Abstract
Background
A polymorphism in a gene may exert its effects on multiple phenotypes. The aim of this study is to explore the association of 10 metabolic syndrome candidate genes with excess weight and adiposity and evaluate the effect of perinatal and socioeconomic factors on these associations.
Methods
The anthropometry, socioeconomic and perinatal conditions and 10 polymorphisms were evaluated in 1081 young people between 10 and 18 years old. Genotypic associations were calculated using logistic and linear models adjusted by age, gender, and pubertal maturation, and a genetic risk score (GRS) was calculated by summing the number of effect alleles.
Results
We found that AGT-rs699 and the IRS2-rs1805097 variants were significantly associated with excess weight, OR = 1.25 (CI 95% 1.01–1.54; p = 0.034); OR = 0.77 (CI 95% 0.62–0.96; p = 0.022), respectively. AGT-rs699 and FTO-rs17817449 variants were significantly and directly associated with body mass index (BMI) (p = 0.036 and p = 0.031), while IRS2-rs1805097 and UCP3-rs1800849 were significantly and negatively associated with BMI and waist circumference, correspondingly. Each additional effect allele in GRS was associated with an increase of 0.020 log(BMI) (p = 0.004). No effects from the socioeconomic and perinatal factors evaluated on the association of the candidate genes with the phenotypes were detected.
Conclusions
Our observation suggests that AGT-rs699 and FTO-rs17817449 variants may contribute to the risk development of excess weight and an increase in the BMI, while IRS2-rs1805097 showed a protector effect; in addition, UCP3- rs1800849 showed a decreasing waist circumference. Socioeconomic and perinatal factors had no effect on the associations of the candidate gene.
Electronic supplementary material
The online version of this article (doi:10.1186/s12263-017-0567-1) contains supplementary material, which is available to authorized users.
Keywords: Body mass index, Adiposity, Polymorphisms, Environmental factors, Youth
Background
The increasingly early onset of overweight and its long-term consequences focus the need to develop interventions for children, teens, and young adults [1]. Obesity, especially the central type, generates conditions that increase the risk of metabolic syndrome (MetS), defined as a set of traits that increase the risk of cardiovascular disease and diabetes mellitus 2 [2].
Being overweight is a result of a continuing imbalance between consumption and energy expenditure of an individual, where food consumption depends on, among others, environmental factors such as availability and genetic factors that influence appetite. Energy expenditure may also be affected by both lifestyle and metabolic efficiency, which, in turn, is influenced by genetic factors [3, 4].
Studies have shown that a polymorphism in a gene related to a given system may exert its effects in other pathways and influence multiple phenotypes [5]. Several loci susceptible to the components of MetS, such as those evaluated in this study, may play a role in the risk of excess weight. Uncoupling Protein-3 (UCP3) is involved in energy expenditure by stimulating thermogenesis, making it an attractive target for studies on the regulation of body weight [6]; Calpain 10 (CAPN10) is involved in numerous cellular functions, including signaling and adipocyte differentiation [7]; fat mass and obesity-associated (FTO) increases energy intake by regulating the expression of genes that control appetite [8]. The Insulin Receptor Substrate 2 (IRS2) and the protein that codes the transcription factor of the 7-like 2 (TCF7L2) gene play an important role in the transduction of insulin signaling [9, 10]; variants in the angiotensin-converting enzyme (ACE) and angiotensinogen (AGT) genes are associated with hypertension, finding the protein expression in adipose tissue [11]; in fact, variations in ATP-binding cassette, sub-family A (ABC1), member 1 (ABCA1), lipoprotein lipase (LPL), and cholesteryl ester transfer protein (CETP), associated with lipoprotein metabolism, contribute to the variation in adipogenesis.
Given these interactions between genetic and environmental factors in the etiology of obesity, increasing evidence reports differences in the frequency of these variants among populations, ethnicities, genders, and socioeconomic stratum [12, 13]. Studies related to the presence of polymorphisms associated with the development of adiposity measures have demonstrated discrepant results among population [14, 15]. Latin American populations, such as those in Colombia, are the result of a recent admixture among three ancestral populations: European, African, and Amerindian. Admixture can result in population stratification and may lead to spurious associations rather than association of genes with disease if the allele frequencies differ among the groups because of systematic differences in ancestry. In addition, socioeconomic stratum and parental education may be related to food availability and the transmission of a food culture [16, 17]; however, their relationship to the development of obesity is not clear enough. Results in a Mediterranean population suggest that education may modify the genetic susceptibility of FTO to obesity, with BMI being higher in non-university subjects compared to university subjects [18]. On the other hand, Pigeyre et al. [19] report the influence of maternal education in the association of neuromedin B rs3809508 and the risk of obesity. Perinatal conditions, such as increased or decreased fetal growth, as measured by weight and height at birth, or low breastfeeding, may also influence the likelihood of obesity later on in life [20].
Our hypothesis is that variants related to the MetS components may be associated with BMI and adiposity measures and this could be modified by socioeconomic and perinatal factors in young people aged 10 to 18 years.
Methods
Study design
The study is a cross-sectional study.
Participants
The sample consisted of 1081 young people between 10 and 18 years of age from Colombia, selected by random sampling and who were participants of the cross-sectional study “Variations in the Prevalence of Metabolic Syndrome in Adolescents According to Different Criteria Used for Diagnosis” [21], a study conducted between 2011 and 2012. The sample size was calculated with a confidence level of 95%, with an estimated prevalence of overweight of 10.3% according to the National Nutritional Situation Survey (2005) [22], a sample error of 2%, and effect on design of 1.2.
Excluded from the study were those young people with a habitual consumption of medications, young people with diabetes and genetic diseases, who are highly competitive athletes, young pregnant women, or those who were breastfeeding.
Measures
Perinatal and diseases history
The history of diseases (presence or absence of the disease) was considered in relatives to the second degree of consanguinity. Additionally, the perinatal history, such as birth weight (low <2500 g and high >4000 g), breastfeeding and duration, was also determined.
Parental education and socioeconomic stratum
Parental education and socioeconomic stratum were determined. The level of education was categorized into primary (0–6 years) and secondary (12 years and subsequent studies (>12 years); the stratum was determined as low (strata 1 and 2), medium (3 and 4), and high (5 and 6), according to the National Administrative Department of Statistics [23].
Pubertal maturation
The stage of pubertal maturation according to the methods established for this purpose [24, 25] was also evaluated.
Anthropometric evaluations
Weight, height, waist circumference, triceps, and subscapular fat fold were measured in all young people with equipment and international techniques before training and standardization of the evaluators; each measurement was assessed and recorded twice. For the classification of the nutritional status, the BMI (weight in kg/height in m2) was calculated; participants were classified according to the 2007 World Health Organization (WHO) BMI-for-age and gender reference standard [26]. Overweight was defined as BMI percentile ≥85.0th; adequate as BMI percentile <85.0th. Waist circumference was considered as being high with a value of more than p90 of the values for Mexican–American young people from the U.S. Third National Health and Nutrition Examination Survey [27]. The percentage of total body fat (%BF) was calculated with the subscapular and triceps fat folds, according to Lohman et al. [28] and was classified as obesity when it was >25% in males and >32% in females, adequate between 12 and 25% in males and 25 and 32% in females, and deficient when it was <12% in males and <15% in females [28].
Genotyping
After excluding individuals related up to the third degree of consanguinity determined through clinical histories and information provided in the General Information questionnaire, ten variants were genotyped in 1005 youth: IRS2-rs1805097, CAPN10-rs3842570, UCP3-rs1800849, FTO-rs17817449, TCF7L2-rs7903146, AGT-rs699, ACE-rs4340, LPL-rs285, CETP-rs708272, and ABCA1-rs2230806; the genotyping was performed by polymerase chain reaction-restriction fragment length polymorphism. The PCR was performed in a 25-μl volume containing 10 ng genomic DNA, 10 × PCR buffer with 1.5 mM MgCl2, 0.5 mM dNTPs, 0.5 units of Taq polymerase, and 5 μmol of each primer. PCR conditions included one step initial desaturation at 95 °C for 3 min, 35 cycles (95 °C for 45 s, 55–62 °C for 45 s, and 72 °C for 45 s), and a final extension at 10-min extension step at 72 °C. The PCR products were checked on 1.5% agarose gel. Amplified PCR products were digested with restriction enzyme overnight. The digestion products were electrophoresed on agarose gel and visualized by staining with ethidium bromide. Primers and restriction enzymes used are reported in the Additional file 1: Table S1. To assess reproducibility, 10% of the samples were doubly genotyped. Negative controls were also added to each 96-well plate. No discordance was detected between the replicated samples, and reproducibility was 98.9%.
Genetic ancestry estimation
Individual admixture proportions were available for 337 out of the 738 participants with normal weight, and for 235 of 267 subjects in the overweight group. For these individuals, European, Amerindian, and African contributions were estimated with the program ADMIXMAP v 3.2 [29], using a set of 40 ancestry informative markers (AIMs) broadly distributed across the genome and according to their significant differences in allele frequencies among two populations. These AIMs have been described previously to accurately estimate ancestry in Latin American populations [30] (Additional file 1: Table S2).
Statistical analysis
A descriptive analysis was performed on the perinatal history and the socioeconomic, health, anthropometric, and genetic variables. The comparison of the variables among the groups according to nutritional status with the Pearson X 2 and Pearson correlation was made; the Student t tests were made for quantitative variables. For the allele and genotype frequencies, the Hardy–Weinberg equilibrium and the associations of the variant phenotypes, PLINK v1.07 [31] were used. Logistic regression analysis was performed to look for associations of polymorphisms with nutritional status (normal weight and overweight groups) using the following models: additive (major allele homozygotes vs. heterozygotes vs. minor allele homozygotes), dominant (major allele homozygotes vs. heterozygotes + minor allele homozygotes), and recessive (major allele homozygotes + heterozygotes vs. minor allele homozygotes). The best model was chosen according to the Akaike information criterion. Logistic regression was employed to estimate the odds ratio (OR) and its confidence interval (CI) of 95%. The association of genotype with BMI, WC, and percentage of BF were evaluated using linear regression; variables with non-normal distributions were log-transformed before analysis. For the anthropometric measurements that were associated to more than two associated variants, an estimation of the individual genetic risk score (GRS) was generated, using the risk alleles. We quantified the unweighted genetic risk score to assess the combined effects of the variants by summing the number of risk alleles; each individual might have 0, 1, or 2 risk alleles in each of the variants. The weighted GRS was calculated by multiplying the number of risk alleles at each locus (0, 1, 2) by the variants’ β coefficient from the predictive model and then summing the product. Effects of covariables (gender, age, pubertal maturation, and BMI) were controlled, as well as the interaction between the genetic variants and the social stratum, maternal education year, birth weight, and maternal breastfeeding. In addition to the previous analysis, we conducted logistic analyses for the subset of the samples for which information ancestry was accessible. The objective of these analyses was to determine if the possible association of the polymorphism with nutritional status (normal weight and overweight groups) or adiposity measurement could be driven by population stratification. Controlling for two out of three admixture estimates avoided collinearity in the model, since the three ancestry components sum up to 1.
Multiple test correction was done via permutation tests; we performed 10,000 permutations to determine empirical significance, which was considered for a value of p < 0.05.
Results
Perinatal history and socioeconomic, health, and anthropometric conditions of the young people
Of the total of 1081 participants, 581 (52.7%) were females; the mean age was 14.2 ± 2.4 years. The prevalence of excess weight (percentile ≥85.0th) was 25.1% (272); of these, 13.7% (149) were overweight and 11.3% (123) had obesity (BMI percentile >95.0th).
No significant differences were found between the socioeconomic stratum and birth weight with the nutritional status; however, excess weight was significantly greater in young people with more-educated parents (p < 0.023), in those who had a family history of hypertension (p = 0.011), dyslipidemia (p = 0.003), and obesity (p = 0.00) and those who presented a minor duration of breastfeeding (p = 0.048) (see Table 1). Characteristics by gender are shown in Additional file 1: Table S3. There were no significant differences in socioeconomic status, parent education, family history of diseases, perinatal history, and BMI. The percentage of individuals who reported prepubertal maturation was significantly higher in men than in women (p = 0.011). In contrast, waist circumference and BF% was significantly higher in women than in men (p < 0.001 for both). A histogram of anthropometric variables, stratified by gender, is shown in Additional file 1: Figure S1.
Table 1.
Characteristics of the study population, stratified according to BMI
| Variable | BMI <p 85.0th (%), n = 809 |
BMI ≥p 85.0th (%), n = 272 |
p value |
|---|---|---|---|
| Socioeconomic statusa | |||
| Low | 43.3 | 39.3 | 0.082 |
| Medium | 37.8 | 36.4 | |
| High | 18.9 | 24.3 | |
| Maternal education, yeara | |||
| 0–6 | 15.3 | 7.7 | 0.008 |
| +6–12 | 47.4 | 49.8 | |
| +12 | 37.3 | 42.4 | |
| Paternal education, yeara | |||
| 0–6 | 14.8 | 10.6 | 0.023 |
| +6–12 | 47.2 | 44.1 | |
| +12 | 38.1 | 45.3 | |
| Pubertal maturationa | |||
| Prepubertal | 18.2 | 18.4 | 0.155 |
| Pubertal | 26.6 | 33.8 | |
| Postpubertal | 55.3 | 47.8 | |
| Family historyb | |||
| Obesity | 34.4 | 60.1 | <0.001 |
| Type 2 diabetes | 55.8 | 62.0 | 0.073 |
| Gestational diabetes | 2.7 | 2.6 | 0.897 |
| Hypertension | 76.0 | 83.4 | 0.011 |
| Dyslipidemia | 59.5 | 69.7 | 0.003 |
| Birth weight, (g)a | |||
| <2500 | 9.2 | 6.3 | 0.056 |
| ≥2500–4000 | 85.7 | 86.1 | |
| >4000 | 5.1 | 7.5 | |
| Maternal breastfeedingb | 93.8 | 90.4 | 0.057 |
| Duration of breastfeeding, (months)a | |||
| 0–1 | 6.2 | 9.7 | 0.048 |
| >1–3 | 21.4 | 22.8 | |
| >3–6 | 21.3 | 21.7 | |
| >6 | 51.1 | 45.7 | |
| Mean ± SD | |||
| Anthropometryc | |||
| Weight, kg | 47.3 (11.0) | 59.8 (15.0) | <0.001 |
| BMI, kg/m2 | 19.0 (2.39) | 24.4 (3.49) | <0.001 |
| BF% | 22,3 (7.8) | 32.2 (8.11) | <0.001 |
| Waist circumference, (cm) | 65,0 (5.8) | 76.2 (8.3) | <0.001 |
Data is shown as percentage (%) or average ± standard deviation The significant p values (p < 0.05) are given in bold
BMI body mass index, BF% body fat percentage
aSpearman correlation
bPearson’s chi-square
cStudent t test
Association between excess weight and adiposity measurements with genetic variants
The genetic analysis considered those subjects unrelated, leaving a total of 1005 participants. Genotype distributions did not deviate from the Hardy–Weinberg expectations. An allelic frequency above 24% was found for the low allele frequency in all variants evaluated, except for UCP3, which was 12%. The sample was divided into two groups: cases with excess weight, with BMI p ≥85.0th (n = 267) and normal-weight controls with BMI p <85.0th (n = 738). A logistic regression analysis was done for each one of the 10 variants under the additive, dominant, and recessive models. The minor allele additive model showed that rs699 in AGT was significantly associated with an increase in the risk of excess weight, whereas rs1805097 in IRS2 was significantly associated with normal weight. The association remained significant after adjusting for age, gender, and pubertal maturation (see Table 2). The AGT-rs699 (additive model p = 0.034, OR = 1.25, 95% CI = 1.01–1.54), the IRS2-rs1805097 (additive model p = 0.022, OR = 0.77 95% CI = 0.62–0.96). An identical association was also observed for the dominant model (Table 2, part A). Focusing on the subset of the samples with information on ancestry, we observed that the ancestral composition is on average 66.3 ± 5.8 (%) European with a range in individuals of 41.0–80.0%; 19.5 ± 4.2 (%) Amerindian with a range of 10.0–35.0%; and 14.2 ± 4.9 (%) African with a range of 3.8–57.9%. Participants did not show any significant differences in ancestry according to nutritional status (normal weight vs. overweight) (Table 3). The odds ratios for the logistic regression analysis, adjusting ancestry as a covariate, were similar to those observed for the total sample, although the p values were weaker because of the smaller sample size (Table 2, part B). In addition, linear regression analyses of the adiposity measures with the 10 variants are shown in Table 4. The AGT-rs699 and IRS2-rs1805097 variants presented an association with BMI, FB%, and WC; UCP3-rs1800849 and FTO-rs17817449 variants presented an association with BMI and WC. After adjustment for age, gender, and pubertal maturation (for FB% and WC adjusted for BMI), the AGT-rs699 and FTO rs17817449 variants showed positive associations with BMI, and the IRS rs1805097 variant showed a negative association with BMI (p = 0.036, p = 0.031, and p = 0.043 respectively), while the UCP3 rs1800849 showed a negative association with WC (p = 0.001) (Table 4). We did a similar analysis on the subset of the sample with information on ancestry. Our results showed that after adjusting for ancestry, associations between IRS2-rs1805097 and FTO-rs17817449 on IMC were not confirmed (Additional file 1: Table S4). However, the association between AGT-rs699 on BMI and UCP3-rs1800849 on WC was still observed after adjusting for ancestry. However, this might be due to the effect of sample size, because IRS2-rs1805097 retained its association in logistic regression analysis after adjusting for ancestry.
Table 2.
Association between 10 selected genetic variants and the risk of excess weight (p ≥85.0th) in young people aged 10 to 18 years
| Part A | Part B | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Genetic variant | GENE | A2/A1 | TEST | OR* | SE | 95% CI | p value | OR† | SE | 95% CI | p value | Risk allele |
| 1q42.21 | rs699 | AGT | C/T | ADD | 1.25 | 0.108 | 1.011, 1.545 | 0.0394 | 1.31 | 0.137 | 1.005, 1.720 | 0.0458 | T |
| DOM | 1.36 | 0.156 | 1.005, 1.855 | 0.0449 | 1.46 | 0.183 | 1.021, 2.097 | 0.0383 | |||||
| REC | 1.29 | 0.205 | 0.865, 1.929 | 0.2108 | 1.42 | 0.248 | 0.873, 2.313 | 0.1575 | |||||
| 2q37.3 | rs3842570 | CAPN10 | IND/DEL | ADD | 0.90 | 0.104 | 0.730, 1.099 | 0.2950 | 0.86 | 0.126 | 0.673, 1.108 | 0.2489 | – |
| DOM | 0.86 | 0.156 | 0.632, 1.165 | 0.3324 | 0.76 | 0.190 | 0.530, 1.117 | 0.1680 | |||||
| REC | 0.87 | 0.189 | 0.601, 1.260 | 0.4612 | 0.87 | 0.221 | 0.564, 1.345 | 0.5333 | |||||
| 8p21.3 | rs285 | LPL | C/T | ADD | 1.11 | 0.102 | 0.910, 1.356 | 0.3009 | 1.19 | 0.124 | 0.934, 1.521 | 0.1572 | – |
| DOM | 1.33 | 0.169 | 0.955, 1.854 | 0.0939 | 1.58 | 0.199 | 1.071, 2.341 | 0.0212 | |||||
| REC | 0.10 | 0.171 | 0.712, 1.394 | 0.9822 | 1.021 | 0.205 | 0.682, 1.526 | 0.9213 | |||||
| 9q31.1 | rs2230806 | ABCA1 | G/A | ADD | 1.11 | 0.109 | 0.899, 1.379 | 0.3308 | 1.03 | 0.139 | 0.788, 1.362 | 0.7985 | – |
| DOM | 1.31 | 0.154 | 0.968, 1.770 | 0.0756 | 1.25 | 0.181 | 0.882, 1.798 | 0.2037 | |||||
| REC | 0.88 | 0.222 | 0.573, 1.369 | 0.5836 | 0.91 | 0.256 | 0.554, 1.515 | 0.7348 | |||||
| 10q25.2 | rs7903146 | TCF7L2 | C/T | ADD | 0.99 | 0.121 | 0.783, 1.260 | 0.9603 | 0.76 | 0.187 | 0.530, 1.106 | 0.1554 | – |
| DOM | 1.04 | 0.149 | 0.775, 1.390 | 0.8100 | 0.87 | 0.175 | 0.618, 1.228 | 0.4318 | |||||
| REC | 0.81 | 0.322 | 0.432, 1.523 | 0.5140 | 0.60 | 0.367 | 0.293, 1.242 | 0.1706 | |||||
| 11q13.4 | rs1800849 | UCP3 | C/T | ADD | 0.96 | 0.165 | 0.697, 1.332 | 0.8220 | 1.24 | 0.419 | 0.545, 2.826 | 0.6061 | – |
| DOM | 0.95 | 0.178 | 0.672, 1.351 | 0.7911 | 1.10 | 0.210 | 0.729, 1.660 | 0.6494 | |||||
| REC | 1.07 | 0.6915 | 0.276, 4.159 | 0.9196 | 1.51 | 0.837 | 0.293, 7.820 | 0.6202 | |||||
| 13q34 | rs1805097 | IRS2 | G/A | ADD | 0.77 | 0.1123 | 0.622, 0.966 | 0.0222 | 0.95 | 0.144 | 0.718, 1.265 | 0.7405 | G |
| DOM | 0.67 | 0.1502 | 0.502, 0.905 | 0.0090 | 0.71 | 0.176 | 0.506, 1.012 | 0.0582 | |||||
| REC | 0.85 | 0.2253 | 0.546, 1.321 | 0.4691 | 1.12 | 0.271 | 0.657, 1.909 | 0.6757 | |||||
| 16q12.2 | rs17817449 | FTO | T/G | ADD | 1.17 | 0.1133 | 0.933, 1.455 | 0.1701 | 1.14 | 0.153 | 0.849, 1.548 | 0.3702 | – |
| DOM | 1.17 | 0.1478 | 0.875, 1.561 | 0.2887 | 1.07 | 0.173 | 0.761, 1.503 | 0.6968 | |||||
| REC | 1.36 | 0.2527 | 0.832, 2.239 | 0.2186 | 1.30 | 0.294 | 0.732, 2.322 | 0.3677 | |||||
| 16q13 | rs708272 | CETP | C/T | ADD | 0.92 | 0.1107 | 0.741, 1.144 | 0.4594 | 0.94 | 0.135 | 0.727, 1.239 | 0.7021 | – |
| DOM | 0.83 | 0.1514 | 0.619, 1.120 | 0.2306 | 0.82 | 0.178 | 0.584, 1.174 | 0.2899 | |||||
| REC | 1.06 | 0.2168 | 0.692, 1.618 | 0.7945 | 1.01 | 0.252 | 0.617, 1.659 | 0.0622 | |||||
| 17q23.3 | rs4340 | ACE | IN/DEL | ADD | 0.95 | 0.1029 | 0.781, 1.169 | 0.6571 | 0.96 | 0.120 | 0.762, 1.222 | 0.7669 | – |
| DOM | 0.98 | 0.1686 | 0.706, 1.367 | 0.9270 | 1.00 | 0.197 | 0.681, 1.475 | 0.9902 | |||||
| REC | 0.90 | 0.1708 | 0.643, 1.257 | 0.5349 | 0.90 | 0.199 | 0.612, 1.339 | 0.6208 | |||||
The ORs are based on the major allele homozygotes as reference. The significant p values (p < 0.05) are given in bold type
A2 major allele, A1 minor allele, ADD additive (major allele homozygotes vs. heterozygotes vs. minor allele homozygotes), DOM dominant (major allele homozygotes vs. heterozygotes + minor allele homozygotes), REC recessive (major allele homozygotes + heterozygotes vs. minor allele homozygotes), OR odds ratio, CI confidence interval
*Adjusted for age, gender, and pubertal maturation
†Adjusted for age, gender, pubertal maturation, and individual ancestry proportions
Table 3.
Mean (standard deviation) of genetic ancestry percentages for nutritional status
| BMI p <85.0th n = 337 |
BMI p ≥85.0th n = 235 |
p value | |
|---|---|---|---|
| European | 66.2 (5.6) | 66.7 (6.3) | 0.327 |
| Amerindian | 19.6 (4.3) | 19.3 (4.2) | 0.439 |
| African | 14.4 (4.7) | 14.1 (5.4) | 0.552 |
Mean and standard deviation values are obtained from inferred individual genetic ancestry components (measured in percentage) for the subset of the samples. Student t test
Table 4.
Association with anthropometric measures at 10 selected genetic variants in young people aged 10 to 18 years
| Log BMI (kg/m2)a | Log BF %b | Log waist circumference (cm)b | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | MAF | TEST | ß coefficient | SE | 95% CI | p value | ß coefficient | SE | 95% CI | p value | ß coefficient | SE | 95% CI | p value |
| rs699 | T | ADD | 0.007 | 0.003 | 0.000, 0.013 | 0.036 | 0.005 | 0.004 | −0.003, 0.013 | 0.210 | 0.0008 | 0.000 | −0.001, 0.002 | 0.365 |
| rs3842570 | D | ADD | −0.000 | 0.003 | −0.006, 0.006 | 0.973 | 0.003 | 0.004 | −0.005, 0.011 | 0.440 | −0.0004 | 0.000 | −0.002, 0.001 | 0.628 |
| rs285 | T | ADD | 0.002 | 0.003 | −0.003, 0.008 | 0.406 | −0.002 | 0.004 | −0.010, 0.005 | 0.577 | −0.0000 | 0.000 | −0.001, 0.001 | 0.994 |
| rs2230806 | A | ADD | 0.003 | 0.003 | −0.003, 0.010 | 0.286 | −0.002 | 0.004 | −0.010, 0.006 | 0.686 | −0.0009 | 0.001 | −0.002, 0.000 | 0.319 |
| rs7903146 | T | ADD | −0.001 | 0.003 | −0.008, 0.006 | 0.851 | 0.002 | 0.005 | −0.007, 0.011 | 0.653 | −0.0003 | 0.001 | −0.002, 0.001 | 0.772 |
| rs1800849 | T | ADD | −0.008 | 0.005 | −0.017, 0.001 | 0.104 | −0.005 | 0.006 | −0.017, 0.006 | 0.373 | −0.0045 | 0.001 | −0.007, −0.001 | 0.001 |
| rs1805097 | A | ADD | −0.006 | 0.003 | −0.013, −0.000 | 0.043 | −0.005 | 0.004 | −0.013, 0.003 | 0.200 | 0.0000 | 0.000 | −0.001, 0.002 | 0.951 |
| rs17817449 | G | ADD | 0.007 | 0.003 | 0.001, 0.014 | 0.031 | −0.000 | 0.004 | −0.009, 0.008 | 0.907 | 0.0011 | 0.001 | −0.000, 0.003 | 0.287 |
| rs708272 | T | ADD | −0.013 | 0.003 | −0.008, 0.005 | 0.672 | −0.004 | 0.004 | −0.012, 0.004 | 0.351 | −0.0007 | 0.001 | −0.002, 0.001 | 0.434 |
| rs4340 | D | ADD | −0.000 | 0.003 | −0.006, 0.006 | 0.954 | −0.001 | 0.004 | −0.008, 0.007 | 0.877 | −0.0009 | 0.000 | −0.002, 0.000 | 0.316 |
| GRS-3 | ADD | 0.020 | 0.007 | 0.006, 0.033 | 0.004 | – | – | – | – | – | – | – | – | |
The significant p values (p < 0.05) are given in bold
MAF minor allele frequency calculated using the data from all the subjects in the analysis, ADD additive, CI confidence interval, GRS genetic risk score
aAdjusted for age, sex, and pubertal maturation
bAdjusted for age, sex, pubertal maturation, and BMI
A GRS was generated for every individual by counting the number of alleles associated with excess weight (AGT-rs699 T and IRS2-rs1805097 G; range 0–4, (GRS-2)) and BMI measurement (AGT-rs699 T, IRS2-rs1805097 G and FTO-rs17817449 G; range 0–6, (GRS-3)). The association of the GRS-2 and the risk of excess weight showed that each additional effect allele was associated with a 1.26-fold increased odds of excess weight (95% CI 1.08–1.47). The GRS-3 was normally distributed. 16.9% of the individuals carried one or fewer risk alleles, and 5% carried ≥6 (Fig. 1). The GRS-3, which examines the cumulative effects of the three SNPs, was significantly associated with BMI (p = 0.004, effect size 0.034 log-transformed/allele 95% CI 0.006–0.033).
Fig. 1.
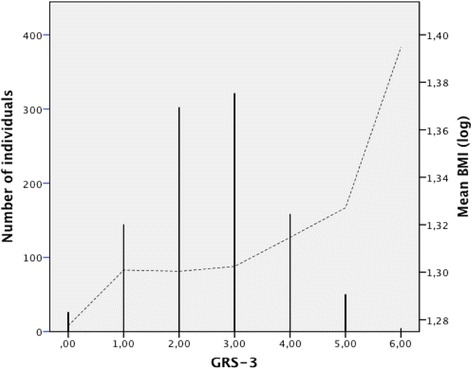
Distribution of the GRS-3 and cumulative effects of the alleles from the three BMI susceptibility variants on log-transformed BMI value
Interaction between genetic variants and socioeconomic and perinatal variables
We did not find any statistically significant association among the rs3842570, rs285, rs2230806, rs7903146, rs708272, and rs4340 with BMI and adiposity measurements in this population. We explored the possible interaction of the socioeconomic status and perinatal history for the 10 variants evaluated and the indicators related to excess weight. This study found no evidence of interaction with these variables. An additional text file shows this in more detail (Additional file 1: Table S5).
Discussion
The clustering of hypertension, dyslipidemia, and insulin resistance with obesity suggests the presence of common factors influenced by both environmental and genetic factors. Previous studies between body fat and common cardiovascular disease risk factors have indicated the presence of some genetic pleiotropism [32]. Additionally, epidemiological studies and clinical settings have established that outcomes often differ between ethnicity, and are modulated by social and environmental conditions. In this study we explore the association of 10 metabolic syndrome candidate genes with excess weight and adiposity and evaluate the effect that socioeconomic and perinatal factors could have on these associations in a group of young people.
Of the 10 variants evaluated, associations with excess weight, or its quantitative measurements, were found for four polymorphisms. A missense SNP in AGT (rs699; M235T) was associated with excess weight and may be a causal candidate variant; AGT II is an important regulator of blood pressure; carriers of the threonine variant have higher values in blood pressure [33], which leads to an increased risk associated with hypertension disorders. Skov and coworkers argued that regulating the renin-angiotensin-aldosterone system (RAAS) was involved in metabolic processes and could explain the origin and complications of some disorders such as MetS [34]. The RAAS components are involved in complex ways in the development of obesity by conditions of satiety, energy expenditure, and growth and differentiation of adipocytes [35]. AGT is also highly expressed in white adipose tissue, second to the liver in terms of mRNA levels; one study showed that overexpression of AGT in adipocytes increased weight in murine models [36]. Several observations suggested a role of AGT in adipose tissue development since this tissue and isolated adipocytes contain RAAS components, leading to AngII [37]. The expression and secretion of AGT increases with differentiation and is higher in adipocytes compared to preadipocytes [38].
We showed that the missense SNP rs1805097 in IRS2 was associated with protection for excess weight, an association not previously described. This SNP leads to a Gly1057Asp. Previous studies showed the role of IRS-2 in the differentiation of preadipocytes from adipocytes through the upregulation of the specific transcriptional factors of expression, such as PPARγ and C/EBP [39]. The variant is associated with diabetes in the adult population. However, studies from Italy and Pima Indians showed that this variant contributed to diabetes risk in obese individuals, but lowered risk for type 2 diabetes in normal-weight individuals, suggesting a possible interaction between gene and the environment [40, 41]. Kilpeläinen et al. [42] reported a locus near IRS1 which was associated with a decrease in BF but with an impaired metabolic profile, including increased insulin resistance, dyslipidemia, risk of diabetes and coronary artery disease, and decreased adiponectin levels.
Different polymorphisms in the FTO-gene intron 1 have been associated with obesity or with BMI in genomic scans as well as in models of simple association in different populations [43, 44]. Recently, it was reported that the intron 1 polymorphisms function in regulatory elements of the expression of the IRX3 gene. IRX3 encodes a transcription factor highly expressed in the brain consistent with the role in regulating energy metabolism and feeding behavior [45]. As in other studies, we found an association with BMI in which the mean BMI was increased 0.33 kg/m2 per allele G [46].
UCP3 participates in thermogenesis; a decrease in the expression or function of this protein may reduce energy expenditure and increase its storage as fat. A promoter region variant −55 CT (rs1800849) is situated 6 bp from the TATA box and 4 bp from a DR1 site, which is a part of a retinoic acid response element [47]. The functions described for UCP3 and the results of variant association have been reported for children and adults in Caucasian and Korean populations [6, 48]. Liu et al. [6] found a statistically significant association between the carriers of the T allele and a lower BMI. Our study did not show an association with BMI, but it did show a statistically significant difference with a lower waist circumference.
With the exception of rs17817449, no association of these variants with BMI or waist circumference was found in the Genetic Investigation of ANthropometric Traits (GIANT) Consortium [49]. A steady genetic effect across population means that genetic variants reflect a common, final biological effect on individuals. However, associations may be altered by age, environmental exposures, interaction with other genes, effects of the differences in the allele frequencies, and the linkage disequilibrium patterns in admixed populations.
Evidence shows an interaction among the variants of obesity with environmental factors, as has been found for FTO, in which the lack of physical activity and less education potentiate the effect of its variants in the increase of obesity [18, 23]. Similarly, a variant of the APOA2 gene increases the association with obesity in those consuming high levels of saturated fatty acids [50]. Another study reported the influence of maternal education on the effect of a variant of the neuromedin B gene on obesity [19]. Based on these studies, we evaluated environments that conducive to possible interaction, such as social status, education, history of breastfeeding, and birth weight. However, no associations between the 10 variants and these factors were found, possibly due to sample size.
However, significant association was found between the education of parents with excess weight in this study similar to that observed in Bangladesh and India, which reported that higher levels of education and greater income were associated with the increase in the rates of obesity [16, 17]. Although not monitored here, families with higher levels of education and better economic incomes may consume higher levels of fast foods and, coupled with the availability of video games, creates sedentary environments that favor the development of obesity.
Birth weight and length are considered to be high-risk variables for excess weight [51]. The study by Loaiza et al. [52] in Chilean children, reported a direct relationship between high birth weight and the risk of obesity in school age. Evidence exists on the effect of the fetal environment and the epigenetic remodeling in fetal genes that regulate or participate in the energy metabolism that will be expressed in obesogenic environments [53]. In this study, birth weight showed a borderline significance (p = 0.056).
Family history of obesity meets both genetic risk factors as well as environmental or cultural factors. Heritability estimates for obesity range between 50 and 80% [54] based on the concordance in monozygotic compared to dizygotic twins.
Several limitations of this study must be considered: the sample size may be relatively small for a predictive clinical-genomic model. In addition, genotype-environment interactions would require a larger population to confirm these results. Furthermore, this study only considered the association of a single variant in each gene with BMI, a widely used but imprecise indicator of adiposity. X-ray dual absorptiometry produces more precise data on adiposity but because of cost its use is limited. A wide variety of other environmental factors and social determinants were not explored in this study which could affect the associations.
Despite these limitations, this study contributes to the literature on understanding childhood obesity, showing associations between the family socioeconomic level and anthropometric, health, and genetic measurements. Although the study reported here, like most association studies, focused on single SNP associations, excess weight is a complex trait produced by combinations of many gene-gene and gene-environment interactions. The pathway for the translation and integration of genomics from the laboratory into everyday medicine remain largely unfulfilled, and the clinical utility has been debated recently [55, 56]. Genetic studies should be considered as a first step in the understanding of the molecular basis of traits or complex diseases but requires a more comprehensive view integrating phenotypic and genotypic factors with environmental and metabolic factors. We believe that the results provide further insights regarding the potential modulating effect of certain genotypes to weight reduction treatments.
Conclusions
We found that the SNPs of AGT rs699 and IRS2 rs1805097 showed significant association with overweight (BMI p >85). The variants rs699, rs1805097, and rs17817449 were significantly associated with BMI and the variant of UCP3 rs1800849, with waist circumference. In addition, we found that the level of parent education, family histories of obesity, hypertension, dyslipidemia, and a minor duration of breastfeeding contribute to excess weight; however, no effect of these socioeconomic and perinatal factors was found on genetic associations.
Although obesity is a complex disease, the implementation of personalized treatments using genotypes could be tested in this condition if our findings are confirmed.
Future research should explore, in a larger sample size, the interaction of other environmental factors such as lifestyles and diet habits, in addition to a larger number of polymorphisms in each gene. This might help explain the relationship between the genotype and phenotype, which may aid clinicians for developing treatments or predicting outcomes.
Acknowledgements
Resources from Colciencias Contract 203–2010; Vidarium Group; and the Committee for the Development of Research (Comité para el Desarrollo de la Investigación, CODI); the University of Antioquia 2013–2014 Sustainability Strategy.
Authors’ contributions
AMM participated in the fieldwork, carried out the molecular genetic studies, analyzed the genetic and environmental information and wrote the article. CMV and GMA director of research, participated in the study design, management of research and data analysis. RMU: conducted the anthropometric component of the study. AE led the statistical analysis. FAP and BEP participated study design. MVP and GB conducted the genetic component. All authors read and approved the final manuscript contributed to the analysis and writing of the article.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
The young people and their parents gave their approval and signed the consent for the protocol, which included the considerations referred to in the Declaration of Helsinki and which was approved by University of Antioquia’s University Research Bioethics Committee (Comité de Bioética de la Sede de Investigación Universitaria – CBEIM-SIU) in Medellín, Colombia.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- %BF
Body fat percentage
- MetS
Metabolic syndrome
- ABCA1
ATP-binding cassette, sub-family A (ABC1), member 1
- ACE
Angiotensin-converting enzyme
- AGT
Angiotensinogen
- AIMs
Ancestry informative markers
- ANOVA
Analysis of variance
- APO2
Apolipoprotein A-II
- BMI
Body mass index
- CAPN10
Calpain 10
- CETP
Cholesteryl ester transfer protein
- CIs
Confidence intervals
- ENSIN
National Nutritional Situation Survey, 2005
- FTO
Fat mass and obesity-associated
- GIANT Consortium
Genetic Investigation of ANthropometric Traits Consortium
- GRS
Genetic risk score
- IRS2
Insulin receptor substrate 2
- LPL
Lipoprotein lipase
- OR
Odds ratio
- SNP
Single nucleotide polymorphism
- TCF7L2
Transcription factor 7-like 2
- UCP3
Uncoupling Protein-3
- WC
Waist circumference
- WHO
World Health Organization
Additional file
Sequence of primer used to PCR. Table S2. Ancestral informative Markers (AIMs), allelic frequencies and delta (δ) of frequencies in the three ancestral populations (European, African and Amerindian). Table S3. Characteristics of the study population, stratified according to gender. Table S4. Association with anthropometric measures at 10 selected genetic variants on the subset of the samples with available ancestry information. Table S5. Interaction between SNP and socioeconomic and perinatal factors in determining body mass index. Figure S1. The histogram of body mass index, body fat percentage and waist circumference by gender. (DOCX 123 kb)
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12263-017-0567-1) contains supplementary material, which is available to authorized users.
References
- 1.Choudhary AK, Donnelly LF, Racadio JM, Strife JL. Diseases associated with childhood obesity. AJR Am J Roentgenol. 2007;188:1118–1130. doi: 10.2214/AJR.06.0651. [DOI] [PubMed] [Google Scholar]
- 2.Lusis AJ, Attie AD, Reue K. Metabolic syndrome: from epidemiology to systems biology. Nat Rev Genet. 2008;9:819–830. doi: 10.1038/nrg2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moraeus L, Lissner L, Yngve A, Poortvliet E, AI-Ansari U, Sjöberg A. Multi-level influences on childhood obesity in Sweden: societal factors, parental determinants and child’s lifestyle. Int J Obes (Lond) 2012;36:969–976. doi: 10.1038/ijo.2012.79. [DOI] [PubMed] [Google Scholar]
- 4.Pankov IA. Genetic variations in energy balance regulation. Biomed Khim. 2010;56:152–167. doi: 10.18097/pbmc20105602152. [DOI] [PubMed] [Google Scholar]
- 5.Pendergrass SA, Brown-Gentry K, Dudek S, Frase A, Torstenson ES, Goodloe R, Ambite JL, Avery CL, Buyske S, Bůžková P, Deelman E, Fesinmeyer MD, Haiman CA, Heiss G, Hindorff LA, Hsu CN, Jackson RD, Kooperberg C, Le Marchand L, Lin Y, Matise TC, Monroe KR, Moreland L, Park SL, Reiner A, Wallace R, Wilkens LR, Crawford DC, Ritchie MD. Phenome-wide association study (PheWAS) for detection of pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network. PLoS Genet. 2013;9:e1003087. doi: 10.1371/journal.pgen.1003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu YJ, Liu PY, Long J, Lu Y, Elze L, Recker RR, Deng HW. Linkage and association analyses of the UCP3 gene with obesity phenotypes in Caucasian families. Physiol Genomics. 2005;22:197–203. doi: 10.1152/physiolgenomics.00031.2005. [DOI] [PubMed] [Google Scholar]
- 7.Patel YM, Lane MD. Role of calpain in adipocyte differentiation. Proc Natl Acad Sci U S A. 1999;96:1279–1284. doi: 10.1073/pnas.96.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karra E, O’Daly OG, Choudhury AI, Yousseif A, Millership S, Neary MT, Scott WR, Chandarana K, Manning S, Hess ME, Iwakura H, Akamizu T, Millet Q, Gelegen C, Drew ME, Rahman S, Emmanuel JJ, Williams SC, Rüther UU, Brüning JC, Withers DJ, Zelaya FO, Batterham RL. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J Clin Invest. 2013;123:3539–3551. doi: 10.1172/JCI44403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takamoto I, Kubota N, Nakaya K, Kumagai K, Hashimoto S, Kubota T, Inoue M, Kajiwara E, Katsuyama H, Obata A, Sakurai Y, Iwamoto M, Kitamura T, Ueki K, Kadowaki T. TCF7L2 in mouse pancreatic beta cells plays a crucial role in glucose homeostasis by regulating beta cell mass. Diabetologia. 2014;57:542–553. doi: 10.1007/s00125-013-3131-6. [DOI] [PubMed] [Google Scholar]
- 10.Withers DJ, Burks DJ, Towery HH, Altamuro SL, Flint CL, White MF. Irs-2 coordinates Igf-1 receptor-mediated beta-cell development and peripheral insulin signaling. Nat Genet. 1999;23:32–40. doi: 10.1038/12631. [DOI] [PubMed] [Google Scholar]
- 11.Darimont C, Vassaux G, Ailhaud G, Negrel Differentiation of preadipose cells: paracrine role of prostacyclin upon stimulation of adipose cells by angiotensin-II. Endocrinology. 1994;135:2030–2036. doi: 10.1210/endo.135.5.7956925. [DOI] [PubMed] [Google Scholar]
- 12.Adeyemo A, Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics. 2010;13:72–79. doi: 10.1159/000218711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson W, Krueger RF. Genetic effects on physical health: lower at higher income levels. Behav Genet. 2005;35:579–590. doi: 10.1007/s10519-005-3598-0. [DOI] [PubMed] [Google Scholar]
- 14.Cardel M, Higgins PB, Willig AL, Keita AD, Casazza K, Gower BA, Fernandez JR. African genetic admixture is associated with body composition and fat distribution in a cross-sectional study of children. Int J Obes (Lond) 2011;35:60–65. doi: 10.1038/ijo.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson MR, Hemani G, Medina-Gomez C, Mezzavilla M, Esko T, Shakhbazov K, Powell JE, Vinkhuyzen A, Berndt S, Gustafsson S, Justice AE, Kahali B, Locke AE, Pers TH, Vedantam S, Wood AR, van Rheenen W, Andreassen OA, Gasparini P, Metspalu A, Berg LH, Veldink JH, Rivadeneira F, Werge TM, Abecasis GR, Boomsma D, Chasman D, de Geus EJ, Frayling TM, Hirschhorn JN, Hottenga JJ, Ingelsson E, Loos RJ, Magnusson PK, Martin NG, Montgomery GW, North KE, Pedersen NL, Spector TD, Speliotes EK, Goddard ME, Yang J, Visscher PM. Population genetic differentiation of height and body mass index across Europe. Nat Genet. 2015;47:1357–1362. doi: 10.1038/ng.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan MM, Kraemer A. Factors associated with being underweight, overweight and obese among ever-married non-pregnant urban women in Bangladesh. Singapore Med J. 2009;50:804–813. [PubMed] [Google Scholar]
- 17.Subramanian SV, Smith GD. Patterns, distribution, and determinants of under- and overnutrition: a population-based study of women in India. Am J Clin Nutr. 2006;84:633–640. doi: 10.1093/ajcn/84.3.633. [DOI] [PubMed] [Google Scholar]
- 18.Corella D, Carrasco P, Sorlí JV, Coltell O, Ortega-Azorín C, Guillén M, González JI, Sáiz C, Estruch R, Ordovas JM. Education modulates the association of the FTO rs9939609 polymorphism with body mass index and obesity risk in the Mediterranean population. Nutr Metab Cardiovasc Dis. 2012;22:651–658. doi: 10.1016/j.numecd.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pigeyre M, Bokor S, Romon M, Gottrand F, Gilbert CC, Valtueña J, Gómez-Martínez S, Moreno LA, Amouyel P, Dallongeville J, Meirhaeghe A, HELENA Study group Influence of maternal educational level on the association between the rs3809508 neuromedin B gene polymorphism and the risk of obesity in the HELENA study. Int J Obes (Lond) 2010;34:478–486. doi: 10.1038/ijo.2009.260. [DOI] [PubMed] [Google Scholar]
- 20.Adair LS, Fall CH, Osmond C, Stein AD, Martorell R, Ramirez-Zea M, Sachdev HS, Dahly DL, Bas I, Norris SA, Micklesfield L, Hallal P, Victora CG, COHORTS group Associations of linear growth and relative weight gain during early life with adult health and human capital in countries of low and middle income: findings from five birth cohort studies. Lancet. 2013;382:525–534. doi: 10.1016/S0140-6736(13)60103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agudelo GM, Bedoya G, Estrada A, Patiño FA, Muñoz AM, Velásquez CM. Variations in the prevalence of metabolic syndrome in adolescents according to different criteria used for diagnosis: Which definition should be chosen for this age group? Metab Syndr Relat Disord. 2014;12:202–209. doi: 10.1089/met.2013.0127. [DOI] [PubMed] [Google Scholar]
- 22.Instituto Colombiano de Bienestar Familiar, Profamilia, Instituto Nacional De Salud, Escuela de Nutrición y Dietética Universidad de Antioquia, Organización Panamericana de la Salud: Encuesta nacional de la situación nutricional en Colombia. Bogotá: Panamericana Formas e Impresos, S.A; 2006.
- 23.Departamento Administrativo Nacional de Estadística (DANE). Modelo de reglamento del comité Permanente de Estratificación socioeconómica. https://www.dane.gov.co/files/geoestadistica/Modelo_de_Reglamento_CPE.pdf. Accessed 9 July 2016.
- 24.Marshall W, Tanner J. Variations in pattern of pubertal changes in girls. Arch Dis Child. 1969;44:291–303. doi: 10.1136/adc.44.235.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall W, Tanner J. Variations in pattern of pubertal changes in boys. Arch Dis Child. 1970;42:13–23. doi: 10.1136/adc.45.239.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Onis M, Onyango AW, Borghi E, Siyam A, Nishida C, Siekmann J. Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Organ. 2007;85:660–667. doi: 10.2471/BLT.07.043497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez JR, Redden DT, Pietrobelli A, Allison DB. Waist circumference percentiles in nationally representative samples of African-American, European-American, and Mexican-American children and adolescents. J Pediatr. 2004;145:439–444. doi: 10.1016/j.jpeds.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 28.Lohman T, Roche A, Martorell RE. Anthropometric standardization reference manual. Champaign: Human Kinetics Books; 1988. [Google Scholar]
- 29.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chacon-Duque JC, Adhikari K, Avendano E, Campo O, Ramirez R, Rojas W, Ruiz-Linares A, Restrepo BN, Bedoya G. African genetic ancestry is associated with a protective effect on Dengue severity in colombian populations. Infect Genet Evol. 2014;27:89–95. doi: 10.1016/j.meegid.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rankinen T, Sarzynski MA, Ghosh S, Bouchard C. Are there genetic paths common to obesity, cardiovascular disease outcomes, and cardiovascular risk factors? Circ Res J Hum. 2015;116:909–922. doi: 10.1161/CIRCRESAHA.116.302888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sethi AA, Nordestgaard BG, Tybjaerg-Hansen A. Angiotensinogen gene polymorphism, plasma angiotensinogen, and risk of hypertension and ischemic heart disease: a meta-analysis. Arterioscler Thromb Vasc Biol. 2003;23:1269–1275. doi: 10.1161/01.ATV.0000079007.40884.5C. [DOI] [PubMed] [Google Scholar]
- 34.Skov J, Persson F, Frokiaer J, Christiansen JS. Tissue Renin-Angiotensin systems: a unifying hypothesis of metabolic disease. Front Endocrinol (Lausanne) 2014;5:23. doi: 10.3389/fendo.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalupahana NS, Moustaid-Moussa N. The renin-angiotensin system: a link between obesity, inflammation and insulin resistance. Obes Rev. 2012;13:136–149. doi: 10.1111/j.1467-789X.2011.00942.x. [DOI] [PubMed] [Google Scholar]
- 36.Massiéra F, Bloch-Faure M, Ceiler D, Murakami K, Fukamizu A, Gasc JM, Quignard-Boulange A, Negrel R, Ailhaud G, Seydoux J, Meneton P, Teboul M. Adipose angiotensinogen is involved in adipose tissue growth and blood pressure regulation. FASEB J. 2001;15:2727–2729. doi: 10.1096/fj.01-0457fje. [DOI] [PubMed] [Google Scholar]
- 37.Schling P, Mallow H, Trindl A, Loffler G. Evidence for a local renin angiotensin system in primary cultured human preadipocytes. Int J Obes Relat Metab Disord. 1999;23:336–341. doi: 10.1038/sj.ijo.0800821. [DOI] [PubMed] [Google Scholar]
- 38.Saint-Marc P, Kozak LP, Ailhaud G, Darimont C, Negrel R. Angiotensin II as a trophic factor of white adipose tissue: stimulation of adipose cell formation. Endocrinology. 2001;142:487–492. doi: 10.1210/endo.142.1.7883. [DOI] [PubMed] [Google Scholar]
- 39.Miki H, Yamauchi T, Suzuki R, Komeda K, Tsuchida A, Kubota N, Terauchi Y, Kamon J, Kaburagi Y, Matsui J, Akanuma Y, Nagai R, Kimura S, Tobe K, Kadowaki T. Essential role of insulin receptor substrate 1 (IRS-1) and IRS-2 in adipocyte differentiation. Mol Cell Biol. 2001;21:2521–2532. doi: 10.1128/MCB.21.7.2521-2532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mammarella S, Romano F, Di Valerio A, Creati B, Esposito DL, Palmirotta R, Capani F, Vitullo P, Volpe G, Battista P, Della Loggia F, Mariani-Costantini R, Cama A. Interaction between the G1057D variant of IRS-2 and overweight in the pathogenesis of type 2 diabetes. Hum Mol Genet. 2000;9:2517–2521. doi: 10.1093/hmg/9.17.2517. [DOI] [PubMed] [Google Scholar]
- 41.Stefan N, Kovacs P, Stumvoll M, Hanson RL, Lehn-Stefan A, Permana PA, Baier LJ, Tataranni PA, Silver K, Bogardus Metabolic effects of the Gly1057Asp polymorphism in IRS-2 and interactions with obesity. Diabetes. 2003;52:1544–1550. doi: 10.2337/diabetes.52.6.1544. [DOI] [PubMed] [Google Scholar]
- 42.Kilpelainen TO, Zillikens MC, Stančákova A, Finucane FM, Ried JS, Langenberg C, Zhang W, Beckmann JS, Luan J, Vandenput L, Styrkarsdottir U, Zhou Y, Smith AV, Zhao JH, Amin N, Vedantam S, Shin SY, Haritunians T, Fu M, Feitosa MF, Kumari M, Halldorsson BV, Tikkanen E, Mangino M, Hayward C, Song C, Arnold AM, Aulchenko YS, Oostra BA, Campbell H, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 44.Villalobos-Comparan M, Flores-Dorantes T, Villarreal-Molina T, Rodríguez-Cruz M, García-Ulloa AC, Robles L, Huertas-Vázquez A, Saucedo-Villarreal N, López-Alarcón M, Sánchez-Muñoz F, Domínguez-López A, Gutiérrez-Aguilar R, Menjivar M, Coral-Vázquez R, Hernández-Stengele G, Vital-Reyes VS, Acuña-Alonzo V, Romero-Hidalgo S, Ruiz-Gómez DG, Riaño-Barros D, Herrera MF, Gómez-Pérez FJ, Froguel P, García-García E, Teresa Tusié-Luna M, Aguilar-Salinas CA, Canizales-Quinteros S. The FTO gene is associated with adulthood obesity in the Mexican population. Obesity (Silver Spring) 2008;16:2296–2301. doi: 10.1038/oby.2008.367. [DOI] [PubMed] [Google Scholar]
- 45.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung HK, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui CC, Gomez-Skarmeta JL, Nóbrega MA. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi Q, Kilpeläinen TO, Downer MK, Tanaka T, Smith CE, Sluijs I, Sonestedt E, Chu AY, Renström F, Lin X, Ängquist LH, Huang J, Liu Z, Li Y, Asif Ali M, Xu M, Ahluwalia TS, Boer JM, Chen P, Daimon M, Eriksson J, Perola M, Friedlander Y, Gao YT, Heppe DH, Holloway JW, Houston DK, Kanoni S, Kim YM, Laaksonen MA, et al. FTO genetic variants, dietary intake and body mass index: insights from 177,330 individuals. Hum Mol Genet. 2014;23:6961–6972. doi: 10.1093/hmg/ddu411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schrauwen P, Xia J, Walder K, Snitker S, Ravussin E. A novel polymorphism in the proximal UCP3 promoter region: effect on skeletal muscle UCP3 mRNA expression and obesity in male non-diabetic Pima Indians. Int J Obes Relat Metab Disord. 1999;23:1242–1245. doi: 10.1038/sj.ijo.0801057. [DOI] [PubMed] [Google Scholar]
- 48.Jun HS, Kim IK, Lee HJ, Lee HJ, Kang JH, Kim JR, Shin HD, Song J. Effects of UCP2 and UCP3 variants on the manifestation of overweight in Korean children. Obesity (Silver Spring) 2009;17:355–362. doi: 10.1038/oby.2008.531. [DOI] [PubMed] [Google Scholar]
- 49.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corella D, Arnett DK, Tucker KL, Kabagambe EK, Tsai M, Parnell LD, Lai CQ, Lee YC, Warodomwichit D, Hopkins PN, Ordovas JM. A high intake of saturated fatty acids strengthens the association between the fat mass and obesity-associated gene and BMI. J Nutr. 2011;141:2219–2225. doi: 10.3945/jn.111.143826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ. Weight in infancy and death from ischaemic heart disease. Lancet. 1989;2:577–580. doi: 10.1016/S0140-6736(89)90710-1. [DOI] [PubMed] [Google Scholar]
- 52.Loaiza S, Coustasse A, Urrutia-Rojas X, Atalah E. Birth weight and obesity risk at first grade in a cohort of Chilean children. Nutr Hosp. 2011;26:214–219. [PubMed] [Google Scholar]
- 53.Lillycrop KA, Burdge GC. Epigenetic mechanisms linking early nutrition to long term health. Best Pract Res Clin Endocrinol Metab. 2012;26:667–676. doi: 10.1016/j.beem.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Santos JL, Martínez JA, Pérez F, Albala C. Genetic epidemiology of obesity: family studies. Rev Med Chil. 2005;133:349–361. doi: 10.4067/S0034-98872005000300012. [DOI] [PubMed] [Google Scholar]
- 55.Delaney SK, Hultner ML, Jacob HJ, Ledbetter DH, McCarthy JJ, Ball M, Beckman KB, Belmont JW, Bloss CS, Christman MF, Cosgrove A, Damiani SA, Danis T, Delledonne M, Dougherty MJ, Dudley JT, Faucett WA, Friedman JR, Haase DH, Hays TS, Heilsberg S, Huber J, Kaminsky L, Ledbetter N, Lee WH, Levin E, Libiger O, Linderman M, Love RL, Magnus DC, et al. Toward clinical genomics in everyday medicine: perspectives and recommendations. Expert Rev Mol Diagn. 2016;16:521–532. doi: 10.1586/14737159.2016.1146593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferguson LR, De Caterina R, Görman U, Allayee H, Kohlmeier M, Prasad C, Choi MS, Curi R, de Luis DA, Gil Á, Kang JX, Martin RL, Milagro FI, Nicoletti CF, Nonino CB, Ordovas JM, Parslow VR, Portillo MP, Santos JL, Serhan CN, Simopoulos AP, Velázquez-Arellano A, Zulet MA, Martinez JA. Guide and Position of the International Society of Nutrigenetics/Nutrigenomics on Personalised Nutrition: Part 1 - Fields of Precision Nutrition. J Nutrigenet Nutrigenomics. 2016;9:12–27. doi: 10.1159/000445350. [DOI] [PubMed] [Google Scholar]


