Abstract
INTRODUCTION:
The emergence of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli has become an important challenge among pediatric patients with community-acquired urinary tract infection (UTI).
OBJECTIVES:
The aim of this study was to assess the antimicrobial susceptibility patterns, associated risk factors and to survey the frequency of bla cefotaximase (CTX-M), bla temoneira (TEM), and bla sulfhydryl variable (SHV) genotypes in ESBL-producing E. coli isolated from children with community-acquired UTI.
METHODS:
This was a prospective study conducted from November 2012 to March 2016 in a tertiary care center. E. coli isolated in urine cultures from children aged ≤18 years was identified and confirmed for ESBL production. ESBL-positive strains were screened for ESBL encoding genes. Chi-square test and Fisher's exact test were used to compare the difference in antibiotic susceptibility with respect to ESBL positive and negative, and binary logistic regression was used to identify the risk factors associated with ESBL production.
RESULTS:
Among 523 E. coli isolates, 196 (37.5%) were ESBL positive, >90% were resistant to cephalosporins, and 56% were resistant to fluoroquinolones. Least resistance was observed for imipenem, netilmicin, and nitrofurantoin (2%, 8.6%, 15.3%). Association between ESBL production and drug resistance was significant for ceftazidime (P < 0.001), cefixime (P < 0.001), cefotaxime (P = 0.010), ceftazidime-clavulanic acid (P < 0.001), levofloxacin (P = 0.037), and gentamicin (P = 0.047) compared to non-ESBL E. coli. CTX-M gene was the most prevalent (87.5%), followed by TEM (68.4%) and SHV (3.1%). Previous history of UTI and intake of antibiotics were the common risk factors.
CONCLUSION:
ESBL-producing E. coli from community-acquired pediatric UTI carries more than one type of beta-lactamase coding genes correlating their increased antibiotic resistance. Aggressive infection control policy, routine screening for detecting ESBL isolates in clinical samples, and antimicrobial stewardship are the keys to prevent their dissemination in community settings.
Keywords: Cefotaximase, community-acquired pediatric urinary tract infection, extended-spectrum beta-lactamase, sulfhydryl variable, temoneira
Introduction
Escherichia coli is a common pathogen causing community-acquired urinary tract infections (UTIs) in adults as well as in pediatric age groups. In recent studies, E. coli is the most frequently isolated extended-spectrum beta-lactamases (ESBL) carrying bacterium.[1] ESBL are enzymes capable of hydrolyzing the oxyimino-cephalosporins and monobactams. They are encoded by plasmids generally derived from cefotaximase (CTX-M), temoneira (TEM), or sulfhydryl variable (SHV).[2] The emergence of ESBL as an important cause of transferable drug resistance in E. coli is a serious problem in both hospital and community settings.[3]
Extensive use of extended-spectrum antibiotics is one of the most important factors associated with high prevalence of ESBL. Since the antibiotic use varies in different geographical regions, it can cause variation in the prevalence of ESBL genotypes. During the 1990s, SHV and TEM types were dominant among ESBL all over the world and CTX-M-producing organisms were rarely isolated. ESBL production in E. coli has significantly increased in numerous countries including India.[3,4] Studies have shown that awareness of risk factors in ESBL-producing E. coli UTI infections can minimize the complications associated with UTI, reduce the burden of medical expenses and development of antibiotic resistance, and enhance the clinical outcome among the pediatric patients.
To the best of our knowledge, no long-term prospective epidemiological surveillance studies have been carried on ESBL-producing E. coli strains among pediatric population in the community settings in India. Very few studies have described the risk factors and antibiotic resistance patterns of community-acquired pediatric UTI caused by ESBL-producing bacteria in India.[3,4] Thus, the aim of this study was to evaluate the resistance of the ESBL producing E. coli strains to commonly used antibiotics, to assess the risk factors associated with ESBL production, and to detect the presence of the three common ESBL genes: TEM, CTX-M, and SHV using polymerase chain reaction (PCR) method. The results of the present study might be valuable to both health professionals and the scientific community and may aid the current understanding of the trends in UTI caused by E. coli.
Methods
Setting
The present study was carried out in the clinical microbiology laboratory of a tertiary care center in Kerala, India. The study was conducted after obtaining approval from the Institutional Ethics Committee and informed consent from either of their parents.
Study design
It is a prospective microbiological study of E. coli isolates from urine specimens of pediatric patients with suspected community-acquired UTI who attended the outpatient department between November 2012 and March 2016.
Inclusion criteria
Children in the age group 3 months to 18 years with culture-proven UTI and satisfying the definition of community-acquired UTI were included in the study. Community-acquired UTI was defined as infection detected at hospital admission or within the first 48 h of admission.
Exclusion criteria
Children who satisfy definitions of health care-associated infection as outlined below:[5,6]
(1) Children who develop UTI during hospitalization (>48 h) or within 48 h of hospital discharge. (2) Children hospitalized in any hospital or nursing home during the 90 days preceding admission. (3) Children having a chronic medical condition requiring frequent medical visits as outpatient or inpatient. (4) Children receiving intravenous therapy or specialized wound care at home. (5) Patient receiving hemodialysis treatment or antineoplastic chemotherapy in 30 days before the infection. (6) Patient residing in a nursing home or transferred from another hospital.
Sample collection and processing
Urine samples were obtained by either of the following methods, i.e., midstream catch or bladder catheterization and cultured. E. coli on isolates was identified by conventional microbiological methods and according to the criteria of the Clinical and Laboratory Standards Institute (CLSI 2010) guidelines.[7,8]
Antibiotic sensitivity test was performed for E. coli by standard Kirby-Bauer disc diffusion method.[9] The following standard antibiotic discs (mcg) were used: ampicillin (10), cefixime (30), gentamicin (10), netilmicin (30), co-trimoxazole (25), ciprofloxacin (5), norfloxacin (10), levofloxacin (5), doxycycline (30), nitrofurantoin (300), and imipenem (10). Screening test for ESBL production was performed using cefotaxime (Ctx) and ceftazidime (CAZ) tested in combination with clavulanic acid. ESBL production was confirmed by phenotypic confirmatory double disc test and triple ESBL detection Ezy MIC™ Strip.[10]
Imipenem resistance was confirmed by E-strip method (imipenem with and without ethylene diamine tetraacetic acid [EDTA] Ezy MIC™ Strips EM078). All culture media and antibiotic discs were procured from HiMedia Laboratories, Mumbai, India. Results were interpreted according to the CLSI 2010 guidelines.[7] Quality control was performed using CLSI recommended E. coli ATCC 25922 and E. coli 35218 strains. The confirmed ESBLs positive E. coli isolates were kept in Luria-Bertani broth with glycerol and preserved at − 80°C for molecular studies.
DNA extraction and identification of bla cefotaximase, bla temoneira, and bla sulfhydryl variable genes by polymerase chain reaction
Confirmed ESBL-producing isolates were characterized for the presence of bla CTX-M, bla TEM, and bla SHV genes by PCR. Preparation of template DNA: E. coli isolate preserved in Luria-Bertani broth with glycerol was subcultured onto MacConkey agar. After overnight incubation, a single colony of each organism was inoculated into 5 ml of Luria-Bertani broth (HiMedia diagnostic, Mumbai) and incubated for 24 h at 37°C. 50 μl of this overnight culture was added to 450 μl of 1X TE (Tris-HCl and EDTA) buffer. This was incubated at 95–97°C in dry bath for 15 min and then kept in ice for 10 min. The DNA thus obtained were kept at −20°C.
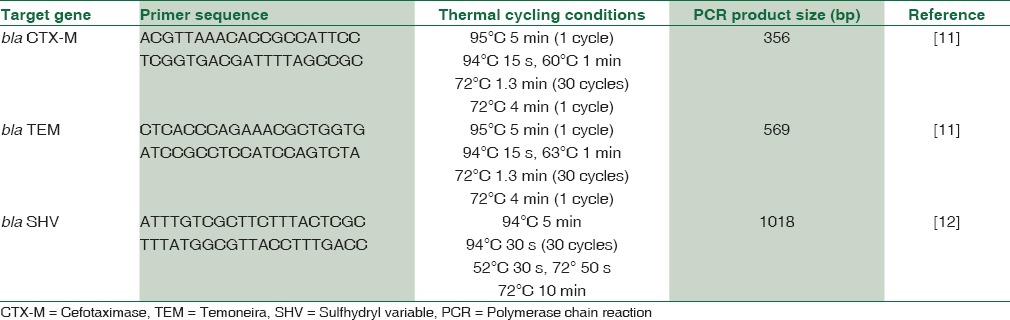
Bacterial genes associated with ESBL production were detected by PCR amplification of target genes by using specific primers in programmable thermal cycler (MJ Research PTC-200, Bengaluru).[11,12] Table 1 shows the primer sequences and specific thermal cycling conditions of CTX, TEM, and SHV genes. PCR mixtures were prepared using 2 μL template DNA, 28 μL PCR master mix (sterile distilled water 22.2 μL, Taq DNA polymerase 0.2 μL, Taq buffer with MgCl2 3 μL [GeNei, India], dNTP 0.6 μL, 1 μL of each 10 pm primer). Primers were obtained from BioServe Technologies, Hyderabad. 15 μl of the resulting PCR products were mixed with 10 μl of loading dye and analyzed by electrophoresis in 1.5% agarose gels (HiMedia, India) containing 1x Tris-acetate EDTA (1X TAE buffer) and 10 μl of ethidium bromide and visualized with the help of gel documentation system (Bio-Rad, India). The PCR band observed at 569 bp (CTX), 356 bp (TEM), and 1018bp (SHV).
Table 1.
Primer sequences and specific thermal cycling conditions of cefotaximase, temoneira, and sulfhydryl variable genes
In addition, clinical data and risk factors were analyzed for their predictability for colonization by an ESBL-producing organism.
Statistical analysis
The data were analyzed using Statistical Package for Social Sciences software version 16 (Chicago, SPSS Inc). The collected information was summarized using frequency, percentage, mean, and standard deviation (descriptive statistics). To compare the difference in antibiotic susceptibility with respect to ESBL positive and negative, Chi-square test and Fisher's exact test were used. To find the strength of association between antibiotic sensitivity and ESBL positive and negative groups, odds ratio (OR) and 95% confidence interval (CI) were computed. Binary logistic regression was used to identify the risk factors associated with ESBL production. A P < 0.05 was considered statistically significant.
Results
A total of 523 children with culture-proven E. coli community-acquired UTI were included in the study. Of this, 319 (60.9%) were boys and 204 (39.1%) girls, with a mean age of 31.19 ± 1.44 months. ESBL positivity was seen in 196 (37.5%), of which 131 (66.8%) were boys and 65 (33.2%) girls. The mean ages of ESBL producers and nonproducers were 30.62 ± 2.21 and 30.92 ± 1.68 months, respectively, with no statistical significance (P = 0.537). Statistical difference between male and female gender with regard to ESBL production was significant (Chi-square 4.49; P = 0.034).
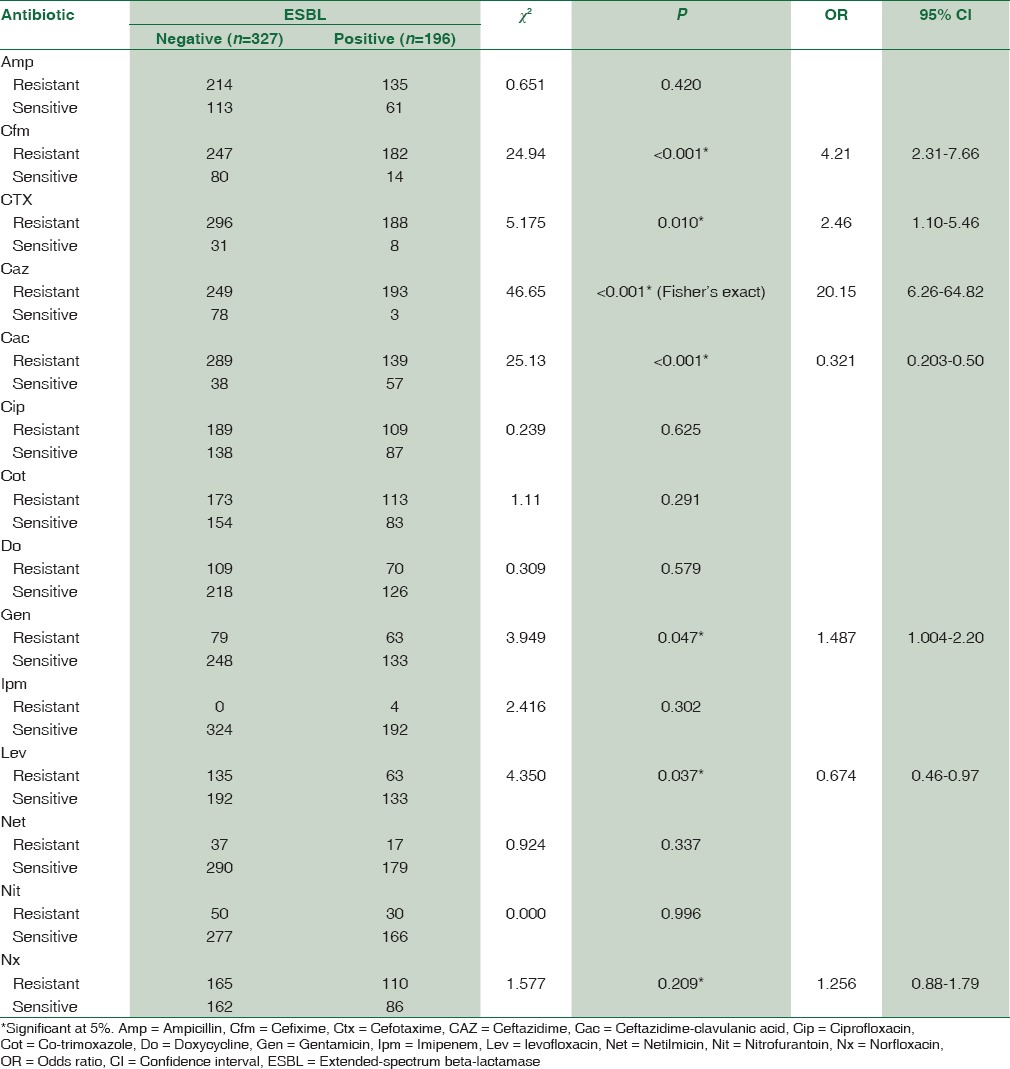
Table 2 allows the comparison of antibiotic resistance patterns of ESBL positive and negative community-acquired E. coli. The association between ESBL production and drug resistance was significant for CAZ (OR 20.15, P < 0.001), cefixime (OR 4.21, P < 0.001), Ctx (OR 2.46, P = 0.010), ceftazidime-clavulanic acid (OR 0.321, P < 0.001), levofloxacin (OR 0.674, P = 0.037), and gentamicin (OR 1.487, P = 0.047) when compared to non-ESBL E. coli. ESBL-positive E. coli showed more than 90% of resistance to cephalosporin group of antibiotics and 56% against ciprofloxacin and norfloxacin. Resistance against doxycycline, gentamicin, and levofloxacin was less than 36%. Least resistance was observed for imipenem, netilmicin, and nitrofurantoin.
Table 2.
Comparison of antibiotic resistance patterns of extended-spectrum beta-lactamase producing and nonextended-spectrum beta-lactamase Escherichia coli isolates (n=523) in community-acquired pediatric urinary tract infection
Disc diffusion test revealed that 4 (2%) of the ESBL-positive isolates were resistant to imipenem. This was confirmed using imipenem-EDTA strip method. MIC range was 4–8 μg/ml for three of the isolates. One isolate showed MIC >256 μg/ml.
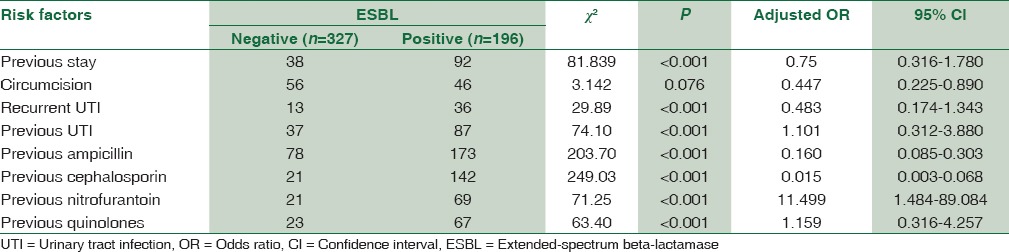
Multivariate analysis by binary logistic regression showed the following factors being significantly associated with E. coli resistance: history of previous UTI (OR 1.10, 95% CI, 0.312–3.880, P < 0.001), history of previous intake of antibiotics such as nitrofurantoin (OR 11.49, 95% CI, 1.484–89.084, P < 0.001) and quinolones (OR 1.15, 95% CI, 0.316–4.257, P < 0.001). The same is shown in Table 3 which includes other risk factors studied.
Table 3.
Risk factors associated with extended-spectrum beta-lactamase production in Escherichia coli isolates (n=523) from community-acquired pediatric urinary tract infections by logistic regression
CTX-M was the most prevalent gene (87.5%), followed by TEM (68.4%) and SHV (3.1%) among the ESBL producers. About 48.5% of ESBL isolates were shown to have both TEM and CTX-M genes, 2.6% possessed CTX and SHV, and 2% had TEM and SHV. Two percent of the isolates were shown to have CTX, TEM, and SHV [Figure 1a–c and Table 4].
Figure 1.
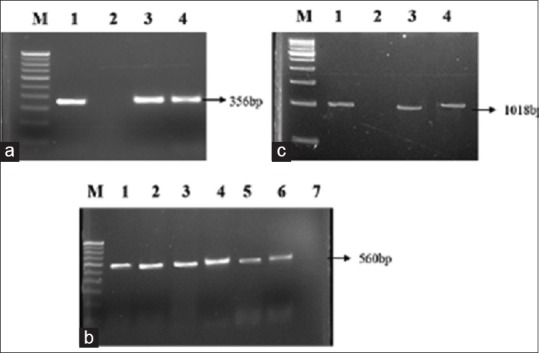
(a) Polymerase chain reaction amplification picture of cefotaximase gene in extended-spectrum beta lactamase producing Escherichia coli strains after agarose (1.5%) gel electrophoresis. Lane M: Molecular marker 100 bp DNA ladder; lane 1: Positive control; lane 2: Negative control; lane 3 and 4: Escherichia coli clinical isolates. (b) Polymerase chain reaction amplification of temoneira gene. Lane M: 100 bp DNA ladder; lane 1: Positive control; lane 2–6: Escherichia coli clinical isolate; lane 7: Negative control. (c) Polymerase chain reaction amplification of sulfhydryl variable gene. Lane M: 500 bp DNA ladder; lane 1: Positive control; lane 2: Negative control; lane 3 and 4: Escherichia coli clinical isolates
Table 4.
Frequency of extended-spectrum beta-lactamase genes isolated in Escherichia coli isolates (n=523) from community-acquired pediatric urinary tract infections
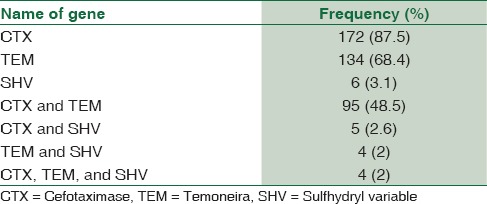
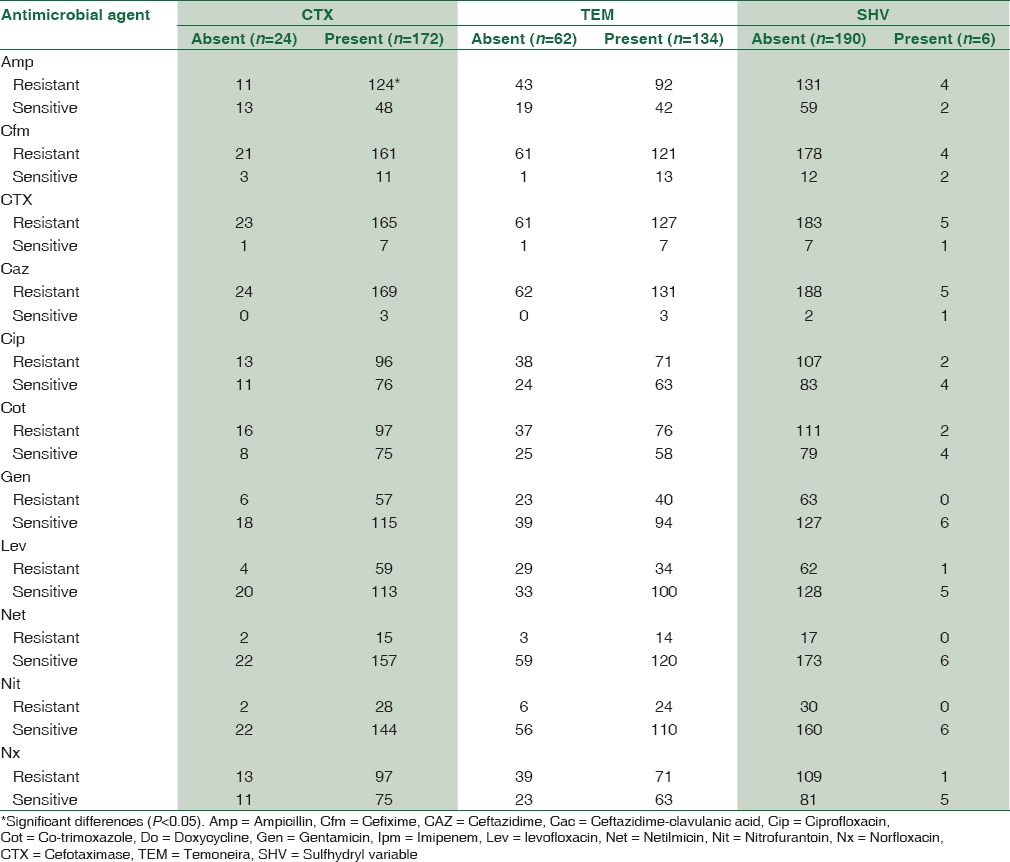
Table 5 shows association between antimicrobial resistance and gene expression in ESBL producing E. coli. In most of the cases, there were not any significant differences regarding genes expression and antibiotic resistance profile. Higher resistance to ampicillin, cefixime, Ctx, and Caz was found in CTX-positive group. Resistance to cefixime, Ctx, Caz, ciprofloxacin, and co-trimoxazole was observed more in TEM-positive isolates.
Table 5.
Association between antimicrobial resistance and gene expression in extended-spectrum beta-lactamase producing Escherichia coli
Discussion
Worldwide, the incidence of community-onset adult UTI caused by ESBL-producing bacteria is rising, but very few studies have described the epidemiology of community-acquired pediatric UTI caused by ESBL-producing bacteria in India.[3,4] The present study (n = 523) has shown that 37.5% of community-acquired E. coli isolates were ESBL producers. Similar results were reported from Turkey (n = 344) and Saudi Arabia (n = 207), in which the prevalence of ESBL-positive E. coli was 41.4% and 44%, respectively, in community-acquired UTI among children.[13,14]
Community-acquired ESBL E. coli showed high resistance rate to most of the currently used antimicrobial agents such as ampicillin (68.8%), cephalosporins including third-generation (>90%), quinolones specifically ciprofloxacin and norfloxacin (>50%), and co-trimoxazole (57.6%) but comparatively less resistance rate to gentamicin (32.1%), levofloxacin (32.1%), nitrofurantoin (15.3%), netilmicin (8.6%), and imipenem (2%). The data are comparable with a study from Switzerland, in which ESBL-producing E. coli urinary isolates showed high resistance rates to amoxicillin/clavulanate, quinolones, and co-trimoxazole and least for nitrofurantoin.[15] Two percent of ESBL E. coli showed imipenem resistance which limits the treatment option. It is reported that non-ESBL producing E. coli isolates also showed high resistance to antibiotics. In this study, the resistance rate shown by non-ESBL E. coli is much higher compared to Kizilca et al.'s data, in which non-ESBL producing isolates showed less than 10% resistance rate for nitrofurantoin, fluoroquinolones, and aminoglycosides.[13]
ESBL positivity was significant in male gender (P = 0.034) when compared to females in this study. Colodner et al. from Israel also reported male gender as a risk factor associated with community-acquired ESBL-positive UTI.[16] There were no statistical differences between the ESBL- and non-ESBL-producing E. coli with regard to age in this study. Previous history of UTI, previous intake of nitrofurantoin and quinolones were identified as risk factors associated with ESBL community-acquired UTI in this study. Other studies from Spain and Turkey have identified previous exposure to antibiotics including third-generation cephalosporins and quinolones and prior UTI history as risk factors associated with community-acquired UTI by ESBL-producing E. coli.[1,13]
Regarding molecular characterization of ESBL-producing E. coli isolates, high prevalence of CTX-M gene was found in 87.5% of E. coli isolates consistent with other studies (Sana et al. from Lebanon).[17] In contrast to our results, Rezai et al. from Iran observed low prevalence of CTX-M (28%).[18] The frequency of TEM in this study was 68.4% which is comparable to a study by Rezai et al. from Iran (49%) and Omar et al. from Sudan (55%), respectively.[18,19] The low prevalence of SHV gene (3.1%) reported is similar to the study carried by Sana et al. from Lebanon (4%).[17] A study by Jemima et al. from Chennai reported SHV genes in 14% and bla CTX-M genes in 50% of E. coli.[12]
The present study highlights the association between the presence of CTX and TEM gene and resistance to ampicillin and cephalosporin group of antibiotics, which is comparable with the study by Rezai et al.[18]
This study was conducted in a single tertiary care center; it may reflect the local demographics and antibiotic resistance patterns and this may be a limitation. In addition, plasmid profile and multilocus sequence typing were not done to measure the DNA sequence variation.
Conclusion
This study demonstrates an increasing trend in the emergence of ESBL E. coli in community-acquired UTI in children in our region warranting the need for ongoing antimicrobial stewardship and surveillance and development of antibiotic guidelines for the management of these complex multidrug-resistant infections. All simple UTIs and complex UTIs should have a culture done and the organisms identified. Overexposure and inappropriate use of antibiotics should be minimized to prevent emergence of ESBL.
It is of great concern that E. coli strains carrying bla CTX-M and bla TEM genes are widespread in Kerala. It is worthwhile that molecular characterization of clinical isolates should be carried out to prevent dissemination of resistant isolates in our community. PCR is a valuable tool for characterization of ESBL producers in clinical and research settings. Determination of CTX-M, TEM, and SHV genes by molecular techniques in ESBL-producing bacteria may give useful data about their epidemiology.
The results emphasize the need for screening ESBL producers and antibiotic profiling along with an assessment of risk factors for managing the children with UTI appropriately to enhance their clinical outcome.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Rodriguez-Bano J, Alcala JC, Cisneros JM, Grill F, Oliver A, Horcajada JP, et al. Community infections caused by extended-spectrum β-lactamase–producing Escherichia coli. Arch Intern Med. 2008;68:1897–902. doi: 10.1001/archinte.168.17.1897. [DOI] [PubMed] [Google Scholar]
- 2.Feizabadi MM, Delfani S, Raji N, Majnooni A, Aligholi M, Shahcheraghi F, et al. Distribution of bla(TEM), bla(SHV), bla(CTX-M) genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad Hospital, Tehran, Iran. Microb Drug Resist. 2010;16:49–53. doi: 10.1089/mdr.2009.0096. [DOI] [PubMed] [Google Scholar]
- 3.Akram M, Shahad M, Khan AU. Etiology and antibiotic resistance patterns of community acquired urinary tract infections in JNMC Hospital, Aligarh, India. Ann Clin Microbiol Antimicrob. 2007;23:4–10. doi: 10.1186/1476-0711-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma S, Kaur N, Malhotra S, Madan P, Ahmad W, Hans C. Serotyping and antimicrobial susceptibility pattern of Escherichia coli isolates from urinary tract infections in pediatric population in a tertiary care hospital. J Pathog. 2016;2016:2548517. doi: 10.1155/2016/2548517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subcommittee on Urinary Tract Infection, Steering Committee on Quality Improvement and Management, Roberts KB. Urinary tract infection: Clinical practice guideline for the diagnosis and management of the initial UTI in febrile infants and children 2 to 24 months. Pediatrics. 2011;128:595–610. doi: 10.1542/peds.2011-1330. [DOI] [PubMed] [Google Scholar]
- 6.Ha YE, Kang CI, Joo EJ, Park SY, Kang SJ, Wi YM, et al. Clinical implications of healthcare-associated infection in patients with community-onset acute pyelonephritis. Scand J Infect Dis. 2011;43:587–95. doi: 10.3109/00365548.2011.572907. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-First Informational Supplement M100-S20. Vol. 30. Wayne, PA: CLSI; 2010. p. 46. [Google Scholar]
- 8.Collee JG, Miles RS, Watt B. Tests for identification of bacteria. In: Collee JG, Fraser AG, Marmion BP, Simmons A, editors. Mackie and McCartney Practical Medical Microbiology. 14th ed. Singapore: Churchill Livingstone; 2006. pp. 131–49. [Google Scholar]
- 9.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–6. [PubMed] [Google Scholar]
- 10.Geser N, Stephan R, Kuhnert P, Zbinden R, Kaeppeli U, Cernela N, et al. Fecal carriage of extended-spectrum ß-lactamase-producing Enterobacteriaceae in swine and cattle at slaughter in Switzerland. J Food Prot. 2011;74:446–9. doi: 10.4315/0362-028X.JFP-10-372. [DOI] [PubMed] [Google Scholar]
- 11.Colomer-Lluch M, Jofre J, Muniesa M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One. 2011;6:e17549. doi: 10.1371/journal.pone.0017549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jemima SA, Verghese S. Multiplex PCR for bla(CTX-M) & bla(SHV) in the extended spectrum beta lactamase (ESBL) producing gram-negative isolates. Indian J Med Res. 2008;128:313–7. [PubMed] [Google Scholar]
- 13.Kizilca O, Siraneci R, Yilmaz A, Hatipoglu N, Ozturk E, Kiyak A, et al. Risk factors for community-acquired urinary tract infection caused by ESBL-producing bacteria in children. Pediatr Int. 2012;54:858–62. doi: 10.1111/j.1442-200X.2012.03709.x. [DOI] [PubMed] [Google Scholar]
- 14.El-Kersh TA, Marie MA, Al-Sheikh YA, Al-Kahtani SA. Prevalence and risk factors of community-acquired urinary tract infections due to ESBL-producing gram negative bacteria in an Armed Forces Hospital in Sothern Saudi Arabia. Glob Adv Res J Med Sci. 2015;4:321–30. [Google Scholar]
- 15.Meier S, Weber R, Zbinden R, Ruef C, Hasse B. Extended-spectrum b-lactamase producing gram-negative pathogens in community-acquired urinary tract infections. An increasing challenge for antimicrobial therapy. Infection. 2011;39:333–40. doi: 10.1007/s15010-011-0132-6. [DOI] [PubMed] [Google Scholar]
- 16.Colodner R, Rock W, Chazan B, Keller N, Guy N, Sakran W, et al. Risk factors for the development of extended-spectrum b-lactamase-producing bacteria in non-hospitalized patients. Eur J Clin Microbiol Infect Dis. 2004;23:163–7. doi: 10.1007/s10096-003-1084-2. [DOI] [PubMed] [Google Scholar]
- 17.Sana T, Rami K, Racha B, Fouad D, Marcel A, Hassan M, et al. Detection of genes TEM, OXA, SHV and CTX-M in 73 clinical isolates of Escherichia coli producers of extended spectrum Beta-lactamases and determination of their susceptibility to antibiotics. Int Arab J Antimicrob Agent. 2011;1:1–6. [Google Scholar]
- 18.Rezai MS, Salehifar E, Rafiei A, Langaee T, Rafati M, Shafahi K, et al. Characterization of multidrug resistant extended-spectrum beta-lactamase-producing Escherichia coli among uropathogens of pediatrics in North of Iran. Biomed Res Int 2015. 2015:309478. doi: 10.1155/2015/309478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Omar BA, Alfadel OO, Atif HA, Mogahid ME. Prevalence of TEM, SHV and CTXM genes in Escherichia coli and Klebsiella spp urinary isolates from Sudan with confirmed ESBL phenotype. Life Sci J. 2013;10:191–5. [Google Scholar]


