Abstract
Ceramide plays a crucial role as a basic building block of sphingolipids, but also as a signalling molecule mediating the fate of the cell. Although Lac1p and Lag1p have been shown recently to be involved in acyl-CoA-dependent ceramide synthesis, ceramide synthase is still poorly characterized. In this study, we expressed tagged versions of Lac1p and Lag1p and purified them to near homogeneity. They copurified with ceramide synthase activity, giving unequivocal evidence that they are subunits of the enzyme. In purified form, the acyl-CoA dependence, fatty acyl-CoA chain length specificity, and Fumonisin B1/Australifungin sensitivity of the ceramide synthase were the same as in cells, showing that these are properties of the enzyme and do not depend upon the membrane environment or other factors. SDS–PAGE analysis of purified ceramide synthase revealed the presence of a novel subunit of the enzyme, Lip1p. Lip1p is a single-span ER membrane protein that is required for ceramide synthesis in vivo and in vitro. The Lip1p regions required for ceramide synthesis are localized within the ER membrane or lumen.
Keywords: C26-fatty acyl-CoA; Fumonisin B1; LAC1, LAG1; sphingolipids; yeast
Introduction
Recently, sphingolipids and their precursors have attracted great attention because these compounds have been shown to play roles in cell signalling, heat stress response, calcium homeostasis, and have been implicated in the formation of specialized membrane microdomains. Sphingolipids also function in membrane trafficking, influencing the intracellular targeting of glycosylphosphatidylinositol-anchored proteins and regulating the internalization step of endocytosis (Dickson, 1998; Dickson and Lester, 2002; Merrill, 2002; Futerman and Hannun, 2004). Although our knowledge of sphingolipid metabolism, in particular in yeast, has increased greatly over the past years (Dickson and Lester, 1999, 2002; Funato et al, 2002; Figure 1), many important questions remain to be answered concerning the regulation of their synthesis and its response to environmental cues. To address these issues, it is necessary to characterize the sphingolipid biosynthetic machinery as well as the enzymes that modify them.
Figure 1.
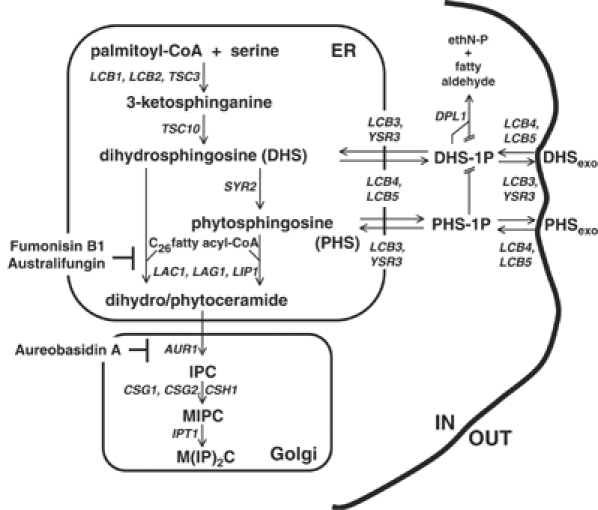
Sphingolipid biosynthetic pathway in yeast. DHS: dihydrosphingosine, PHS: phytosphingosine, IPC: inositolphosphorylceramide, MIPC: mannose inositolphosphorylceramide, M(IP)2C: mannose di(inositolphosphoryl)ceramide, ER, endoplasmic reticulum. Yeast genes required for the individual enzymatic steps are shown.
Among sphingolipid precursors, ceramide is a central molecule. Structurally critical for cell growth, ceramide also mediates different cellular events, such as apoptosis, growth arrest, endocytosis and stress response (Perry and Hannun, 1998; Hannun and Luberto, 2000; Hannun and Obeid, 2002; Acharya et al, 2003). Moreover, the balance between sphingoid bases (DHS/PHS/sphingosine), their phosphorylated counterparts and ceramide seems to control cell fate (Merrill, 2002; Spiegel and Milstien, 2002). The term ‘sphingolipid rheostat' has been used to describe this critical balance (Cuvillier et al, 1996; Mandala et al, 1998; Maceyka et al, 2002; Kobayashi and Nagiec, 2003). It is vital to our understanding of these signalling events to characterize the enzymes that convert sphingoid bases into ceramide.
Ceramide is synthesized mainly from the reaction of a fatty acyl-CoA with a sphingoid base by an acyl-CoA-dependent ceramide synthase (Morell and Radin, 1970; Akanuma and Kishimoto, 1979). In yeast, C26 fatty acyl-CoA is the major acyl chain donor (Smith and Lester, 1974; Lester and Dickson, 1993; Lester et al, 1993; Oh et al, 1997), while in animal cells various acyl chain lengths are used (Morell and Radin, 1970). However, the origin of the fatty acyl chain length specificity is unknown. In addition to acyl-CoA-dependent ceramide synthesis, YPC1 and YDC1 have been shown to encode for a minor acyl-CoA-independent ceramide synthase activity due to their reverse ceramidase action (Mao et al, 2000a, 2000b). CoA-dependent ceramide synthase activity is specifically inhibited by Fumonisin B1. Two genes, LAG1 and LAC1, have been shown to be required for ceramide synthase activity in Saccharomyces cerevisiae (Guillas et al, 2001; Schorling et al, 2001). Lag1p and Lac1p are homologous multispanning transmembrane proteins of the endoplasmic reticulum (ER). Homologs have been found in a wide variety of eukaryotes (Jiang et al, 1998; Brandwagt et al, 2000). Mutant lag1Δ lac1Δ cells have reduced sphingolipid levels due to a loss of the Fumonisin B1-sensitive and acyl-CoA-dependent ceramide synthase reaction. This activity was characterized using crude microsomal membranes (Guillas et al, 2001; Schorling et al, 2001).
Here, we have purified the ceramide synthase and show that it has the same properties as in membranes. These data provide direct and definitive evidence that Lag1p and Lac1p are subunits of the acyl-CoA-dependent ceramide synthase. Moreover, we have identified another essential component of the ceramide synthase, Lip1p, which forms a heteromeric complex with Lac1p and Lag1p. Lip1p is required for ceramide synthesis in vivo and in vitro. Like Lac1p and Lag1p, it is localized to the ER. It is also an integral membrane protein, with one transmembrane domain. Its short N-terminal part is cytoplasmic and not required for ceramide synthesis.
Results
Purified Lac1p and Lag1p show ceramide synthase activity
Lac1p and Lag1p are integral membrane proteins with six to eight predicted transmembrane domains. We tagged Lac1p and Lag1p at their N-termini to preserve their ER retention signals. The resulting fusion proteins are functional, because they restored growth and sphingolipid synthesis to the lac1Δlag1Δ strain (data not shown). We used different epitope-tagged versions of the proteins: Flag, c-myc, double GST-Flag or HA epitopes. We found the same results whichever epitope was used. We show the data obtained with the Flag-tagged proteins, unless specified otherwise.
We assayed different detergents (25 mM CHAPS, 1% decylmaltoside, 1% digitonin, 1% dodecylmaltoside, 1% octylglycoside, 1% Triton X-100) and 2 M urea to assess their solubilization ability towards Lac1p and Lag1p. Decylmaltoside, digitonin, dodecylmaltoside and Triton X-100 showed the best solubilization efficiency (data not shown). Triton X-100 and digitonin were chosen to test ceramide synthase activity of the proteins purified by Flag pulldown of microsomes solubilized with either 1% Triton X-100 or 1% digitonin. Digitonin was the more suitable detergent, because it preserved the ceramide synthase activity of the purified proteins in contrast to Triton X-100 (Figure 2A). Therefore, the enzymatic activity of ceramide synthase was characterized with proteins purified in the presence of digitonin.
Figure 2.
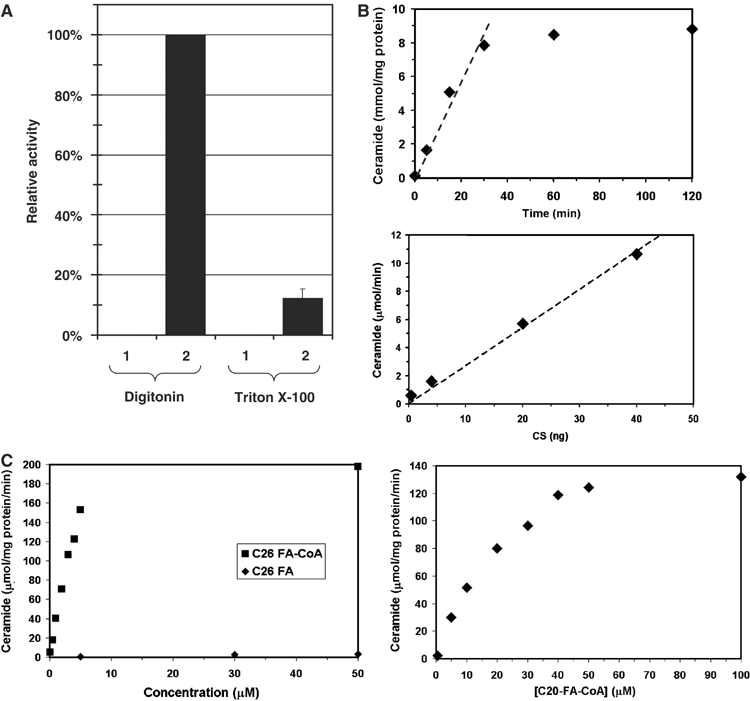
Enzymatic characterization of purified ceramide synthase. Microsomal membranes were solubilized with either digitonin or Triton X-100, and Flag-Lac1p-containing protein complexes were purified by immunoisolation and eluted from the beads. Ceramide synthase activity was measured. (A) Lanes 1, 2: membranes were solubilized with 1% digitonin, lanes 3, 4: membranes were solubilized with 1% Triton X-100. Lane (1) corresponds to experiments with strain RH 5665 and lane (2) to strain RH 5666. (B) Time course (upper panel) and protein concentration dependence (lower panel) of ceramide synthase activity. (C) Fatty acyl-CoA concentration and CoA dependence of ceramide synthase activity. C26 fatty acid substrates are shown in the left panel and C20 fatty acyl-CoA in the right panel.
Microsomal membranes were solubilized by incubation with the nonionic detergent digitonin. After high-speed centrifugation, the supernatant, corresponding to solubilized membranes, was recovered and incubated with anti-Flag antibody-coupled beads. Proteins bound to the beads were then eluted with a buffer containing the Flag peptide. The immunoisolation step was very effective for purification of ceramide synthase, because it resulted in about a 3500-fold enrichment of the specific activity (Table I). The purification procedure was efficient because we could obtain 68% yield of ceramide synthase activity from the detergent-treated membranes.
Table 1.
Purification of the ceramide synthase
| Fraction | Protein |
Ceramide synthase activity |
Yield |
Enrichment |
|---|---|---|---|---|
| mg/ml | nmol/mg of protein/min | Percentage | Fold | |
| Digitonin-treated membranesa | 5.6 | 6.1 | 100 | 1 |
| Solubilized membranesb | 4.6 | 6.0 | 80 | 0.98 |
| Elution of anti-Flag M2 affinity beads | 5.4 × 10−3c | 20 678 | 68 | 3420 |
| Ceramide synthase was purified from membranes as described in Materials and methods. Data are the average of five sets of experiments. Each time similar results were found. | ||||
| aMembranes resuspended in the presence of 1% digitonin prior to incubation. | ||||
| bSupernatant after centrifugation (100 000 g for 1 h) of the digitonin-treated membranes. | ||||
| cProtein concentrations were estimated by densitometric comparison of silver-stained proteins of the fraction with stained calibration bands of bovine serum albumin of known concentrations in SDS–PAGE. |
Characterization of ceramide synthase activity
A mixture of [4,5-3H]D-erythro-dihydrosphingosine ([3H]DHS)/DHS was dried under a stream of nitrogen and the residue was resuspended in BSA solution to prevent spontaneous reaction of the activated fatty acyl-CoA with DHS. Membrane extracts or eluates containing purified Lag1p and Lac1p were added to this mixture, followed by C26 fatty acyl-CoA to initiate the ceramide synthase reaction.
The time course of ceramide formation by purified Lac1p and Lag1p was virtually linear for the first 30 min, and then reached a plateau (Figure 2B, upper panel). The activity was proportional to the amount of enzyme to at least 50 ng (Figure 2B, lower panel), was acyl-CoA-dependent (Figure 2C, left panel) and showed strong specificity for very-long-chain fatty acyl-CoA with an apparent Km of ∼3 μM for C26 FA-CoA, and ∼30 μM for C20 FA-CoA (Figure 2C). This is in agreement with previous data showing that the major ceramide species in yeast carries a C26 fatty acid moiety (Smith and Lester, 1974; Lester and Dickson, 1993; Oh et al, 1997) and that microsomal fractions had higher activity towards long-chain rather than short-chain acyl-CoA (Guillas et al, 2003). Purified ceramide synthase was highly sensitive to Fumonisin B1 (Ki≈400 μM) and Australifungin (Ki≈15 μM) (data not shown), potent inhibitors of ceramide synthase activity (Wang et al, 1991; Mandala et al, 1995; Wu et al, 1995).
Characterization of the ceramide synthase complex
Protein patterns of the purified Flag-Lac1p were analyzed by SDS–PAGE. The elution fraction displayed four bands visible by silver staining, two major bands at Mr 48 000 and 28 000, and two minor bands at Mr ∼70 000 (A) and Mr ∼40 000 (B) (Figure 3A, lane 2, digitonin). Protein patterns of purified Flag-Lag1p were similar (data not shown). Band (A) was nonspecific as it also appeared in isolations performed on cells without Flag-tagged proteins (Figure 3A, lane 1, digitonin and Triton X-100). Band (B) was identified by mass spectrometry as a degradation product of Lac1p. The major band at Mr 48 000 corresponded to Lac1p. The other major band at Mr 28 000 was co-immunoprecipitated specifically with Lac1p upon digitonin solubilization, but not when we used Triton X-100 as a detergent (Figure 3A, lane 2, digitonin and Triton X-100). The loss of this specific partner coincided with the loss of ceramide synthase activity (Figures 2A and 3A). Quantitation of protein bands after silver staining allowed us to detect at least 10 ng of a standard protein, BSA. No other bands than the ones seen in the ceramide synthase preparation (Figure 3A) could be detected while visualizing as much as 170 ng of Lac1p and 200 ng of Lip1p, suggesting that, apart from the shown contaminant (Band A), the enzyme is greater than 95% pure. To ascertain whether proteins escaped detection by silver staining, we used Sypro Ruby as an alternative, as well as purification from 35S-labelled cells, visualizing the radioactivity by a phosphorimager. No other bands than the ones observed by silver staining appeared (data not shown).
Figure 3.
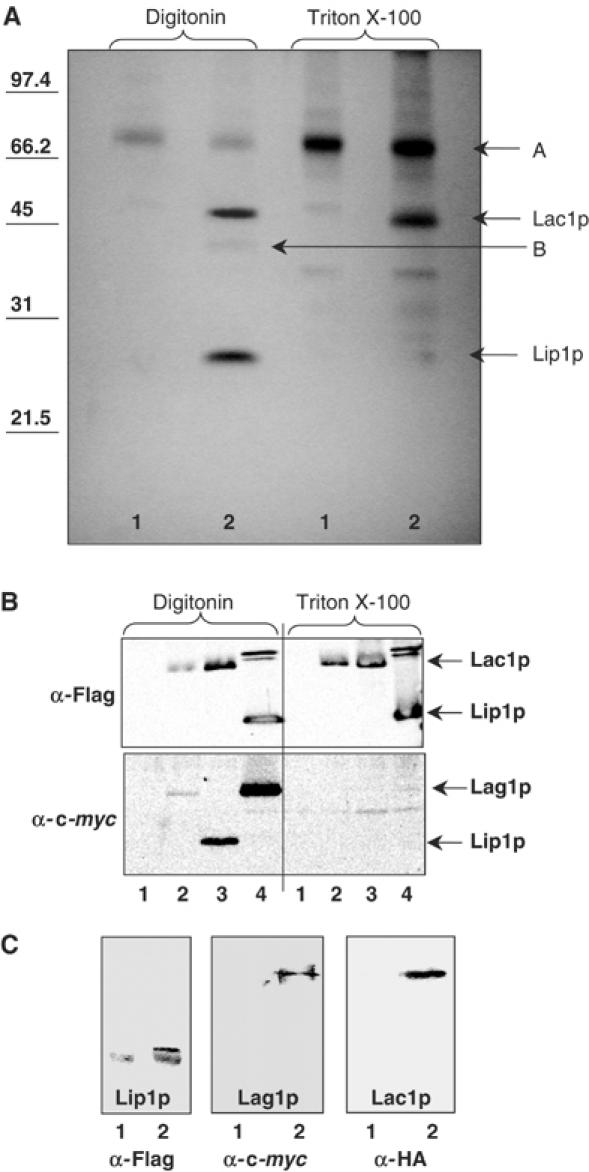
Ceramide synthase complex. (A) Eluates after immunoisolation from a strain RH5665 (1) or RH5668 (2) were separated by SDS–PAGE. Membranes were solubilized with 1% digitonin (first two lanes) or with 1% Triton X-100 (last two lanes). (B) Strains co-expressing Flag-tagged and c-myc tagged proteins. Immunoprecipitation against the Flag-tagged protein was performed and the eluate was submitted to Western blotting against Flag epitope (upper panel) and against c-myc epitope (lower panel). (1) Assay performed with strain RH5665, (2) strain RH5666, (3) strain RH6047, (4) strain RH6067. Membranes were solubilized with 1% digitonin (first four lanes) or with 1% Triton X-100 (last four lanes). (C) Flag immunoprecipitation of digitonin-solubilized membranes of a strain expressing HA-Lac1p, c-myc-Lag1p and Flag-Lip1p (RH6344, lane 2) was performed. Western blotting against Flag, c-myc and HA epitopes. Lane (1) corresponds to the control experiment, performed with strain RH6345.
The new partner (Mr 28 000) was termed Lip1p, standing for Lag1p/Lac1p interacting protein. Lip1p was identified by mass spectrometry as Ymr298wp. The function of this protein had not previously been identified, although its interaction with Lac1p and Lag1p had already been suggested from systematic two-hybrid studies (Ito et al, 2001). The predicted molecular mass of Lip1p is 17 kDa, but its sequence contains many prolines, and the protein has a predicted acidic pI, which could explain the higher apparent molecular mass (∼28 kDa). Furthermore, it is possible that Lip1p is modified by O-linked glycosylation, but this has not been tested. One transmembrane domain is predicted near to the amino-terminal end of the protein by different programs (HMMTOP, SOSUI, TMHMM, TMpred and TopPred2). No strong homologs in animals or plants were found using sequence alignments (Clustal W protein alignment), but were found in other fungi (Figure 4).
Figure 4.
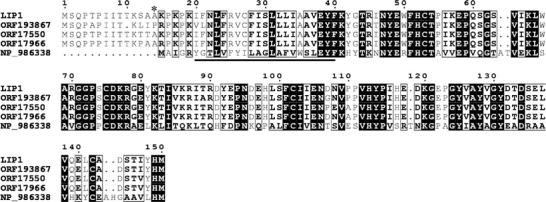
Multiple alignment of LIP1 with fungal homologs. The alignment includes the deduced protein sequence of S. cerevisiae LIP1 (YMR298w), S. bayanus predicated ORF19386, S. mikatae predicated ORF17550, S. paradoxus predicated ORF17966 and Asbia gossypii predicated NP_986338. The alignment was generated using the T-coffee (Notredame et al, 2000) and ESPript (Gouet et al, 1999) programs. Black boxes indicate identical residues, black borders show similar amino acids. The predicted transmembrane domain is underlined. The beginning of the truncated Lip1p (ΔNT-Lip1p) is labelled by a star.
In order to provide additional evidence for Lac1p/Lag1p/Lip1p interaction, we used the pESC plasmid co-expressing two proteins, one Flag tagged and the other c-myc tagged, and carried out co-immunoprecipitation experiments. Immunoprecipitations were performed against the Flag-tagged versions of the different proteins, and beads were eluted with Flag peptide. Immunoprecipitation efficiency was checked by Western blotting of the eluates against Flag epitope (Figure 3B, upper panel). Eluates were then tested for c-myc-tagged partners by Western blotting (Figure 3B, lower panel). We could confirm that Lag1p interacts with Lac1p and Lip1p, that Lac1p interacts with Lag1p and Lip1, that Lip1p interacts with Lac1p and Lag1p (Figure 3B), and that Lip1p interacts with itself (data not shown) when membranes were solubilized by digitonin. These interactions were lost if membranes were solubilized by Triton X-100 (Figure 3B). Using a similar approach, we confirmed the interaction between the three partners, Lac1p, Lag1p and Lip1p, by Flag immunoprecipitation, and anti-c-myc and anti-HA Western blotting in a strain expressing Flag-Lip1p, c-myc-Lag1p and HA-Lac1p (Figure 3C).
Next, we determined the apparent molecular mass of ceramide synthase in a strain expressing Flag-Lac1p and Lip1p, but not Lag1p, by glycerol gradient centrifugation. Eluates from a Flag pulldown were separated by centrifugation on glycerol gradients containing 1% digitonin, fractions were collected and analyzed by Western blotting. Flag-Lac1p was found in fractions 4–8, peaking in fraction 6 (Figure 5A). We analyzed the fractions by silver staining (Figure 5B). The protein pattern was similar to the one observed in Figure 3A (lane 2 digitonin). The nonspecific band (A) did not cofractionate exactly with the ceramide synthase, whereas band (B) (degradation product of Lac1p) and Lip1p exhibited the same distribution as Lac1p. The glycerol gradient fractions were also tested for ceramide synthase activity, which cofractionated with the Lac1p peak (data not shown). Similar results were obtained with strains expressing Lag1p and Lac1p, or either of the two individually always giving a complex of apparent mass of approximately 250–260 kDa. The complex is large enough to accommodate two of the homologous large subunits and two Lip1p included in a digitonin micelle of 73–86 kDa (aggregation number 60–70, Calbiochem catalog).
Figure 5.
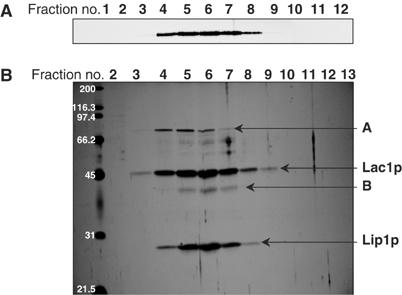
Glycerol gradient centrifugation of the ceramide synthase complex. Eluate from Flag immunoisolation of digitonin-solubilized membranes of the strain RH5668 was separated by glycerol gradient centrifugation and fractions were collected from the top of the tube. (A) Western blotting against the Flag tag of Lac1p. (B) Silver staining.
Lip1p is an integral membrane protein, localized to the ER
Analysis of the sequence of Lip1p predicted one transmembrane domain (Figure 4). To investigate the nature of its membrane association, we treated cell lysates with different reagents able to extract proteins from the membranes, and then centrifuged the samples at 100 000 g for 1 h. The presence of proteins in the supernatant (S) and pellet (P) was determined by Western blotting. The Lip1p pattern was similar to the one of Lac1p, and Wbp1p (an integral ER protein taken as a control) (Figure 6A). Lip1p was found in the membrane pellet (Figure 6A, last two columns). When the lysates were treated with either 1% Triton X-100 or 1% SDS, Lip1p was partially released into the soluble fraction. When lysates were treated with either 1 M NaCl or 0.1 M Na2CO3 (pH 11.5), Lip1p remained in the pellet fraction. These results show that Lip1p, like Lac1p, behaves like an integral membrane protein.
Figure 6.
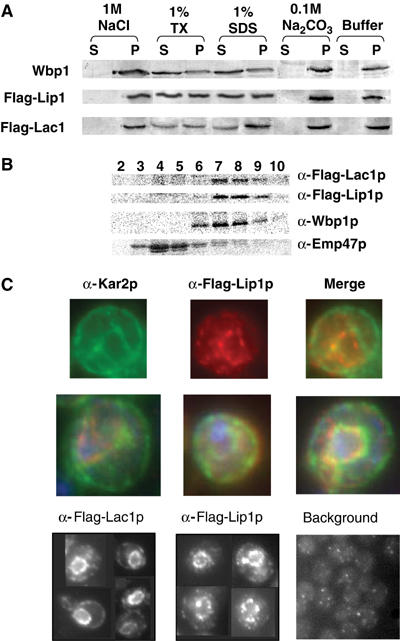
Lip1p is an integral ER membrane protein. The strains used were RH 5666 and RH 6068. (A) Cell lysates were treated with different chemical reagents as indicated, centrifuged, and pellet (P) and soluble (S) fractions were analyzed by Western blotting. (B) Cell homogenates were separated on sucrose density gradients. Fractions were collected from the top of the tube and analyzed by Western blotting. Wbp1p is a marker of the ER, Emp47p of the Golgi apparatus. (C) Colocalization of Kar2p and Lip1p by indirect immunofluorescence (upper panels), additional merged images with DAPI (middle panels). Immunofluorescence of Flag-Lac1p and Flag-Lip1p (lower panels).
Lac1p and Lag1p and ceramide synthase activity have been shown to be localized to the ER (Barz and Walter, 1999; Funato and Riezman, 2001). We verified that Lip1p is also localized to the ER by subcellular fractionation using sucrose gradient centrifugation followed by Western blotting. Lip1p and Lac1p both appeared in fractions 7–9, which were enriched in the ER marker Wbp1p, whereas Emp47p, a Golgi marker, was present in fractions 3–6 (Figure 6B). We further verified the ER localization of the different proteins by indirect immunofluorescence microscopy against their Flag tag. Flag-Lip1p colocalized with Kar2p, an ER marker (Figure 6C, upper panel). Flag-Lip1p and Flag-Lac1p had a characteristic ER staining, that is, a ring around the nucleus. However, when compared to the Kar2p pattern, the staining was more perinuclear and less in the peripheral ER. Such a perinuclear staining was also observed for Lcb1p, a protein involved in the first step of sphingolipid metabolism (Yasuda et al, 2003). Some dots were also present in the cytoplasm, but they were not experimentally significant as they could also be found in the cells carrying an empty plasmid and visualized under the same conditions (Figure 6C, lower panel). Lip1p staining in the perinuclear ER seemed to be concentrated in specific areas of the nuclear envelope/ER that are not enriched for Kar2p, but this could be an effect of its overproduction. Additional evidence will be required to determine if the ceramide synthase is found in specialized ER microdomains.
Lip1p is required for ceramide synthase activity
Our experiments described above suggest that Lip1p is required for ceramide synthase activity because its loss from the complex coincides with a loss of activity (Figures 2A and 3). To definitively address if Lip1p is essential for the ceramide synthase activity, we created lip1Δ mutant cells. In a systematic high-throughput study, deletion of LIP1 was found to be lethal (Winzeler et al, 1999; Uetz et al, 2000). However, as the lag1Δlac1Δ double mutant grows extremely slowly, we suspected that the same could be true for lip1Δ mutant, and that this slow growth had been missed. We disrupted one of the alleles of LIP1 gene in a diploid strain, sporulated the heterozygous diploids and dissected tetrads. After short growth periods of 2–3 days, there was a 2/0 segregation pattern, indicating that the two lip1Δ strains were either inviable or grew very slowly (Figure 7A, left panel). However, after more than 1 week, we obtained lip1Δ mutant colonies (Figure 7A, middle panel) whose growth was very slow even after colonies were restreaked onto rich medium plate (Figure 7A, right panel).
Figure 7.
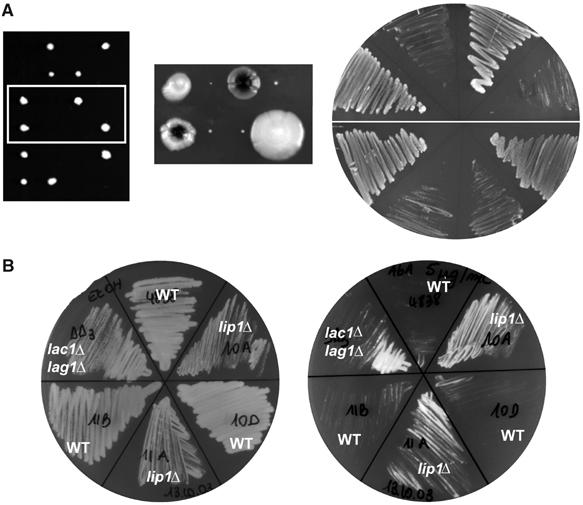
Growth of the lip1Δ strain. (A) Tetrads were dissected from a heterozygous lip1Δ diploid. Left panel: tetrads after 3 days. Middle panel: two tetrads after 10 days. Right panel: growth on rich medium after re-streaking these two tetrads. (B) Aureobasidin A (final concentration ≈5 μg/ml, right) or ethanol (left) was spread onto YPD agar plates. These strains were streaked on the plates and incubated at 24°C. RH4838, RH5308, RH6013, RH6016, RH5994, RH5995.
Previously, we developed a quick and easy test to assess if a strain is likely to be deficient in ceramide synthesis (Schorling et al, 2001). Cells are streaked on a plate containing aureobasidin A, an inhibitor of inositolphosphorylceramide (IPC) synthesis, known to be highly toxic (Endo et al, 1997; Nagiec et al, 1997). If cells are deficient in ceramide synthesis, they are resistant to aureobasidin A, as they cannot accumulate ceramide. Wild-type (WT) cells were sensitive to aureobasidin A, but lip1Δ strains grew on plates containing aureobasidin A, as did the double lac1Δlag1Δ mutant, suggesting strongly that Lip1p is involved in ceramide synthesis (Figure 7B).
Next, we measured ceramide synthase activity by performing in vivo labellings on the lip1Δ strain. Strains were labelled with [3H]-myo inositol and their lipids were analyzed by TLC. A lip1Δ strain had a pattern similar to the one of lac1Δlag1Δ strain (Figure 8A). The complex sphingolipids were absent (see quantification; Figure 8A, lower panel) and lyso-PI accumulated. Moreover, an extra product (*) was observed as for lac1Δlag1Δ strain. This spot has been found and identified in extracts from the lac1Δlag1Δ strain as a PI with a very-long-chain fatty acid (Guillas et al, 2001). We also labelled cells with [3H]DHS and analyzed ceramide production by TLC. Mutant lip1Δ cells were defective in synthesis of ceramide from DHS (Figure 8B). These results show that LIP1 is required for ceramide synthesis in vivo. However, in vivo labellings would still be consistent with an indirect role of Lip1p in ceramide synthesis, including preparation or delivery of one of the substrates to the enzyme or biogenesis of the complex. To address this point, we assayed ceramide synthesis using Flag–Lac1p complexes purified from a lip1Δ strain. Complexes isolated from this strain were not able to synthesize ceramide (Figure 8C), even though Lac1p and Lag1p were expressed, could interact together and were purified by immunoisolation (data not shown).
Figure 8.
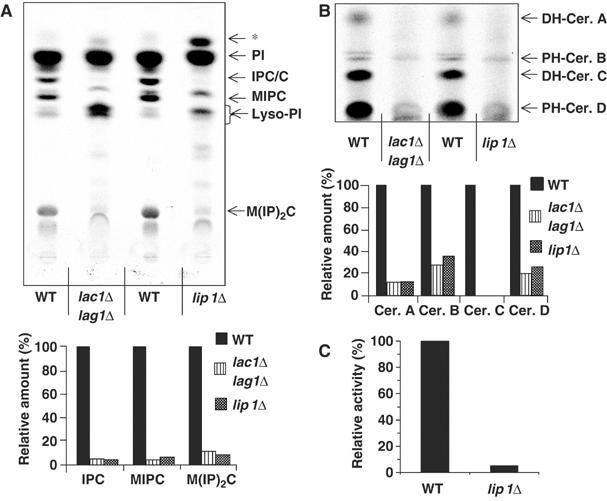
Labellings of Lip1Δ mutant. (A) Cells (strains RH4838, RH5308 and RH 6013) were labelled with [3H]myo inositol, lipids were analyzed on TLC and detected using a phosphorimager. Radioactivity in sphingolipids was quantified, and the relative amount of each species was determined as a percentage of the amount in WT cells. PI, phosphatidylinositol; IPC/C: inositolphosphorylceramide C; MIPC: mannose inositolphosphorylceramide; M(IP)2C: mannose di(inositolphosphoryl)ceramide; * indicates a spot that migrates identically to that previously identified as a C26 fatty acid containing PI. (B) The same cells were labelled with [3H]DHS and lipids were analyzed as above. Radioactivity in sphingolipids was quantified as in (A). (C) Ceramide synthesis activity of eluates from a Flag immunoisolation of digitonin-solubilized membranes from strains RH5666 and RH6075.
The N-terminal part of Lip1p is cytoplasmic and not required for sphingolipid synthesis
Lip1p is an integral membrane protein (Figure 6A) predicted to have one transmembrane domain close to its N-terminus. Due to the distribution of negative and positive charges surrounding the transmembrane domain (Goder et al, 2004), we predicted that the N-terminus would be cytoplasmic (Figure 4). To assess Lip1p topology, we performed a protease protection assay. Lip1p as well as Lac1p were tagged with a Flag epitope at their N-terminus, and Lip1p was also tagged with a Flag epitope at its C-terminus. Microsomal membranes were incubated with or without Triton X-100 in the presence or absence of proteinase K. Kar2p, an ER lumenal protein, was used to check the integrity of the ER membrane. In the absence of Triton X-100, the N-termini of Lip1p and Lac1p were sensitive to proteinase K, whereas the C-terminus of Lip1p was preserved (Figure 9A). Kar2p was unaffected, showing that the ER had remained intact. In the presence of Triton X-100, both Lip1p-Flag and Kar2p were degraded (Figure 9A). These results show that the N-terminus of Lip1p is cytoplasmic and its C-terminus is lumenal.
Figure 9.
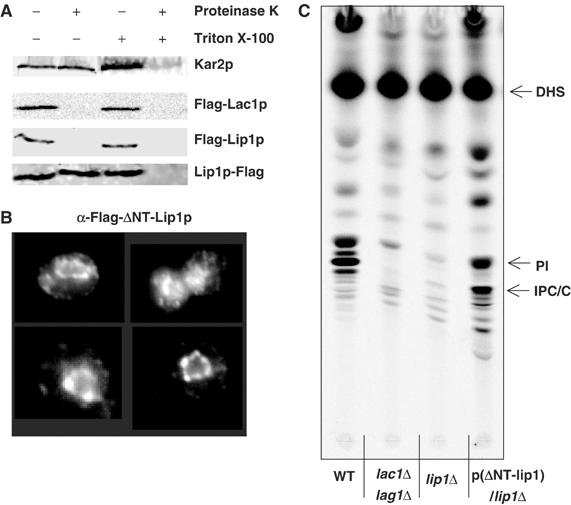
Lip1p topology. (A) Microsomal membranes from strains RH6068, RH5666 or RH6348 were incubated with or without Triton X-100 in the presence or absence of Proteinase K. The samples were analyzed by Western blotting to reveal the Flag tag and Kar2p. (B) Flag-Lip1p-ΔNT (RH6069) was visualized by indirect immunofluorescence. (C) Cells (strains RH4838, RH5309, RH6013 and RH6069) were labelled with [3H]DHS and analyzed as in Figure 8.
Next, we deleted the first 14 amino acids of Lip1p, comprising most of its cytoplasmic domain. A few charged amino acids were kept prior to the transmembrane domain to preserve its integration into the membrane (Figure 4). The localization of the truncated protein by indirect immunofluoresence microscopy was the same as the tagged WT protein (Figure 9B), showing that the N-terminal part of Lip1p is not required for its correct localization. Next, we checked sphingolipid composition of the Lip1p-ΔNT strain by [3H]DHS labelling in vivo. The lipid pattern was very similar to that of a WT strain (Figure 9C). Therefore, the cytoplasmic N-terminal part of Lip1p is not necessary for ceramide synthesis. These results suggest that the functional domain of Lip1p is most likely located in the lumen of the ER or in its transmembrane domain.
Discussion
In recent years, sphingolipids have attracted the attention of many researchers and striking advances in the field have been made. While most of the enzymes involved in this biosynthetic pathway have been identified genetically, only serine palmitoyltransferase from Chinese hamster ovary cells and from Sphingomonas paucimobilis (Hanada et al, 2000; Ikushiro et al, 2004) and yeast 3-ketosphinganine reductase expressed in Escherichia coli (Beeler et al, 1998) have been purified. All the other enzymes of this pathway, in particular the integral membrane enzymes, have not yet been purified and fully characterized, most likely due to technical difficulties. As most of the molecules of this pathway are not only biosynthetic intermediates, but also signalling molecules, characterization of these enzymes is crucial for our understanding of cell physiology.
Using tagged versions of Lac1p and Lag1p, we purified an active ceramide synthase complex to near homogeneity with more than a 3400-fold increase in specific activity starting from a detergent-treated microsomal fraction (Table I). The fold purification from whole cells is much greater, but very difficult to determine exactly, because of the difficulty in measuring the enzyme activity quantitatively in crude extracts. The activity of the purified ceramide synthase was similar to the ceramide synthesis activity detected in intact cells or isolated microsomal membranes (Morell and Radin, 1970; Akanuma and Kishimoto, 1979; Wu et al, 1995; Guillas et al, 2003; Kobayashi and Nagiec, 2003). Purified yeast ceramide synthase activity was CoA dependent, preferred very-long-chain fatty acyl-CoA, which is consistent with the acyl-CoA specificity of the activity characterized in cells or membranes (Guillas et al, 2003; Kobayashi and Nagiec, 2003) and shows that this specificity does not depend upon the membrane environment. This is consistent with the fact the human homologs of Lag1p expressed in yeast show ceramide synthase activity with slightly different preferences for fatty acid length (Guillas et al, 2003).
After SDS–PAGE and silver-staining analysis, the active purified ceramide synthase containing Lac1p and Lag1p displayed an additional protein, Lip1p, which was shown to be essential for ceramide synthase activity in vivo and in vitro (Figures 2A, 3A, and 8). Lip1p can be found in complexes with both Lac1p and Lag1p, and with itself (Figure 3B and C). These results show that the ceramide synthase consists of Lac1p, and/or Lag1p, and Lip1p. Lip1p, corresponds to Ymr298wp, a protein of previously unknown function. LIP1 has numerous homologs among fungi, but no obvious homologs were found in mammals (Figure 4). However, as the open reading frame is quite short (450 bp), it may not have been properly annotated, in particular if a mammalian ortholog contains several introns. Since human homologs of LAC1 and LAG1 have been recently isolated and can complement the lac1 and lag1 mutations (Venkataraman et al, 2002; Guillas et al, 2003; Riebeling et al, 2003), it would be surprising if no functional homologs of Lip1p were found.
To definitively determine if Lip1p is required for ceramide synthase activity, we created a deletion mutant. The lip1Δ strain behaved almost identically to the lag1Δlac1Δ deletion strain. Lip1p is necessary for ceramide synthase activity, but its precise role is not known. Several hypotheses can be raised: Lip1p could be necessary for stability of the ceramide synthase or for an upstream step in ceramide synthesis. These are both unlikely because Lac1p and Lag1p were copurified from a strain lacking Lip1p (data not shown), but the complex was inactive (Figure 8). Lip1p could be a regulatory subunit of the ceramide synthase complex or be required for the enzymatic activity of the ceramide synthase complex. If the latter were the case, this would suggest that the ceramide synthase reaction requires a component in the ER lumen or within the membrane.
To get the first idea about the subunit stoichiometry of purified ceramide synthase complex, we estimated its molecular mass by glycerol gradient centrifugation. The ceramide synthase complex that we have purified could be mixtures of Lag1p–Lac1p heteromers with at least two Lip1p or Lag1p or Lac1p homodimers with at least two Lip1p. The different complexes could have different enzymatic characteristics, such as slightly different specificities for chain length or hydroxylation rate of the fatty acyl-CoAs or for the sphingoid bases (C16 versus C18). It has been suggested that LAG1 proteins function in substrate recognition (Guillas et al, 2003) and several mammalian homologs of Lag1p exist, making them ideal candidates to explain the wide variety of fatty acid chains that are incorporated into ceramide in animal cells.
Like Lac1p and Lag1p, Lip1p is localized to the ER where the synthesis of ceramide occurs (Funato and Riezman, 2001). Ceramide synthase has been proposed to have its active site on the cytoplasmic surface of the ER membrane (Hirschberg et al, 1993) because its activity is protease sensitive in isolated microsomal membranes. Now that we have shown that Lag1p, Lac1p and Lip1p comprise the ceramide synthase, these results must be reinterpreted. Lag1p and Lac1p span the ER membrane multiple times and their susceptibility to protease digestion cannot be taken as an indication of the topology of the active site. To synthesize ceramide from exogenous sphingoid bases, these have to be phosphorylated by Lcb4p and dephosphorylated by Lcb3p, sphingoid base kinase and phosphatase, respectively (Funato et al, 2003). Lcb4p is a peripheral membrane protein mainly found on the ER membrane (Funato et al, 2003) and Lcb3p is an integral ER membrane protein (Mao et al, 1999) whose active site has been shown to be in the lumen of the ER (Kihara et al, 2003). Therefore, after the phosphorylation/dephosphorylation cycle, the sphingoid bases should be found in the lumen of the ER. On the other hand, the 3-ketosphinganine reductase that generates dihydroshingosine seems to have its active site localized to the cytoplasmic face of the ER (Kihara and Igarashi, 2004), suggesting that one of the substrates, dihydrosphingosine, can be made available from either side of the ER membrane. Lip1p is an integral membrane protein with one predicted transmembrane domain. We found that its short, highly charged N-terminus is cytoplasmic and dispensible for ceramide synthase activity. These data suggest that the functional region of Lip1p is localized in the lumen of the ER, or in its transmembrane domain. Lac1p and Lag1p initially attracted interest because of significant sequence similarity with TRAM (translocating chain associating membrane protein), a mammalian protein thought to be involved in protein translocation across the ER membrane (Barz and Walter, 1999). Even if they do not share functional homologies, these proteins could share common structural features. Based on TRAM characteristics, we speculate that ceramide synthase complex would form a ‘channel' across the ER membrane and this may permit the enzyme active site to accept dihydrosphingosine from either side of the membrane. Predictions (Jazwinski and Conzelmann, 2002) and our preliminary evidence (N Yahara and H Riezman) suggest that the conserved Lag domain, presumably containing the active site, is indeed mostly embedded in the membrane. Given the multiple spanning membrane structure of ceramide synthase, only a high-resolution structure is likely to resolve these important topology questions.
Materials and methods
Yeast strains, media and reagents
The strains and plasmids used in this study are listed in Tables II and III. To construct deletion strains, entire open reading frames were deleted and replaced with the designated genes. Deletions were confirmed by PCR.
Table 2.
Plasmids used in this study
| Plasmid number | Plasmids | Inserts | Source |
|---|---|---|---|
| 372 | pEG(KT) | None | Deschenes |
| 1118 | pESC-URA3 | None | Dieter Oesterhelt |
| 1119 | pSTS30a | LAG1 (N-term. c-myc tagged) | Dieter Oesterhelt |
| 1120 | pSTS30 | LAG1 and LAC1 (N-term. c-myc and Flag tagged, resp.) | Dieter Oesterhelt |
| 1121 | pSTS30b | LAC1 (N-term. Flag tagged) | Dieter Oesterhelt |
| 1285 | pWB94 | LAC1 (N-term. HA tagged) | This study |
| 1479 | pEG(KT)-(GST-Flag-Lac1p) | LAC1 (N-term. GST and Flag tagged) | Barz and Walter (1999) |
| 1548 | pSTS30c | LAG1 (N-term. Flag tagged) | This study |
| 1551 | pESC(Flag-Lip1p) | LIP1 (N-term. Flag tagged) | This study |
| 1552 | pESC(Flag-ΔNT-Lip1p) | LIP1 (N-term. Flag tagged) without the first 14 amino acids | This study |
| 1559 | pESC(c-mycLag1p/Flag-Lip1p) | LAG1 and LIP1 (N-term. c-myc and Flag tagged, resp.) | This study |
| 1560 | pESC(c-mycLip1p/Flag-Lac1p) | LIP1 and LAC1 (N-term. c-myc and Flag tagged, resp.) | This study |
| 1700 | pESC(Lip1p-Flag) | LIP1 (C-term. Flag tagged) | This study |
| 1701 | pESC(c-mycLip1p/Flag-Lip1p) | LIP1 and LIP1 (N-term. c-myc and Flag tagged) | This study |
Table 3.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| RH4838 | Mat a his3 leu2 ura3 trp1 bar1 AUR1∷3HA∷HIS3 | Lab strain |
| RH5308 | Mat a his3 leu2 ura3 trp1 bar1 AUR1∷3HA∷HIS3 lac1∷LEU2 lag1∷TRP1 | Schorling et al (2001) |
| RH5660 | Strain RH5308 transformed with the plasmid no. 1285 (HA-LAC1) | This study |
| RH5664 | Strain RH5308 transformed with the plasmid no. 1479 (GST-Flag-LAC1) | This study |
| RH5665 | Strain RH4838 transformed with the plasmid no. 1118 (Vector control) | This study |
| RH5666 | Strain RH5308 transformed with the plasmid no. 1120 (c-myc-LAG1, Flag-LAC1) | This study |
| RH5668 | Strain RH5308 transformed with the plasmid no. 1121 (Flag-LAC1) | This study |
| RH5994 | Mat alpha leu2 ura3 trp1 bar1 lip1∷HIS3 | This study |
| RH5995 | Mat a leu2 ura3 trp1 his3 lys2 ade2 bar1 | This study |
| RH6013 | Mat alpha leu2 ura3 trp1 bar1 lip1∷HIS3 | This study |
| RH6016 | Mat alpha leu2 ura3 trp1 his3 lys2 bar1 | This study |
| RH6044 | Strain RH5308 transformed with the plasmid no. 1548 (Flag-LAG1) | This study |
| RH6046 | Strain RH5308 transformed with the plasmid no. 1559 (c-myc-LAG1, Flag-LIP1) | This study |
| RH6047 | Strain RH5308 transformed with the plasmid no. 1560 (c-myc-LIP1, Flag-LAC1) | This study |
| RH6067 | Strain RH6013 transformed with the plasmid no. 1559 (c-myc-LAG1, Flag-LIP1) | This study |
| RH6068 | Strain RH6013 transformed with the plasmid no. 1551 (Flag-LIP1) | This study |
| RH6069 | Strain RH6013 transformed with the plasmid no. 1552 (Flag-LIP1-ΔNT) | This study |
| RH6075 | Strain RH6013 transformed with the plasmid no. 1120 (c-myc-LAG1, Flag-LAC1) | This study |
| RH6344 | Strain RH2881 transformed with the plasmids no. 1285 and no. 1559 (HA-LAC1, c-myc-LAG1, Flag-LIP1) | This study |
| RH6345 | Strain RH2881 transformed with the plasmids no. 1285 and no. 1118 (HA-LAC1, vector control) | This study |
| RH6348 | Strain RH 6013 transformed with the plasmid no. 1700 (LIP1-Flag) | This study |
| RH6349 | Strain RH 6013 transformed with the plasmid no. 1701 (c-myc-LIP1, Flag-LIP1) | This study |
Yeast strains were grown in rich medium (20 g/l glucose, 20 g/l peptone and 10 g/l yeast extract with adenine, uracil and tryptophan at 40 mg/l) at 24°C.
Protein concentration was determined using the detergent compatible Bio-Rad protein assay kit. [3H]DHS was from Anawa Trading SA or American Radiolabeled Chemical Inc. (ARC, St Louis, MO). DHS, Fumonisin B1, C20:0 fatty acyl-CoA, C26:0 fatty acid, Flag beads, Flag antibody and Flag peptide were from Sigma. HA rat antibody was purchased from Roche. Digitonin was from Acros, and high-purity digitonin from Calbiochem. Australifungin was a gift from Merck.
Purification of Lac1p, Lag1p and Lip1p
Cells were grown in rich medium, washed twice and resuspended at 200 OD/ml in TNE buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM PMSF, 1 μg/ml protease inhibitor mixture). Cells were disrupted with glass beads, and the cell debris and glass beads were removed by centrifugation at 1000 g for 10 min at 4°C. The supernatant was then centrifuged at 100 000 g for 1 h at 4°C. The pellet corresponding to microsomal membranes was resuspended in TNE, the appropriate volume of a 10 × stock solution of detergent (Triton X-100 or digitonin (Acros)) was then added to a final concentration of detergent of 1%. Microsomal membranes were solubilized for 1 h at 4°C with a rotating movement. The unsolubilized membranes were removed by centrifugation at 100 000 g for 1 h at 4°C. Immunoprecipitation using the HA epitope was performed using rat antibody and protein G beads for 2–4 h at 4°C with a rocking movement. The immunoprecipitated beads were then washed three times with TNE/1% digitonin or Triton X-100, and eluted with sample buffer. Immunoprecipitation using the Flag epitope was performed using Flag beads at 4°C for 2–4 h with rocking. The immunoprecipitated beads were then washed three times with TNE/1% digitonin or Triton X-100, and eluted with a 2 mg/ml solution of Flag peptide in TNE/1% digitonin or Triton X-100 on ice for 20–30 min. In the case of double-tagged protein (GST-Flag), the eluate from the Flag beads was further incubated with glutathione beads at 4°C for 3–5 h with rocking. The beads were washed three times with TNE/1% digitonin or Triton X-100, and eluted with a solution of 20 mM reduced glutathione in TNE (pH 9.4)/1% digitonin or Triton X-100 on ice for 30 min.
In vitro ceramide synthesis assay
The enzyme was purified as described above, but the beads were washed three times with TNE and twice with B88 (20 mM HEPES, pH 6.8, 150 mM KAc, 5 mM MgAc, 250 mM sorbitol). In case of HA-tagged proteins, the assay was performed directly on the beads. For the Flag-tagged version, the immunoprecipitated beads were eluted with 2 mg/ml of Flag peptide in B88 and detergent.
For the assay, 5 μl of a mixture of [3H]DHS/DHS (1/4, 20 μM) in ethanol was dried under a stream of nitrogen, the residue was resuspended in 5 μl of a 40 μM solution of BSA. Eluates or beads in B88 were added to this mixture, followed by C26 fatty acyl-CoA. The final volume of the reaction was 100 μl, with a 2 μM final concentration of BSA, 1 μM final concentration for [3H]DHS/DHS and 0.5 μM for C26 fatty acyl-CoA, unless specified otherwise. The samples were incubated at 24°C for 30 min, and the reaction was stopped by adding 600 μl of chloroform/methanol (1/1, v/v). The lipids were extracted by n-butanol/water partitioning as previously described (Schorling et al, 2001), and analyzed by TLC using the solvent system I, chloroform/acetic acid (9/1, v/v). The TLC plates were visualized and quantified by using tritium-sensitive screens and a Cyclone phosphorimager (Packard, Meriden, CT). The radioactivity extracted from enzyme-negative controls was regarded as the background level and subtracted. On each TLC, a known amount of [3H]DHS was loaded, and used as a standard.
Glycerol gradient
In all, 200 μl of proteins were loaded onto a glycerol gradient. A 4 ml-step gradient from 15% (v/v) to 40% (v/v) glycerol in TNE+1% digitonin (Calbiochem) or Triton X-100 was prepared and centrifuged at 200 000 g in a SW60 rotor (Beckman) for 6 h at 4°C. Fractions of 250 μl were collected from the top. The sample buffer was added, and fractions were analyzed either by Western blotting or by silver staining (according to either Mann or Blum protocols (Blum et al, 1987; Shevchenko et al, 1996)).
In vivo lipid labelling
In vivo lipid labellings were performed as previously described (Zanolari et al, 2000). Sphingolipids were extracted by chloroform/methanol (1/1) in presence of glass beads and then by butanol/water. For ceramide analysis, lipids were extracted first by ethanol/diethylether/pyridine/4.2 N ammonium hydroxide (15/5/1/0.018), and then by butanol/water partitioning. Lipids were analyzed on TLC using solvent system II, chloroform/methanol/0.25% KCl (55/45/10) for sphingolipid labelled with [3H]myo-inositol, solvent system III, chloroform/methanol/4.2 N ammonium hydroxide (9/7/2) for sphingolipid labelled with [3H]DHS, and solvent system IV, chloroform/methanol/acetic acid (190/9/1) for ceramide.
Protein extraction and subcellular fractionation
To characterize the nature of membrane association of proteins, cells were grown in rich medium, washed and resuspended in TNE buffer. Cells were disrupted with glass beads, and the cell debris and glass beads were removed by centrifugation at 720 g for 5 min at 4°C. One volume of TNE buffer containing 2 M NaCl, 2% Triton X-100, 2% SDS, or 200 mM Na2CO3 (pH 11.5) was added to the supernatants. The mixture was incubated on ice for 30 min and centrifuged at 100 000 g for 1 h at 4°C. The resulting pellet and supernatant fractions were subjected to SDS–PAGE and then analyzed by Western blotting using a rabbit polyclonal antibody against Wbp1p, or a mouse mAb M2 against Flag. Subcellular fractionation was performed as described (Funato et al, 2003). Samples were subjected to SDS–PAGE, followed by Western blotting using a mouse Ab against Flag epitope, a rabbit polyclonal Ab against Wbp1p or Emp47p.
Indirect immunofluorescence microscopy
Indirect immunofluorescence on whole fixed yeast cells was performed as described (Beck et al, 1999). A mouse Ab against Flag followed by an indocarbocyanine, Cy-3-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) and a rabbit polyclonal Ab against Kar2 (kindly provided by R Schekman) followed by a fluorescein isothiocyanate donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories) were used. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma) at a concentration of 1 μg/ml. Cells were visualized with a Axiophot 2 microscope (× 100).
Protease protection assay
Spheroplasts prepared as described above were lysed by Dounce homogenization in B88. The 3000 g supernatant was centrifuged at 10 000 g and then at 100 000 g for 1 h. The pellet was washed with B88 and centrifuged at 100 000 g twice more. Finally, the microsome pellet was resuspended at a concentration of 4000 OD/ml. The protein concentration was determined using BioRad protein assay kit. In all, 80 μg of proteins were incubated with or without 0.1% of Triton X-100 in the presence or absence of proteinase K (final concentration, 30 μg/ml) on ice for 30 min. The reaction was stopped by addition of PMSF (final concentration, 2 mM). The samples were subjected to SDS–PAGE, followed by Western blotting using a mouse Ab against Flag epitope, a rabbit polyclonal Ab against Kar2p.
Supplementary Material
Supplementary Material
Acknowledgments
We thank Dieter Oesterhelt, Jeanette Holenstein, Véronique Koerin and Brigitte Bernadets for reagents and help, as well as the Riezman laboratory, in particular Kouichi Funato and Reika Watanabe, for stimulating discussions and suggestions. Many thanks to Hélène Bénédetti for her patience and support. This work was funded by a FEBS fellowship (BV) and a grant from the Office Fédérale de la Santé (EC network grant HPRN-CT-2000-00077; HR).
References
- Acharya U, Patel S, Koundakjian E, Nagashima K, Han X, Acharya JK (2003) Modulating sphingolipid biosynthetic pathway rescues photoreceptor degeneration. Science 299: 1740–1743 [DOI] [PubMed] [Google Scholar]
- Akanuma H, Kishimoto Y (1979) Synthesis of ceramides and cerebrosides containing both alpha-hydroxy and nonhydroxy fatty acids from lignoceroyl-CoA by rat brain microsomes. J Biol Chem 254: 1050–1060 [PubMed] [Google Scholar]
- Barz WP, Walter P (1999) Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins. Mol Biol Cell 10: 1043–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T, Schmidt A, Hall MN (1999) Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol 146: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler T, Bacikova D, Gable K, Hopkins L, Johnson C, Slife H, Dunn T (1998) The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Delta mutant. J Biol Chem 273: 30688–30694 [DOI] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross H (1987) Improved silver staining of plant proteins, RNA, and DNA in polyacrylamide gels. Electrophoresis 8: 93–99 [Google Scholar]
- Brandwagt BF, Mesbah LA, Takken FL, Laurent PL, Kneppers TJ, Hille J, Nijkamp HJ (2000) A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc Natl Acad Sci USA 97: 4961–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind S, Spiegel S (1996) Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 381: 800–803 [DOI] [PubMed] [Google Scholar]
- Dickson RC (1998) Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu Rev Biochem 67: 27–48 [DOI] [PubMed] [Google Scholar]
- Dickson RC, Lester RL (1999) Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1438: 305–321 [DOI] [PubMed] [Google Scholar]
- Dickson RC, Lester RL (2002) Sphingolipid functions in Saccharomyces cerevisiae. Biochim Biophys Acta 1583: 13–25 [DOI] [PubMed] [Google Scholar]
- Endo M, Takesako K, Kato I, Yamaguchi H (1997) Fungicidal action of aureobasidin A, a cyclic depsipeptide antifungal antibiotic, against Saccharomyces cerevisiae. Antimicrob Agents Chemother 41: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K, Lombardi R, Vallee B, Riezman H (2003) Lcb4p is a key regulator of ceramide synthesis from exogenous long chain sphingoid base in Saccharomyces cerevisiae. J Biol Chem 278: 7325–7334 [DOI] [PubMed] [Google Scholar]
- Funato K, Riezman H (2001) Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J Cell Biol 155: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K, Vallee B, Riezman H (2002) Biosynthesis and trafficking of sphingolipids in the yeast Saccharomyces cerevisiae. Biochemistry 41: 15105–15114 [DOI] [PubMed] [Google Scholar]
- Futerman AH, Hannun YA (2004) The complex life of simple sphingolipids. EMBO Rep 5: 777–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V, Junne T, Spiess M (2004) Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol Biol Cell 15: 1470–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308 [DOI] [PubMed] [Google Scholar]
- Guillas I, Jiang JC, Vionnet C, Roubaty C, Uldry D, Chuard R, Wang J, Jazwinski SM, Conzelmann A (2003) Human homologues of LAG1 reconstitute Acyl-CoA-dependent ceramide synthesis in yeast. J Biol Chem 278: 37083–37091 [DOI] [PubMed] [Google Scholar]
- Guillas I, Kirchman PA, Chuard R, Pfefferli M, Jiang JC, Jazwinski SM, Conzelmann A (2001) C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J 20: 2655–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Hara T, Nishijima M (2000) Purification of the serine palmitoyltransferase complex responsible for sphingoid base synthesis by using affinity peptide chromatography techniques. J Biol Chem 275: 8409–8415 [DOI] [PubMed] [Google Scholar]
- Hannun YA, Luberto C (2000) Ceramide in the eukaryotic stress response. Trends Cell Biol 10: 73–80 [DOI] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM (2002) The ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J Biol Chem 277: 25847–25850 [DOI] [PubMed] [Google Scholar]
- Hirschberg K, Rodger J, Futerman AH (1993) The long-chain sphingoid base of sphingolipids is acylated at the cytosolic surface of the endoplasmic reticulum in rat liver. Biochem J 290: 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushiro H, Hayashi H, Kagamiyama H (2004) Reactions of serine palmitoyltransferase with serine and molecular mechanisms of the actions of serine derivatives as inhibitors. Biochemistry 43: 1082–1092 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM, Conzelmann A (2002) LAG1 puts the focus on ceramide signaling. Int J Biochem Cell Biol 34: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Jiang JC, Kirchman PA, Zagulski M, Hunt J, Jazwinski SM (1998) Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res 8: 1259–1272 [DOI] [PubMed] [Google Scholar]
- Kihara A, Igarashi Y (2004) FVT-1 is a mammalian 3-ketodihydrosphingosine reductase with an active site that faces the cytosolic side of the endoplasmic reticulum membrane. J Biol Chem 279: 49243–49250 [DOI] [PubMed] [Google Scholar]
- Kihara A, Sano T, Iwaki S, Igarashi Y (2003) Transmembrane topology of sphingoid long-chain base-1-phosphate phosphatase, Lcb3p. Genes Cells 8: 525–535 [DOI] [PubMed] [Google Scholar]
- Kobayashi SD, Nagiec MM (2003) Ceramide/long-chain base phosphate rheostat in Saccharomyces cerevisiae: regulation of ceramide synthesis by Elo3p and Cka2p. Eukaryot Cell 2: 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester RL, Dickson RC (1993) Sphingolipids with inositolphosphate-containing head groups. Adv Lipid Res 26: 253–274 [PubMed] [Google Scholar]
- Lester RL, Wells GB, Oxford G, Dickson RC (1993) Mutant strains of Saccharomyces cerevisiae lacking sphingolipids synthesize novel inositol glycerophospholipids that mimic sphingolipid structures. J Biol Chem 268: 845–856 [PubMed] [Google Scholar]
- Maceyka M, Payne SG, Milstien S, Spiegel S (2002) Sphingosine kinase, sphingosine-1-phosphate, and apoptosis. Biochim Biophys Acta 1585: 193–201 [DOI] [PubMed] [Google Scholar]
- Mandala SM, Thornton RA, Frommer BR, Curotto JE, Rozdilsky W, Kurtz MB, Giacobbe RA, Bills GF, Cabello MA, Martin I, Pelaez F, Harris GH (1995) The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J Antibiot (Tokyo) 48: 349–356 [DOI] [PubMed] [Google Scholar]
- Mandala SM, Thornton R, Tu Z, Kurtz MB, Nickels J, Broach J, Menzeleev R, Spiegel S (1998) Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc Natl Acad Sci USA 95: 150–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Saba JD, Obeid LM (1999) The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem J 3: 667–675 [PMC free article] [PubMed] [Google Scholar]
- Mao C, Xu R, Bielawska A, Obeid LM (2000a) Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J Biol Chem 275: 6876–6884 [DOI] [PubMed] [Google Scholar]
- Mao C, Xu R, Bielawska A, Szulc ZM, Obeid LM (2000b) Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J Biol Chem 275: 31369–31378 [DOI] [PubMed] [Google Scholar]
- Merrill AH Jr (2002) De novo sphingolipid biosynthesis: a necessary, but dangerous, pathway. J Biol Chem 277: 25843–25846 [DOI] [PubMed] [Google Scholar]
- Morell P, Radin NS (1970) Specificity in ceramide biosynthesis from long chain bases and various fatty acyl coenzyme A′s by brain microsomes. J Biol Chem 245: 342–350 [PubMed] [Google Scholar]
- Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, Lester RL, Dickson RC (1997) Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem 272: 9809–9817 [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins D, Heringa J (2000) T-coffee: a novel method for multiple sequence alignments. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Oh CS, Toke DA, Mandala S, Martin CE (1997) ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem 272: 17376–17384 [DOI] [PubMed] [Google Scholar]
- Perry DK, Hannun YA (1998) The role of ceramide in cell signaling. Biochim Biophys Acta 1436: 233–243 [DOI] [PubMed] [Google Scholar]
- Riebeling C, Allegood JC, Wang E, Merrill AH Jr, Futerman AH (2003) Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J Biol Chem 278: 43452–43459 [DOI] [PubMed] [Google Scholar]
- Schorling S, Vallee B, Barz WP, Riezman H, Oesterhelt D (2001) Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisae. Mol Biol Cell 12: 3417–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Smith SW, Lester RL (1974) Inositol phosphorylceramide, a novel substance and the chief member of a major group of yeast sphingolipids containing a single inositol phosphate. J Biol Chem 249: 3395–3405 [PubMed] [Google Scholar]
- Spiegel S, Milstien S (2002) Sphingosine 1-phosphate, a key cell signaling molecule. J Biol Chem 277: 25851–25854 [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Venkataraman K, Riebeling C, Bodennec J, Riezman H, Allegood JC, Sullards MC, Merrill AH Jr, Futerman AH (2002) Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J Biol Chem 277: 35642–35649 [DOI] [PubMed] [Google Scholar]
- Wang E, Norred WP, Bacon CW, Riley RT, Merrill AH Jr (1991) Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J Biol Chem 266: 14486–14490 [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Vlockaert G, Ward TR, Wysocki R, Yen GS, Yu KX, Zimmermann K, Philippsen P, Johnston M, Davis RW (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Wu WI, McDonough VM, Nickels JT Jr, Ko J, Fischl AS, Vales TR, Merrill AH Jr, Carman GM (1995) Regulation of lipid biosynthesis in Saccharomyces cerevisiae by fumonisin B1. J Biol Chem 270: 13171–13178 [DOI] [PubMed] [Google Scholar]
- Yasuda S, Nishijima M, Hanada K (2003) Localization, topology, and function of the LCB1 subunit of serine palmitoyltransferase in mammalian cells. J Biol Chem 278: 4176–4183, (Epub 2002 December 4102) [DOI] [PubMed] [Google Scholar]
- Zanolari B, Friant S, Funato K, Sutterlin C, Stevenson BJ, Riezman H (2000) Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J 19: 2824–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


