004).
As their animal counterparts, all known plant RBR proteins contain two blocks of conserved sequences, which form the so-called A/B pocket domain. This pocket domain is the docking place for the E2F transcription factors. Plant RBR proteins interact with D cyclins through a conserved LXCXE motif at the N-terminus of the latter (Huntley et al, 1998). Recently, a loss-of-function of the Arabidopsis RBR gene has been shown to be gametophytically lethal (Ebel et al, 2004).
Arabidopsis has six E2Fs (E2Fa, E2Fb, E2Fc, E2Fd/DEL2, E2Fe/DEL1 and E2Ff/DEL3) and two DPs (Vandepoele et al, 2002). Two of the E2Fs (E2Fa and E2Fb) act as activators and E2Fc as repressor (del Pozo et al, 2002; Kosugi and Ohashi, 2002; Mariconti et al, 2002; Rossignol et al, 2002). Overexpression of E2Fa/DPa or E2Fb/DPa shortens cell cycle duration (unpublished data). E2Fa, E2Fb, E2Fc and DPs only contain one DNA-binding domain and, therefore, require dimerization to interact with the canonical E2F motif. On the other hand, E2Fd/DEL2, E2Fe/DEL1 and E2Ff/DEL3 proteins contain two DNA-binding domains, allowing them to bind as a monomer to the E2F site (Kosugi and Ohashi, 2002; Mariconti et al, 2002; Vandepoele et al, 2002). Additionally, these proteins do not contain a transactivation domain. Interestingly, only very recently a mammalian homolog, designated E2F7, has been discovered in mammals (reviewed by Bracken et al, 2004). This discovery highlights that cell cycle research in plants can yield novel insights into cell cycle research in animals. E2F proteins with two DNA-binding motifs act as competitors of bona fide E2F/DP proteins and, because they lack an activation domain, they repress E2F/DP-regulated genes (Mariconti et al, 2002; Bracken et al, 2004). Recently, E2Ff/DEL3 has been shown to play a possible role in repressing cell wall biosynthesis during cell elongation in differentiated cells (Ramirez-Parra et al, 2004) and E2Fe/DEL1 appears to control endoreduplication (Vlieghe et al, 2005).
Cell cycle and development
The role of the cell cycle machinery in plant development has been subject to a great deal of debate. Obviously, division is essential in generating the cells that constitute tissues and organs. However, the question whether cell division is the driver of growth and development (known as the cellular theory) or, alternatively, merely follows a developmental plan (known as the organismal theory) is more difficult to answer. Thanks to the ability to alter cell cycle parameters in transgenic plants, this long-standing question has been addressed. At least for leaves, development seems to follow the concept laid down in the organismal theory. In other words, the size and shape of leaves is, to some extent, predetermined and cell division merely serves to fill the predetermined space by cells. Experimental data support this viewpoint: transgenic plants constitutively overproducing any of the CDK inhibitors (KRPs) have smaller leaves that consist of 10-fold fewer cells whose average size is six-fold greater than that of control cells (Figure 2A–D). The reduction in cell number is thus compensated by an increased cell size (Wang et al, 2000; De Veylder et al, 2001; Jasinski et al, 2002). Similarly, overexpression of a dominant-negative CDKA or a nondegradable CYCB1;1 in tobacco retards the cell cycle and causes the formation of larger cells (Hemerly et al, 1995; Weingartner et al, 2004). On the other hand, constitutive overexpression of positive regulators of the cell cycle, such as E2Fa or CYCD3;1, results in more smaller cells (De Veylder et al, 2002; Dewitte et al, 2003; Figure 2E–H). Again, an increase in cell number is compensated by a decrease in cell size. Plants overexpressing E2Fa have larger cotyledons (leaf-like structures formed in the developing embryo) with three-fold more cells that are approximately twice as small as the control cells (De Veylder et al, 2002). Transgenic plants overexpressing CYCD3;1 show a massive increase in cell proliferation, which is, to some extent, balanced by a decrease in cell size (Dewitte et al, 2003). Also in animals, cell division and cell expansion can compensate each other to achieve an optimal species-specific organ size (Potter and Xu, 2001).
Figure 2.
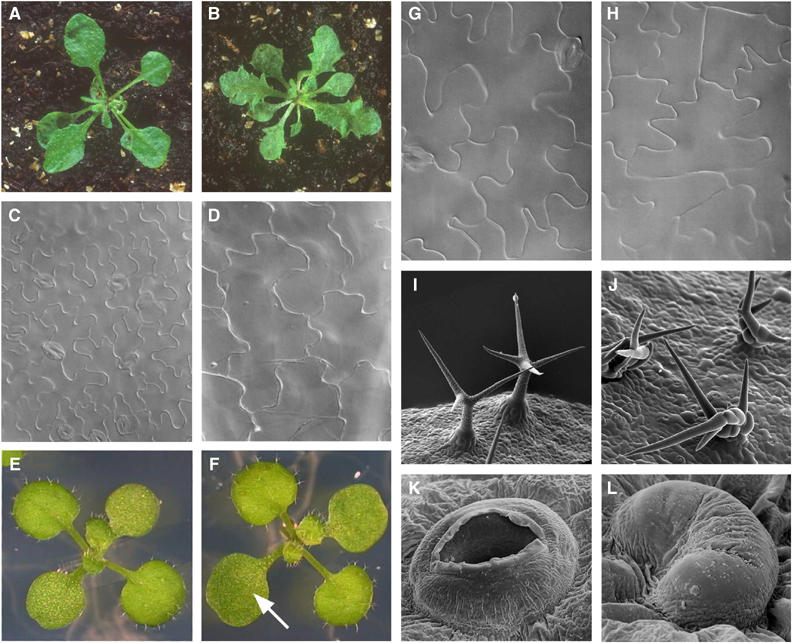
Examples of phenotypes caused by ectopic overexpression of cell cycle genes. (A, C, E, G, I, K) Wild-type plants and structures. (B, D) Plants constitutively overexpressing KRP2, a CDK inhibitor. Note the serrated leaves (B) composed of fewer, but much larger cells (D) (De Veylder et al, 2001; ©2001, American Society of Plant Biologists, reprinted with permission). (F, H) Overexpression of E2Fa leading to the formation of larger cotyledons (F, arrow), consisting of a larger number of cells with a smaller average size (H) (De Veylder et al, 2002). (J) Trichome-specific overexpression of CYCD3;1 causing the formation of multicellular hairs, whereas the wild-type hairs are unicellular (I) (Schnittger et al, 2002; © 2002, National Academy of Sciences, USA, reprinted with permission). (L) Constitutive overexpression of a dominant-negative CDKB1;1 interfering with the development of stomata. Whereas the wild-type stomata are built of two guard cells (K), the transgenic stomata often consist of only one kidney-shaped cell (L) (Boudolf et al, 2004a; © 2004, American Society of Plant Biologists, reprinted with permission).
Evidence also suggests that cell division acts independently of cell differentiation. Neither inhibition (by using, for instance, dominant-negative CDKs or KRPs) nor stimulation (by using overexpression of CYCB1;1, CYCD2;1, CYCD3;1 or EFa/DPa) severely affect cell differentiation (Hemerly et al, 1995, 2000; Doerner et al, 1996; Cockcroft et al, 2000; De Veylder et al, 2001, 2002). For example, trichome-specific expression of CYCD3;1 in Arabidopsis converts unicellular trichomes (leaf hairs) in multicellular hairs, without affecting differentiation (Figure 2I–J; Schnittger et al, 2002). Another example highlighting the independence of cell division from cell differentiation is found in transgenic Arabidopsis plants expressing a dominant-negative CDKB1;1. In these plants, many stomata, normally built of two guard cells, consist of only one kidney-shaped cell (Figure 2K–L; Boudolf et al, 2004a).
Endoreduplication
The normal cell cycle mode is characterized by a round of DNA replication (S phase) followed by mitosis and cytokinesis (M phase) and separated by two gap phases (G1 and G2). However, many plant and animal cells have a different cell cycle mode in which cells undergo iterative DNA replications without any subsequent mitosis and cytokinesis. This endoreduplication is frequently observed in some, but not all, plants and the level of ploidy varies between species and tissues (Sugimoto-Shirasu and Roberts, 2003). The physiological role of endoreduplication is subject to debate and several hypotheses have been proposed. For a long time, endoreduplication has been believed to be essential to allow cell growth and to maintain an optimal balance between cell volume and nuclear DNA. However, this concept is currently challenged. First, cell size and ploidy level are not correlated in root cells of different Arabidopsis ecotypes (Beemster et al, 2002). Secondly, recent experiments in which the level of endoreduplication is modified also do not support this hypothesis. For example, overproduction of KRPs causes a remarkable overall decrease in ploidy level but, at the same time, a large increase in cell size (De Veylder et al, 2001; Jasinski et al, 2002; Zhou et al, 2002). Alternatively, endoreduplication might buffer mutations that plants accumulate during their sessile life. Indeed, many plants are exposed during their life cycle to less favorable conditions and having 8, 16, 32, 64,… copies of the genome would safeguard that the genome retains functional copies. To our knowledge, no experimental data support this hypothesis, but available transgenic plants in which the ploidy level is changed might address this question experimentally. Furthermore, a hypothesis states that endoreduplication might enhance the metabolic capacity of plant cells. The endosperm cells of cereal seeds are highly polyploid and, possibly, the numerous gene copies allow these cells to synthesize very large amounts of storage products, such as starch. However, transgenic maize endosperm producing a dominant-negative form of CDKA had a lower level of endoreduplication, but only a slightly reduced starch and storage protein accumulation (Leiva-Neto et al, 2004).
How are cells triggered to undergo endoreduplication? In maize endosperm and tomato fruit development, the onset of endoreduplication coincides with the inhibition of a M-phase-specific CDK by a yet unknown factor (Sugimoto-Shirasu and Roberts, 2003). The best documented molecular mitosis-to-endocycle switch is the CCS52A protein, a plant ortholog of the yeast and animal CDH1 proteins that acts as a substrate-specific activator of the APC/C. Mitotic cyclins are likely candidate substrates of CCS52A-mediated proteolysis (Kondorosi and Kondorosi, 2004). The recent finding that CDKB1;1 also plays an important role in the decision between mitosis and endocycle makes it also a possible CCS52A substrate (Boudolf et al, 2004b).
Perspectives
Plants, particularly Arabidopsis, will help considerably in gaining a better understanding of the relationship between cell cycle and developmental signals as well as environmental cues. Nowadays, the generation of transgenic plants is very simple and can easily be done high throughput, thus allowing the testing of many genes for function, expression and localization of the corresponding proteins. Furthermore, plants have an astonishing developmental plasticity and, despite the dramatic effect of some cell cycle perturbations, seed set can often be obtained. All these factors call for a green light to further endorse research on the plant cell cycle.
Acknowledgments
I sincerely thank all past and current members of the plant growth and development group of the Department of Plant Systems Biology for their continuous efforts to unravel the plant cell cycle. I am particularly grateful to Lieven De Veylder, Ryan Whitford and Tom Beeckman for their suggestions to improve this review and to Martine De Cock, Karel Spruyt and Hilde Diegenant for helping preparing the manuscript.
References
- Beemster GTS, De Vusser K, De Tavernier E, De Bock K, Inzé D (2002) Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol 129: 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Barrôco R, de Almeida Engler J, Verkest A, Beeckman T, Naudt M, Inzé D, De Veylder L (2004a) B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GTS, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inzé D, De Veylder L (2004b) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K (2004) E2F target genes: unraveling the biology. Trends Biochem Sci 29: 409–417 [DOI] [PubMed] [Google Scholar]
- Castellano MM, del Pozo JC, Ramirez-Parra E, Brown S, Gutierrez C (2001) Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13: 2671–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft CE, den Boer BGW, Healy JMS, Murray JAH (2000) Cyclin D control of growth rate in plants. Nature 405: 575–579 [DOI] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M (2004) CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117: 899–913 [DOI] [PubMed] [Google Scholar]
- Criqui MC, Parmentier Y, Derevier A, Shen W-H, Dong A, Genschik P (2000) Cell cycle-dependent proteolysis and ectopic overexpression of cyclin B1 in tobacco BY2 cells. Plant J 24: 763–773 [DOI] [PubMed] [Google Scholar]
- de Bruin RAM, McDonald WH, Kalashnikova TI, Yates J III, Wittenberg C (2004) Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117: 887–898 [DOI] [PubMed] [Google Scholar]
- de Jager SM, Menges M, Bauer U-M, Murray JAH (2001) Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol Biol 47: 555–568 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, Inzé D (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa/DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCFAtSKP2 pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JMS, Jacqmard A, Kilby NJ, Murray JAH (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner P, Jørgensen J-E, You R, Steppuhn J, Lamb C (1996) Control of root growth and development by cyclin expression. Nature 380: 520–523 [DOI] [PubMed] [Google Scholar]
- Ebel C, Mariconti L, Gruissem W (2004) Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780 [DOI] [PubMed] [Google Scholar]
- Hemerly A, de Almeida Engler J, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P (1995) Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira PCG, Van Montagu M, Engler G, Inzé D (2000) Cell division events are essential for embryo patterning and morphogenesis: studies on dominant-negative cdc2aAt mutants of Arabidopsis. Plant J 23: 123–130 [DOI] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley R, Healy S, Freeman D, Lavender P, de Jager S, Greenwood J, Makker J, Walker E, Jackman M, Xie Q, Bannister AJ, Kouzarides T, Gutiérrez C, Doonan JH, Murray JAH (1998) The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol Biol 37: 155–169 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Riou-Khamlichi C, Roche O, Perennes C, Bergounioux C, Glab N (2002) The CDK inhibitor NtKIS1a is involved in plant development, endoreduplication and restores normal development of cyclin D3;1-overexpressing plants. J Cell Sci 115: 973–982 [DOI] [PubMed] [Google Scholar]
- Joubès J, De Schutter K, Verkest A, Inzé D, De Veylder L (2004) Conditional, recombinase-mediated, expression of genes in plant cell cultures. Plant J 37: 889–896 [DOI] [PubMed] [Google Scholar]
- Khadaroo B, Robbens S, Ferraz C, Derelle E, Eychenié S, Cooke R, Peaucellier G, Delseny M, Demaille J, Van de Peer Y, Picard A, Moreau H (2004) The first green lineage cdc25 dual-specificity phosphatase. Cell Cycle 3: 513–518 [PubMed] [Google Scholar]
- Kondorosi E, Kondorosi A (2004) Endoreduplication and activation of the anaphase-promoting complex during symbiotic cell development. FEBS Lett 567: 152–157 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) E2Ls, E2F-like repressors of Arabidopsis that bind to E2F sites in a monomeric form. J Biol Chem 277: 16553–16558 [DOI] [PubMed] [Google Scholar]
- Landrieu I, da Costa M, De Veylder L, Dewitte F, Vandepoele K, Hassan S, Wieruszeski J-M, Corellou F, Faure J-D, Van Montagu M, Inzé D, Lippens G (2004) A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 13380–13385, (Err. 101: 16391) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiva-Neto JT, Grafi G, Sabelli PA, Dante RA, Woo Y-M, Maddock S, Gordon-Kamm WJ, Larkins BA (2004) A dominant negative mutant of cyclin-dependent kinase A reduces endoreduplication but not cell size or gene expression in maize endosperm. Plant Cell 16: 1854–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Mészáros T, Miskolczi P, Deák M, Fehér A, Brown S, Kondorosi E, Athanasiadis A, Pongor S, Bilgin M, Bakó L, Koncz C, Dudits D (1997) Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9: 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti L, Pellegrini B, Cantoni R, Stevens R, Bergounioux C, Cella R, Albani D (2002) The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem 277: 9911–9919 [DOI] [PubMed] [Google Scholar]
- Planchais S, Samland AK, Murray JAH (2004) Differential stability of Arabidopsis D-type cyclins: CYCD3;1 is a highly unstable protein degraded by a proteasome-dependent mechanism. Plant J 38: 616–625 [DOI] [PubMed] [Google Scholar]
- Porceddu A, Stals H, Reichheld J-P, Segers G, De Veylder L, De Pinho Barrôco R, Casteels P, Van Montagu M, Inzé D, Mironov V (2001) A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J Biol Chem 276: 36354–36360 [DOI] [PubMed] [Google Scholar]
- Potter CJ, Xu T (2001) Mechanisms of size control. Curr Opin Genet Dev 11: 279–286 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Gutierrez C (2000) Characterization of wheat DP, a heterodimerization partner of the plant E2F transcription factor which stimulates E2F-DNA binding. FEBS Lett 486: 73–78 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, López-Matas MA, Fründt C, Gutierrez C (2004) Role of an atypical E2F transcription factor in the control of Arabidopsis cell growth and differentiation. Plant Cell 16: 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Rossignol P, Stevens R, Perennes C, Jasinski S, Cella R, Tremousaygue D, Bergounioux C (2002) AtE2F-a and AtDP-a, members of the E2F family of transcription factors, induce Arabidopsis leaf cells to re-enter S phase. Mol Genet Genomics 266: 995–1003 [DOI] [PubMed] [Google Scholar]
- Schnittger A, Schöbinger U, Stierhof Y-D, Hülskamp M (2002) Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr Biol 12: 415–420 [DOI] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu M, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell DA, Marchbank A, McMaho K, Dickinson JR, Rogers HJ, Francis D (2002) A WEE1 homologue from Arabidopsis thaliana. Planta 215: 518–522 [DOI] [PubMed] [Google Scholar]
- Stals H, Inzé D (2001) When plant cells decide to divide. Trends Plant Sci 6: 359–364 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K (2003) ‘Big it up': endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 1–10 [DOI] [PubMed] [Google Scholar]
- Sun Y, Dilkes BP, Zhang C, Dante RA, Carneiro NP, Lowe KS, Jung R, Gordon-Kamm WJ, Larkins BA (1999) Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc Natl Acad Sci USA 96: 4180–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW (2001) Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev 15: 1652–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GTS, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, Inzé D, De Veylder L (2005) The DP-E2F-like DEL1 gene controls the endocycle in Arabidopsis thaliana. Curr Biol 15: 59–63 [DOI] [PubMed] [Google Scholar]
- Wang G, Kong H, Sun Y, Zhang X, Zhang W, Altman N, dePamphilis CW, Ma H (2004) Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol 135: 1084–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Fowke LC, Crosby WL (1997) A plant cyclin-dependent kinase inhibitor gene. Nature 386: 451–452 [DOI] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15: 501–510 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24: 613–623 [DOI] [PubMed] [Google Scholar]
- Weingartner M, Criqui M-C, Mészáros T, Binarova P, Schmit A-C, Helfer A, Derevier A, Erhardt M, Bögre L, Genschik P (2004) Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of phragmoplast. Plant Cell 16: 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartner M, Pelayo HR, Binarova P, Zwerer K, Melikant B, de la Torre C, Heberle-Bors E, Bögre L (2003) A plant cyclin B2 is degraded early in mitosis and its ectopic expression shortens G2-phase and alleviates the DNA-damage checkpoint. J Cell Sci 116: 487–498 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Fowke LC, Wang H (2002) Plant CDK inhibitors: studies of interactions with cell cycle regulators in the yeast two-hybrid system and functional comparisons in transgenic Arabidopsis plants. Plant Cell Rep 20: 967–975 [Google Scholar]


