Abstract
Background
In order to develop a more effective prophylactic HIV-1 vaccine it is important optimize the components, improve Envelope glycoprotein immunogenicity as well as to explore prime-boost immunization schedules. It is also valuable to include several HIV-1 subtype antigens representing the world-wide epidemic.
Methods
HIVIS-DNA plasmids which include Env genes of subtypes A, B and C together with Gag subtypes A and B and RTmut/Rev of subtype B were modified as follows: the Envelope sequences were shortened, codon optimized, provided with an FT4 sequence and an immunodominant region mutated. The reverse transcriptase (RT) gene was shortened to contain the most immunogenic N-terminal fragment and fused with an inactivated viral protease vPR gene. HIVISopt-DNA thus contains fewer plasmids but additional PR epitopes compared to the native HIVIS-DNA. DNA components were delivered intradermally to young Balb/c mice once, using a needle-free Biojector® immediately followed by dermal electroporation. Vaccinia-based MVA-CMDR boosts including Env gene E and Gag-RT genes A were delivered intramuscularly by needle, once or twice.
Results
Both HIVIS-DNA and HIVISopt-DNA primed humoral and cell mediated responses well. When boosted with heterologous MVA-CMDR (subtypes A and E) virus inhibitory neutralizing antibodies were obtained to HIV-1 subtypes A, B, C and AE. Both plasmid compositions boosted with MVA-CMDR generated HIV-1 specific cellular responses directed against HIV-1 Env, Gag and Pol, as measured by IFNγ ELISpot. It was shown that DNA priming augmented the vector MVA immunological boosting effects, the HIVISopt-DNA with a trend to improved (Env) neutralization, the HIVIS-DNA with a trend to better (Gag) cell mediated immune reponses.
Conclusions
HIVIS-DNA was modified to obtain HIVISopt-DNA that had fewer plasmids, and additional epitopes. Even with one DNA prime followed by two MVA-CMDR boosts, humoral and cell-mediated immune responses were readily induced by priming with either DNA construct composition. Priming by HIV-DNA augmented neutralizing antibody responses revealed by boosting with the vaccinia-based heterologous sequences. Cellular and antibody responses covered selected strains representing HIV-1 subtypes A, B, C and CRF01_AE. We assume this is related to the inclusion of heterologous full genes in the vaccine schedule.
Keywords: Immunology, Infectious disease, Vaccines, Virology
1. Introduction
The best and most impressive preventive measure against infectious diseases has been vaccination. Over twenty different infectious diseases are preventable today by immunization. However, several serious viral infections, such as human immunodeficiency viruses, most of the herpes viruses and the hepatitis C virus lack efficient vaccines. One common property of these viruses is that they rapidly become chronic or persistent before protective immunity is established. Attempts to develop prophylactic or therapeutic vaccines or a cure against HIV have been going on for more than three decades, with lessons learned from several trials [1, 2, 3, 4, 5]. The Immune space presents variables such as immunoprofiles that can be used to assess and select future vaccine components [6].
Genetic/DNA vaccines have shown promise for priming of immune responses, both for antibody production and for cell-mediated immunity. In non-human models the induction of protective immunity to infectious diseases and tumors has been impressive [7] [8] [9] [10]. In humans and larger animals, difficulties were encountered with delivery systems and with expression of correct conformational proteins/peptides. A few protective or therapeutic schedules obtained by genetic vaccination were revealed [8, 11, 12, 13].
To obtain broad antibody and cell mediated immunities, the concept of prime-boost has proven effective. Heterologous vaccine strategies incorporating DNA, MVA and or HIV protein have been shown to augment and improve the quality of the immune response in mice [14] [15]. Both these approaches have met with modest success, and at best one of the prophylactic HIV-1 vaccine trials has obtained 60% early and 31,2% endpoint protective effects [16, 17]. These findings emphasize the importance to induce high quality long-term memory, recently acquired by a prime-boost schedule containing heterogeneous HIV-1 components [18, 19, 20] [21].
An HIV-1 vaccine for prophylactic use should be capable of eliciting potent antibody-mediated protection but also cell-mediated immunity against several HIV-1 subtypes, while a therapeutic vaccine is expected to work mainly by cell-mediated immunity and antibody dependent cellular immunities (ADCC) [22, 23].
HIVIS-DNA represents HIV-1 subtypes A, B and C [21, 24, 25]. The HIVIS-DNA has been the priming part followed by a vaccinia-based boost (MVA-CMDR) in several clinical trials that have induced broad and strong and long-lasting humoral and cellular immune reponses [19] [20] [26]. The priming schedule is however complex since several plasmids are combined. We have reduced the complexity in clinical trials by showing that intradermal needle-free delivery gives better immune responses than intramuscular [27], that higher doses in smaller volumes can improve responses [28] and that, in difference to animal studies, a combination of all plasmids results in as good responses as the separation of Gag from Env plasmids [29]. In order to reduce the complexity of the HIVISDNA and retain the reduced DNA amount to be given intradermally and still represent the various HIV antigens in separate plasmids, we optimized Env plasmids further by codon optimization, reduction in length and introduced an FT4 trimerization unit, thus eliminating the Rev plasmid that is not highly immunogenic. The RT plasmid was shortened to include the most prevalent epitopes and expressed together with an enzymatically destroyed PR which harbors several potent epitopes for cell mediated immunity. We thus obtained HIVISopt-DNA which was compared in an experimental system to reveal better immune responses or non-inferiority to HIVIS-DNA, finally for clinical purposes.
The murine model may be used to assess applicability of several variables that would benefit a human vaccine schedule. Generally we could establish a non-inferiority of the new HIV-DNA priming composition, and since these comparisons were done in parallel we might assume that the optimized HIVISopt-DNA can act to induce broad humoral and cellular immune responses in humans, similar to or better than the HIVIS-DNA, followed by the MVA-CMDR.
Finally, trials in humans need further preparation, such as selection and composition of plasmids in relation to one another, dosing of primes and boosts, and decisions on rapid or protracted immunization schedules to obtain long-term memory. The present aim was to refine immunization components and schedules using a preclinical model.
2. Materials and methods
2.1. Immunogens
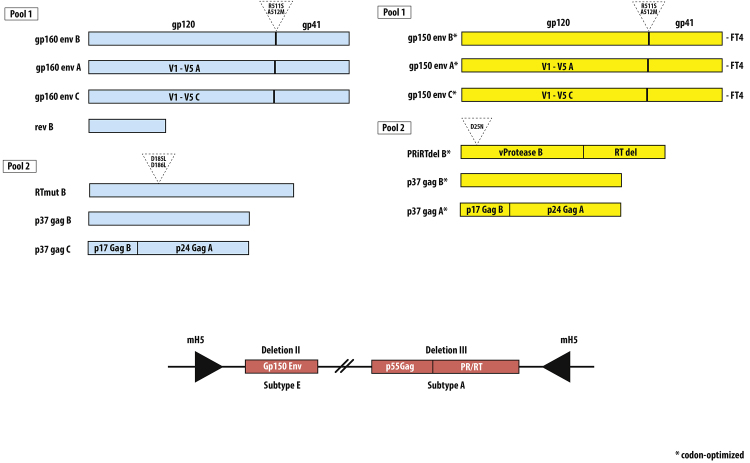
Fig. 1 HIVIS-DNA represents HIV-1 subtypes A, B and C as follows, Table 1: Gag p37 A and B, Env gp160 A, B, C, reverse transcriptase (RT) and Rev B [21] [24] [30] [31]. HIVISopt-DNA plasmids consist of gp150 subtypes A, B and C modified to contain FT4C-terminal ends to improve trimerization by T4 fibritin [32] together with p37 Gag A and B plus PRidelRT (aa mut, 413 aa) [33]. In summary, the original immunogen composition of HIVIS-DNA contains a Rev-expressing plasmid, which was exchanged for shortening of the Env plasmids at the C-terminal site, providing them with FT4 endings and a change of several nucleotide triplets to improve expression. The immunodominant gp41 region was mutated in all constructs. The RT plasmid was shortened to include the most prevalent epitopes inducing cell mediated immunity (Los Alamos) and expressed together with an enzymatically destroyed PR which harbors several potent epitopes for cell mediated immunity. Gag plasmids were not revised. Fig. 1 shows a schematic picture of vaccine compounds.
Fig. 1.
Overview of HIVIS-DNA and HIVISopt-DNA compounds. Blue color indicates components of HIVIS-DNA, yellow color indicates components of HIVISopt-DNA and red color indicates components of MVA-CMDR.
Table 1.
Immunization schedule of HIVIS-DNA, HIVISopt-DNA and HIV-MVA-CMDR.
| Groups of immunized animals | Day 0 | Days 21, 42 |
|---|---|---|
| 1 A. 1 x HIVIS-DNA; 1x MVA | DNA id/EP | 1 x MVA im |
| Content | Env A, B, C/Rev | Env E |
| Content | Gag p37A B, RT B | Gag Pol A |
| 1 B. 1 x HIVIS-DNA; 2 x MVA | DNA id/EP | 2 x MVA im |
| Content | Env A, B, C/Rev | Env E |
| Content | Gag p37A B, RT B | Gag Pol A |
| 2 A. 1 x HIVIS-DNAopt; 1 x MVA | DNAopt id/EP | 2 x MVA im |
| Content | Env A, B, C | Env E |
| Content | Gag p37A B, RTPR B | Gag Pol A |
| 2 B. 1 x HIVIS-DNAopt; 2 x MVA | DNAopt id/EP | 2 x MVA im |
| Content | Env A, B, C | Env E |
| Gag p37A B, RTPR B | Gag Pol A | |
| 3 A. 1 x MVA | MVA im | |
| Content | Env E | |
| Content | Gag A, Pol A | |
| 3 B. 2 x MVA | MVA im | 1 x MVA im |
| Content | Env E | Env E |
| Content | Gag A, Pol A | Gag A, Pol A |
| 4. Naive controls | none | none |
Five Balb/c mice were immunized per group. HIV-DNA doses: 200 μg DNA Gag and RT/PR, 200 μg Env A-C, Rev/mouse/immunization; id Bioject and id EP. 107 pfu of MVA-CMDR/mouse was divided equally between the right and left hind legs, 50 μl/side. Mice were sacrificed at day 30 (groups 1A, 2A, 3 A), or days 63-70 (groups 1B, 2B, 3B).
The recombinant vector virus, MVA-CDMR was designed and produced by the National Institutes of Health and Walter Reed Army Institute of Research (MD, USA) to express the following HIV-1 antigens, Gag p55 subtype A, protease and RT of subtype A and Env gp150 subtype E derived from CRF01_AE [34, 35, 36, 37].
In the prime-boost schedule genes for Env covered HIV-1 subtypes A, B, C and E, genes encoding Gag, Rev and RT/PR represented HIV-1 subtypes A and B (Tables 1, 2 , 3 , 4 , 5 and http://www.hiv.lanl.gov/ contain sequence information).
Table 2.
HIV-1 neutralizing serum titers (medians and ranges) from 1 x HIVIS-DNA or HIVISopt-DNA prime followed by 2 x MVA-CMDR.
| Group |
HIV–1 A 92UG29 R5 |
HIV-1 B 6920 R5 |
HIV-1C J10687 R5 |
HIV-1 AE 1525 |
HIV-1 III B LAI X4 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 50% NT | 80% NT | 50% NT | 80% NT | 50% NT | 80% NT | 50% NT | 80% NT | 50% NT | ||
| 1B | Median | 140 | 60 | 360 | 210 | 85 | 60 | 160 | 120 | 760 |
| Range | (60–320) | (<20–185) | (110- > 540) | (40–300) | (<20–210) | (<20–100) | (110–450) | (75–230) | ||
| Response | 5/5 | 4/5 | 5/5 | 5/5 | 4/5 | 2/5 | 5/5 | 5/5 | 5/5 | |
| 2B | Median | 100 | 20 | 625 | 240 | 220 | 140 | 160 | 75 | 700 |
| Range | (30–220) | (<20–100) | (260–880) | (60–540) | (70–300) | (75–170) | (40- > 540) | (<20–540) | ||
| Response | 5/5 | 2/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 3/5 | 5/5 | |
| 3B | Median | 160 | 55 | 80 | 40 | 40 | <20 | 60 | <20 | 110 |
| Range | (25–180) | (<20–140) | (30–170) | (<20–100) | (<20–55) | (<20) | (<20–320) | (<20–20) | ||
| Response | 5/5 | 2/5 | 5/5 | 1/5 | 2/5 | 0/5 | 4/5 | 0/5 | 5/5 | |
| 4 | Median | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| Range | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | |
| Response | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
HIV-1 isolates were used at 25–30 TCID50 concentration. 50% and 80% neutralization titers are show, as well as titer ranges between individuals. No. of responding individuals are shown for 50% neutralization of each strain.
Group 1B received Env components of subtype A (DNA and MVA), B (DNA), C (DNA), E (MVA).
Group 2B received Env components of subtype A (DNA and MVA), B (DNA), C (DNA), E (MVA).
Group 3B received Env components of subtype E (MVA).
Table 3.
ELISpot IFNγ cellular responses against peptide pools representing HIV-1 RT and PR of subtype B.
| Responder frequencies with HIV RT and PR peptides |
|||
|---|---|---|---|
| RT I | RT II | PR | |
| (aa 38–109) | (aa 110–179) | (aa 12–38) | |
| Groups of immunized animals | |||
| Group 1A | |||
| 1 x HIVIS; 1 x MVA | 3/5 | 5/5 | 2/5 |
| Median IFNγ SFC/106 | 40 | 45 | 144 |
| Range | (30–73) | (35–95) | (55–152) |
| Group 2A | |||
| 1 x HIVISopt; 1 x MVA | 2/5 | 3/5 | 3/5 |
| Median IFNγ SFC/106 | 33 | 105 | 196 |
| Range | (29–112) | (35–95) | (61–1744) |
| Group 3A | |||
| 2 x MVA | 1/5 | 2/5 | 2/5 |
| Median IFNγ SFC/106 | 96 | 167 | 108 |
| Range | 96 | (40–294) | (98–118) |
Abbreviations: RT I = N-terminal peptides of reverse transcriptase protein subtype B, RT II = C-terminal peptides of reverse transcriptase protein subtype B, PR = peptides representing the protease protein of subtype B.
Group 1A received RT components of subtype A (MVA), B (DNA); no PR.
Group 2A received RT components of subtype A (MVA), B (DNA); PR of subtype B (DNA).
Group 3A received RT components of subtype A (MVA); PR of subtype A (MVA).
Table 4.
HIVIS-DNA and HIV-MVA subtypes in prime-boost-boost schedules.
| HIV-1 protein expressed | HIV-1 subtype |
Name, accession nos.* | Vector | Ref. |
|---|---|---|---|---|
|
HIVIS-DNA prime |
||||
| Env gp160 | A1 | A1.UG92031 KY4773268 | pkCMV | Ljungberg et al 2002 |
| Env gp160 | B | B.FR.HXB2, KY4773267 | pkCMV | Ljungberg et al 2002 |
| Env gp160 | C | C.BR.92BR025, KY4773269 | pkCMV | Ljungberg et al 2002 |
| Gag p37 | A1/B | A1.UG92031 B.FR.HXB2 KY4773265 |
pkCMV | Bråve et al 2005 |
| Gag p37 | B | B.FR.HXB2 KY4773266 |
pkCMV | Bråve et al 2005 |
| Rev | B | B.FR.HXB2 KY4773270 |
pkCMV | Kjerrström et al 2001 |
| RTmut | B | B.FR.HXB2 KY4773271 |
pkCMV | Isaguliants et al 2000 |
| HIV-MVA boost | ||||
| Env gp150 | E | CRF01_AE CM235 |
MVA** | Earl et al 2009 |
| Gag p55 | A | CRF01_AE CM240 |
MVA | Earl et al 2009 |
| Pol (RTmut, PRmut) | A | CRF01_AE CM240 |
MVA | Earl et al 2009 |
HIV database: http://www.hiv.lanl.gov and Genbank gb-admin@ncbi.nml.nih.gov.**Modified Vaccinia Ankara MVAp579.
Table 5.
HIVISopt-DNA and MVA-CMDR subtypes in prime-boost schedules.
| HIV-1 proteins expressed | HIV-1 subtype |
Name, accession nos.* | Vector | Ref. |
|---|---|---|---|---|
| HIVISopt-DNA prime | ||||
| Env gp150 | A1opt | A1.UG92031 KY773272 |
pkCMV | Ljungberg et al 2002, Hallengärd 2014 |
| Env gp150 | Bopt | B.FR.HXB2, KY773273 | pkCMV | Ljungberg et al 2002, Hallengärd 2014 |
| Env gp150 | Copt | C.BR.92BR025 KY773274 | pkCMV | Ljungberg et al 2002, Hallengärd 2014 |
| Gag p37 | A1/B | A1.UG92031 B.FR.HXB2 KY773265 |
pkCMV | Bråve et al 2005 |
| Gag p37 | B | B.FR.HXB2 KY773266 |
pkCMV | Bråve et al 2005 |
| PRidelRT | B | B.FR.HXB2 KY773275 |
pkCMV | Hallengärd et al 2014 |
| HIV-MVA boost | ||||
| Env gp150 | E | CRF01_AE CM235 |
MVA** | Earl et al 2009 |
| Gag p55 | A | CRF01_AE CM240 |
MVA | Earl et al 2009 |
| Pol (RTmut, PRmut) | A | CRF01_AE CM240 |
MVA | Earl et al 2009 |
HIV database: http://www.hiv.lanl.gov and Genbank gb-admin@ncbi.nml.nih.gov.
Modified Vaccinia Ankara MVAp579.
2.2. Animals and immunizations
Female BALB/c mice (6–8 weeks old, H-2Dd) were housed at the Astrid Fagraeus animal facility at Karolinska Institutet. HIV-1 DNA-plasmids were dissolved in saline and given intradermally (ID) at two separate injection sites on the shaved flanks of the mice. Plasmids encoding Gag and RT were mixed and given intradermally on the shaved right flank (100 μl volumes, 200 μg, at a DNA concentration of 2 mg/ml). Plasmids encoding Env subtypes A, B and C with (HIVIS-DNA) or without (HIVISopt-DNA) Rev subtype B were mixed and given intradermally on the shaved left flank. Both immunizations were given by Biojector®, immediately followed by electroporation (EP) [15, 38, 39, 40, 41, 42]. EP was performed over the immunization site (DermaVax, Cellectis, Romainville, France; pulse frequency and amplitudes according to Roos et al. [38]). MVA-CMDR was given intramuscularly (IM) twice. Mice immunized with recombinant vaccinia vector were given 107 plaque forming units (PFU) of MVA-CDMR intramuscularly in the hind legs in 50 μl of phosphate buffered saline (PBS) [34]. Sera were sampled before and at 10–20 days after boost immunizations. Spleens were sampled at the end of each study (Table 1). In these, as well as in previous experimental studies, general health and significant and similar growth curves of individual mice participating in the prime-boost studies were noted. Ethical permission was obtained from the Northern Stockholm Board of Animal Ethics dnr N66/13.
2.3. Enzyme-linked immunosorbent assay
ELISA was performed with recombinant HIV-1 antigens: Recombinant proteins (1 μg/ml) Gag p55 (Protein Sciences, CT), Gag p17/p24 (ARP6010, Centre for AIDS Reagents, NIBSC) and CN54 gp140C (Polymun, AU), baculovirus recombinant protein subtype B gp160/LAI (Protein Sciences) and synthetic peptides (10 μg/ml) representing the HIV-1 gp120 subtype B V3-epitope (aa 308–323) or gp41 subtypes A, B, C). Gp41 peptides were synthesized as homologs of the antiretroviral compound T20 (Enfuvirtide, Roche, NL) represent the ELDKWAS epitope of broadly neutralizing human monoclonal antibody 2F5 from subtypes A (aa 652–661), B (aa 661–676) and C (aa 551–566). The CCR5 peptide represents the cellular HIV-1 co-receptor (Los Alamos database, http://www.hiv.lanl.gov/and Thermohybaid, DE). Microplates (Nunc Maxisorb, DK) were coated with 100 μl/well of each antigen and sera assayed as described [33, 43]. For peptide ELISAs, absorbance values are given for serum dilutions of 1:20.
2.4. Neutralization assay
Viral isolates used for neutralization derived from subtype A 92UG29/WHO strain, subtype B laboratory strain IIIB LAI, HIV-1 B/6920 R5, HIV-1 B/6794 X4, HIV-1C J10687 and HIV-1CRF01_AE 1525 (http://www.hiv.lanl.gov/, Table 2), selected from strains close to the DNA plasmid constructs [37] [31]. The III B LAI is the comparator sequence for subtype B (http://www.hiv.lanl.gov). Sera were studied individually or pooled and inactivated at 56 °C for 1 h to prevent complement-mediated neutralization. Sera were diluted in RPMI 1640 supplemented with 5% FCS and antibiotics (Invitrogen Life Technologies, SE) in 96-well tissue culture plates (Nunc Microwell plates, Pierce Thermo Scientific, Walthman, MA). Sera were assayed individually or pooled from mice with the highest serum IgG titers against the envelope proteins, and a second pool with sera from the animals with the lowest anti-Env IgG titers (Fig. 4). Each dilution was mixed with virus (25–80 TCID50) and incubated at 37 °C for 1 h followed by the addition of 105 human PBMCs activated by phytohemagglutinin (PHA) and rIL-2 (PeproTech, Rocky Hill, NJ) or CCR5 receptor-rich C8166 rR5 cells. The cells were incubated at 37 °C in 5% CO2 in air over night, washed twice with RPMI 1640 and received new medium. After 6 further days of culture, the presence of HIV-1 p24 antigen in the culture medium was measured by ELISA for HIV-1 subtypes A and B [43] or the CavidiTech Lenti-RT assay (Uppsala, Sweden) for HIV-1 subtypes C and CRF01_AE. The background of the p24 ELISA was determined for each plate and subtracted from all wells before the percentage neutralization was determined as [1-(mean p24 OD in the presence of test serum/mean p24 OD in the absence of test serum)] × 100. CavidiTech results were calculated as pg of RT/ml.
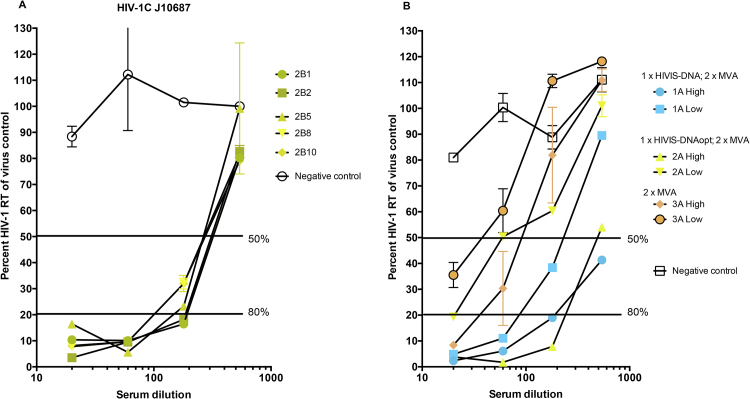
Fig. 4.
Virus neutralization results with sera from mice with high or low ELISA binding titers. A. Sera pooled from HIVIS-DNA, HIVISopt-DNA or MVA groups and naive mice were tested at dilutions 20, 60, 180 and 540 against HIV-1CRF01_AE strain 1525. B. Individual sera from HIVISopt-DNA boosted twice by MVA-CMDR were tested at dilutions 20, 60, 180 and 540 against HIV-1 subtype C strain J10687. Numbers 2B1-2B10 indicate single mouse identifications. RT measurements define viral replication, lower concentrations reflect a better neutralization. Blue color indicates components of HIVIS-DNA, yellow color indicates components of HIVISopt-DNA and red color indicates components of MVA-CMDR.
2.5. ELISpot assays
Cell-mediated immune responses were analyzed by interferon gamma (IFNγ) ELISpot assays (MabTech, Nacka, SWEDEN). Anti-IFNγ pre-coated ELISpot plates were used, and another monoclonal antibody (Mab) AN18 for IFNγ detection. A total of 2 × 105 cells were plated per well and stimulated for 24 h with pools of overlapping peptides of HIV-1 representing MVA-CMDR peptides representing Env (subtype E), Gag (subtype A) and Pol (subtype A) of CRF01_AE (2.5 μg/ml) (Nilsson Wahren 2015) (a kind gift from Dr. J. Cox, WRAIR, MD). Concanavalin A (Con A, 5 μg/ml, Sigma-Aldrich, S:t Louis, MA) was used as a positive control, peptide CCR5 as a background control and culture medium (MDEM with 2 mM L-glutamine, 1% Penicillin-Streptomycin and 5% bovine calf serum) as negative control. ELISpots were developed with biotinylated detection Mab R4-6A2 (1 μg/ml) followed by streptavidine alkaline phosphatase (ALP) and 5-bromo-4-chloro-3-indolyl phosphate-nitroblue tetrazolium (BCIP/NBT) substrate. The number of spot-forming cells (SFCs) was determined using an ELISpot reader (BioSys, BioReader Autoplate 5000, Karben, Germany). The HIV-1CRF01_AE peptides contain subtype A for Gag and Pol (RT and PR), subtype E for Env (Table 3 and Tables 4 and 5).
2.6. Statistical analysis
Statistical analyses were performed using Prism 5 GraphPad Software, CA. Statistical methods used were the non-parametric Mann-Whitney U test and Kruskal- Wallis non-parametric analysis with Dunńs correction.
3. Results
3.1. Binding antibodies
An overview of antigens used is given in Fig. 1. Groups of Balb/c mice were primed once by HIV-1HIVIS-DNA (Groups 1 A and B) or HIVISopt-DNA (Groups 2 A and B), followed by one or two MVA-CMDR boosts, or given only MVA-CMDR once or twice (Groups 3 A and B) (Table 1).
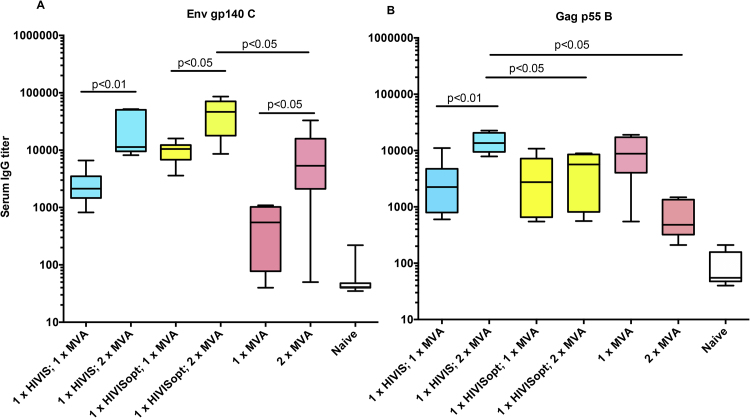
Anti-Env gp140C and anti-Env gp160 B (not shown) binding antibodies were primed by both HIVIS-DNA and HIVISopt-DNA and were considerably boosted by the second MVA-CMDR boost (Fig. 2A, p < 0.01, p < 0.05 and p < 0.05). The group primed by HIVISopt-DNA following the first MVA-CMDR boost obtained slightly higher (ns) median titers than the group primed by the native HIVIS-DNA. Following the second MVA-CMDR boost, both DNA primed groups obtained high and similar titers (p = 0.29, median titers 10 000–50 000). Without DNA priming, the MVA-CMDR second immunization resulted in significant boosting of anti-Env (p < 0.05) with an endpoint median titer of around 5 000. The highest median endpoint titer of around 50 000 against Env was obtained in the HIVISopt-DNA primed group. It is notable that the group primed with HIVISopt-DNA (both primes contain a plasmid expressing Env C) primed group gave higher reactivity to Env gp140C antigen than the non-primed group after two MVA-CMDR immunizations (MVA-CMDR does not contain Env C) (Fig. 2A, p < 0.05).
Fig. 2.
Antibody titers. A.Serum IgG ELISA titers against HIV-1 Env gp140C. B. Serum IgG ELISA titers against HIV-1 Gag p55 B were compared after one and two MVA-CMDR boost immunizations. Median titers and ranges are shown as box plots. Significant differences after one and two boosts are shown, as analyzed by the non-parametric Mann-Whitney U test. Blue color indicates components of HIVIS-DNA, yellow color indicates components of HIVISopt-DNA and red color indicates components of MVA-CMDR.
Antibody measurements to Gag p55 B (Fig. 2B) shows that HIVIS-DNA and HIVISopt-DNA induce similar levels of antibodies after the first boost with MVA-CMDR. After the second boost with MVA-CMDR, the HIVIS-DNA had primed antibody titers to Gag to higher levels than HIVISopt-DNA (p < 0.05). There is no difference in Gag plasmid composition between HIVIS and HIVISopt immunizations (Table 1). The highest median binding endpoint titer against Gag was obtained in the HIVIS-DNA primed group, around 15 000. A substantial reduction in antibody titers to Gag was observed after the second injection of MVA-CMDR.
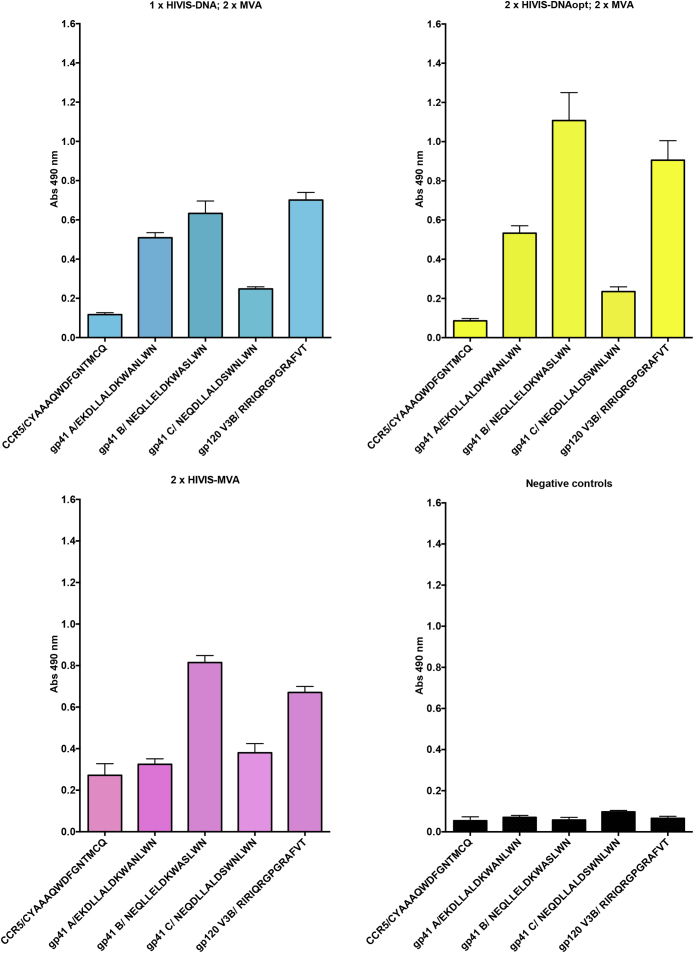
Sera from five animals were pooled within groups in an attempt to identify anti-peptide serological immune responses. The gp120 V3 peptide was recognized by all groups (Fig. 3A-C). The Gp41 peptides were selected from sites of HIV-1 subtypes A, B and C strains assumed to induce broad neutralization (see Methods). Here, the gp41 subtype B peptides were clearly recognized by all immunized groups, while the subtype A gp41 peptide was weakly recognized only by HIV-DNA primed animals. Control peptides representing the human HIV co-receptor CCR5 of cellular origin and not present in the vaccines were used to estimate background antibody levels. Pre-immune sera had very low responses to any peptide (Fig. 3D).
Fig. 3.
Anti-peptide gp41 and anti-V3 titers. A. Serum IgG ELISA reactivity of HIVIS-DNA against HIV-1 envelope peptides representing HIV-1 subtypes A, B and C epitopes tested after HIVIS-DNA and two MVA-CMDR boost immunizations. B. Serum IgG ELISA reactivity of HIVISopt-DNA against HIV-1 envelope peptides representing HIV-1 subtypes A, B and C epitopes tested after HIVISopt-DNA and two MVA-CMDR boost immunizations. C. Serum IgG ELISA reactivity of MVA-CMDR against HIV-1 envelope peptides representing HIV-1 subtypes A, B and C epitopes was tested after two MVA-CMDR boost immunizations. D. Serum IgG ELISA reactivity in non-immunized mice against HIV-1 envelope peptides representing HIV-1 subtypes A, B and C epitopes. Absorbance values are given for serum dilutions of 120. + Blue color indicates components of HIVIS-DNA, yellow color indicates components of HIVISopt-DNA and red color indicates components of MVA-CMDR.
3.2. Neutralizing antibodies
Neutralizing titers were assayed against viruses of HIV-1 subtypes A, B, C and CRF01_AE strains (Table 2). Neutralization titers were assessed after HIVIS-DNA or HIVISopt-DNA priming and twice boosted by MVA-CMDR. There was no major difference between the neutralization titers between HIVIS-DNA and HIVISopt-DNA primed mice after 2 MVA-CMDR boosts. Two immunizations with MVA-CMDR also resulted in neutralizing titers to all strains, with generally lower median titers. The HIV-1CRF01_AE strain was neutralized by all groups at titers of 60–160.
DNA priming generally increased median neutralizing titers to subtypes B and C. Prime-boosted groups had the highest median neutralization titers of around 700–760 against the consensus HIV-1 III B strain. Sera from non-immunized controls (and pre-immunization sera, not shown) were all negative (Table 2).
Fig. 4A shows a detailed picture of virus subtype CRF01_AE neutralization, indicating that pooled sera from mice with high binding titers from the HIVISopt-DNA group had IC50 titers of 220 while pooled sera with high binding titer from the HIVIS-DNA group had titers of around 200, indicating that two MVA-CMDR (AE) boosts induce similar titers to AE strains irrespective of priming. Fig. 4B shows an overview of subtype C neutralization with individual sera from animals receiving a single HIVISopt-DNA prime followed by two MVA-CMDR boosts (group 2 B see Table 1). All five animal sera neutralized HIV-1 subtype C, with a median neutralization titer of 220.
3.3. Cell mediated immune responses
After one HIV-DNA priming followed by a first and second MVA-CMDR boost we noted cell mediated responses to peptides derived from Env (subtype E), Gag (subtype A) and Pol (subtype A) (Fig. 5A and B).
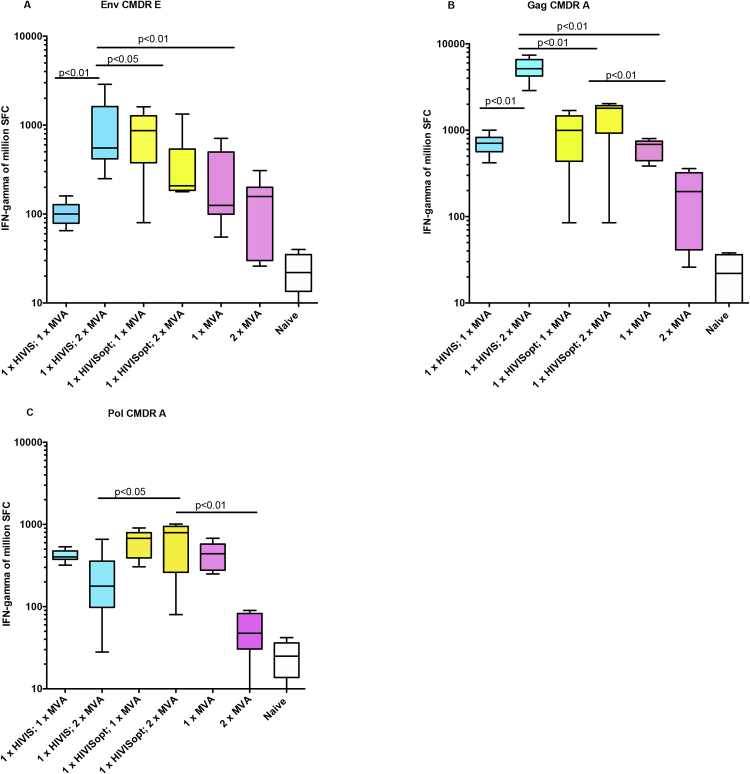
Fig. 5.
Cell mediated immune responses. A. Median and ranges of IFNγ ELISpot reactivity against HIV-1 peptide pools representing Env E. B. Median IFNγ ELISpot reactivity against HIV-1 peptide pools representing Gag A. C. Median IFNγ ELISpot reactivity against HIV-1 peptide pools representing Pol A. The figures show responses obtained after DNA prime and one or two MVA-CMDR boost immunizations. Blue color indicates components of HIVIS-DNA, yellow color indicates components of HIVISopt-DNA and red color indicates components of MVA-CMDR.
Fig. 5A compares the Env-specific cell mediated responses by spot forming spleen cells (SFC) secreting IFNγ following HIVIS-DNA or HIVISopt-DNA boosted with MVA-CMDR once or twice. HIVIS-DNA primed mice had lower reactivity against Env CMDR after the 1st MVA-CMDR boost than HIVISopt-DNA primed group. The second MVA-CMDR boost increased reactivity considerably in the HIVIS-DNA primed group (p < 0.01). After HIVISopt-DNA priming and MVA-CMDR boosting there was no further increase.
Gag reactivity after HIVIS-DNA or HIVISopt-DNA priming was boosted by one MVA-CMDR immunization (Fig. 5B). Two MVA-CMDR boosts raised IFNγ spots significantly for the HIVIS-DNA primed group (p < 0.01). The cell mediated responses were lower when the group that received MVA-CMDR only, had not been primed (p < 0.01).
Pol peptides of the CMDR subtype A origin were used for a similar experiment, Fig. 5C. Here all three animal groups showed 400–800 SFC/million spleen cells after one MVA-CMDR boost. Two MVA-CMDR immunizations did not increase the anti-Pol cellular immunity (Fig. 5C), similar to findings with the Env or Gag peptide antigens (Fig. 5A and B). It is likely that addition of PR sequences to HIVISopt-DNA (Table 3) contributed to the higher (p < 0.05) cellular reactivity following HIVISopt-DNA priming and MVA boosting.
The Con A activation of spleen cells from immunized mice showed a median of around 8 000–10 000 SFC/106 cells, the medium control 40–70 SFC/106 cells (not shown). Control non-immunized mice had a median of around 2 000 SFC/106 cells after Con A activation, and 30 SFC/106 cells with Env, Gag and Pol peptides.
Peptide pools representing segments of the HIV-1 subtype B Pol region were used in an attempt to map cellular responses (Table 3). The addition of PRidelRT plasmid to HIVISopt-DNA gave rise to cell mediated responses to both RT and PR peptides. In total, these responses were similar after priming with HIVIS-DNA or HIVISopt-DNA, however the variation of reactivity to these peptide pools was considerable. The highest median spot level was obtained for PR peptides after HIVISopt-DNA priming and one MVA-CMDR boost (196, range 61–1744 spots).
Thus, for cell mediated immunity, MVA-CMDR appears to be a strong boosting agent for both types of HIV-DNA priming whether HIVIS-DNA or HIVISopt-DNA (Fig. 5A and B), as measured by stimulation with CMDR-analogous peptides. The highest IFNγ spot forming units for Gag A peptides were discovered after one HIVIS-DNA prime and two HIV-MVA boosts. Gag A sequences are present in both the HIV-DNA plasmid compositions and in MVA-CMDR (Table 1).
4. Discussion
Recent clinical HIV-1 vaccine trials or studies in primates have used prime-boost schedules, often with combinations of DNA plasmids with viral vectors such as Pox- or Adenovirus–based vectors [1, 20, 44, 45, 46, 47, 48]. It is also anticipated that the novel idea of using a latency-forming vector, based on replicating Cytomegalovirus, may be helped by HLA-E/KIR in inducing novel CD8+ cellular responses [49, 50].
Broadly neutralizing humoral antibodies have been described following multi-clade HIV-DNA plasmids [51, 52] and recombinant protein booster immunizations in primates [47], rabbits [53], and rodents [21, 52, 54, 55]. In clinical studies with HIV-DNA prime, the virus-reducing activities of antibodies have been characterized as neutralization [56] or prominent ADCC reactivity [23], for a review see [22].
In a series of clinical studies we documented broad and durable immune responses to an HIV-DNA prime and modified Vaccina Ankara-based MVA-CMDR boost schedule, which warrants further exploration. DNA-MVA based HIV-1 vaccines induce both binding, neutralizing and high frequencies of ADCC antibodies to relevant conserved and variable sites of the HIV-1 subtypes A, B, C and AE [23] [57]. Likewise, cell mediated immune responses of long duration have been induced to several subtypes as represented by peptides overlapping the HIV-1 Env and Gag sequences [19, 20]. The studies have demonstrated dose sparing and simplification of DNA administration and priming in human trials [27, 28].
For larger clinical studies it is important to reduce the number of vaccine components while still not compromising the excellent broad cell mediated immunities and functional antibody responses of long duration. For this purpose we modified the HIV-DNA, MVA-based immunization schedule. Our rational was to retain representation of long Env and Gag proteins from multiple HIV subtypes, while reducing the number of plasmids and still add immunogenic epitopes. An overview of the subtype-specific antigens represented in the prime and boost immunizations is given in Table 1.
The relative immunological responses to two prime-boost schedules were evaluated in a murine model. HIVIS-DNA prime resulted in serum IgG binding titers against Env gp140 C, Env gp160 B, Gag p55 B, RT and PR B antigens. Mice receiving the HIVISopt-DNA prime followed by one MVA-CDMR boost responded with similar or higher IgG titers compared to those obtained by HIVIS-DNA and MVA-CDMR boost. Binding antibodies were seen to different subtypes of HIV-1, although it is difficult to determine whether such antibody binding cross-reactivities would extend to inhibit primary HIV-1 infection in humans.
Virus-neutralizing antibodies were found to several HIV-1 subtypes following both HIVIS-DNA and HIVISopt-DNA priming. Both DNA primed groups, which received B and C Env components in addition to the E Env component, acquired good neutralizing titers to the selected B and C HIV-1 strains. Thus, neutralizing activity to HIV-1 subtypes A, B, C and CRF01_AE was induced, indicating a broad response, similar to that found in clinical studies [2] [19]. We ascribe these results to the heterogenous composition of the vaccine, including full sequences from strains of subtypes A, B and C in the prime and E (AE) in the boost.
This was a first attempt to compare neutralizations between the two immunization schedules. We have therefore started to assay against strains of several subtypes with which we are familiar and also represent the subtypes included in the vaccine schedules. Human sera derived from clinical studies with HIVIS-DNA priming have been assayed with the TZM.bl as well as PBMC assays (Joachim et al. PlosOne 2016).
By measuring cell-mediated immune responses, multi-subtype reactivities were found related to the peptides of both A and E subtypes used for the stimulation of IFNγ secretion. Our previous studies have shown cellular reactivities by ELISpot and lymphoproliferation to peptide and proteins of subtypes A, B, C, D and E reagents of mice and humans [20, 58] (and unpublished). Both HIVIS-DNA and HIVISopt-DNA primed anti-Gag cellular responses as measured by ELISpot.
The HIVISopt-DNA primed a larger number of epitope responses to Pol, also anticipated since the antigen PR was added and RT modified in these plasmids. Expression of RT has [57] been shown to be significantly higher in the PRidRT construct of HIVISopt than that of RT from the RTmut construct of HIVIS-DNA [33]. In that study, PR related cell mediated immune responses occurred of around 2 000 SFC/106 cells, a reactivity that was similar or higher when PRidRT was combined with HIVIS-DNA plasmids [33].
The reasons for the MVA-CMDR not continuing to boost cell mediated immunity when given the second time as seen in the present study, cannot be easily explained. Similar phenomena occur with relative antigen overload or when timing between two strong antigens is too short. Two immunizations with MVA-CMDR as given here may require a longer time interval than 3 weeks between these potent boost immunizations to obtain the best contraction of primarily induced B- and T-cell immune responses. The MVA-CMDR humoral Env responses were boosted by the second HIV-MVA immunization, but Gag antibody and Gag cellular responses were not boosted. It is known that kinetics for humoral and cell mediated responses as well as for different antigens differ, and we may not have been able to access the optimal time point for both [59]. Here, this was evident for antibody induction to Gag and cell mediated reactivities to Gag and Pol. This work, as well as other studies, highlight that DNA vaccines potently prime potent immunogenicity in mice although this may not translate in nonhuman primates and human studies. In fact, the DNA prime and MVA boost model has shown variable levels of immunogenicity in humans [60] [20] [19] [1].The reasons for the variability between studies are multiple. Except for the species selected, the age and sex play roles [19]. The expression of plasmids and vectors may vary related to tissue in which they are delivered, facts that might be modulated with new formulations of DNA to better enhance immunogenicity.
Correct delivery and dosing of DNA are key questions for good expression of plasmid DNA. Both needle-less Biojector® and electroporation have been proven efficient means to increase numbers of transfected cells expressing antigens [2, 40, 41, 61, 62]. Delivery of DNA to mice appears to be optimal by Biojector® or electroporation targeted in the skin, which is resident to large numbers of dendritic cells [15]. In humans, the needle-less Biojector® works well after delivery of DNA plasmids to the skin, comparable to vaccine delivery by electroporation [2].
It is well known that mice respond well to DNA vaccines in contrast to many larger animals. The limitations of the presented results generated in Balb/c mice do not take into account dosing or delivery systems for humans, which we have shown to be important for the HIVIS-DNA priming in clinical trials.However, we feel that the small animal system can be used for comparisons between vaccine schedules related to certain criteria such as general trends for indicating whether B-cell or T-cell responses would be improved or similar.
We intended to show non-inferiority of the reduction and compression of antigens in the HIVIS-DNA vaccination schedule, which would be useful for clinical trials. This was successful and allows continuation with either the 6 or 7 component DNA vaccine.
Late boosts are essential to retain good immunological memory. Extended identification of remaining B- and T-cell memories have been revealed after DNA prime, MVA boosts in the clinic [20] [26]. The clinical study RV144 experienced a higher rate for protection early, which waned until the pre-decided endpoint [17]. It was notable in the present study, that priming once with DNA followed by one MVA boost induced higher immune responses than one MVA alone and at times even two MVA boosts. In a coming clinical trial it might therefore be optimal to start immunizations by one high dose composite DNA prime and one high dose MVA boost, followed at 2–3 yearly intervals with the same schedule. This would provide good initial priming and very long-term memory responses.
Declarations
Author contribution statement
Jorma Hinkula, Stefan Petkov: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Karl Ljungberg: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
David Hallengärd, Sumit Sharma: Performed the experiments; Wrote the paper.
Andreas Bråve, Merlin Robb, Bernard Moss: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Maria Isaguliants, Charlotta Nilsson: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Tina Falkeborn: Performed the experiments; Wrote the paper.
Valentina Liakina: Performed the experiments.
Mike Eller: Analyzed and interpreted the data; Wrote the paper.
Gunnel Biberfeld, Britta Wahren: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Eric Sandström: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Kerstin Markland, Pontus Blomberg: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by Doctors against AIDS association, Stockholm, and EU projects Vactrain, EAVI and Epiical.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We wish to acknowledge Richard Stout for providing Biojector® delivery devices for mice, Dietmar Katinger for provision of gp140C and advice, Gunnel Engström for excellent laboratory preparations of HIV-DNAs, Erik Rollman Waara, Sofia Stenler and Bo Hejdeman for interesting discussions on gp41 boosting effects. Stefan Petkov is a fellow of Vactrain, Maria Isaguliants and Britta Wahren are members of EU programs Vactrain and BAN, Britta Wahren is a member of EU programs EAVI and Epiical.
References
- 1.Goepfert P.A. Specificity and 6-month durability of immune responses induced by DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J. Infect. Dis. 2014;210(1):99–110. doi: 10.1093/infdis/jiu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nilsson C. HIV-DNA Given with or without Intradermal Electroporation Is Safe and Highly Immunogenic in Healthy Swedish HIV-1 DNA/MVA Vaccinees: A Phase I Randomized Trial. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0131748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deeks S.G. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat. Med. 2016;22(8):839–850. doi: 10.1038/nm.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Safrit J.T. Status of vaccine research and development of vaccines for HIV-1. Vaccine. 2016;34(26):2921–2925. doi: 10.1016/j.vaccine.2016.02.074. [DOI] [PubMed] [Google Scholar]
- 5.Stephenson K.E., D'Couto H.T., Barouch D.H. New concepts in HIV-1 vaccine development. Curr. Opin. Immunol. 2016;41:39–46. doi: 10.1016/j.coi.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manrique A. The immune space: a concept and template for rationalizing vaccine development. AIDS Res. Hum. Retroviruses. 2014;30(11):1017–1022. doi: 10.1089/aid.2014.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulmer J.B., Wahren B., Liu M.A. Gene-based vaccines: recent technical and clinical advances. Trends Mol. Med. 2006;12(5):216–222. doi: 10.1016/j.molmed.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Ault A. Immunogenicity and clinical protection against equine influenza by DNA vaccination of ponies. Vaccine. 2012;30(26):3965–3974. doi: 10.1016/j.vaccine.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis B.S. West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J. Virol. 2001;75(9):4040–4047. doi: 10.1128/JVI.75.9.4040-4047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fynan E.F. DNA vaccines: protective immunizations by parenteral, mucosal, and gene-gun inoculations. Proc. Natl. Acad. Sci. USA. 1993;90(24):11478–11482. doi: 10.1073/pnas.90.24.11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dayan G.H. Preclinical and clinical development of a YFV 17 D-based chimeric vaccine against West Nile virus. Viruses. 2013;5(12):3048–3070. doi: 10.3390/v5123048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trimble C.L. Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: a randomised, double-blind, placebo-controlled phase 2b trial. Lancet. 2015;386(10008):2078–2088. doi: 10.1016/S0140-6736(15)00239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahren B., Liu M. DNA vaccines: Recent developments and the future. Vaccines (Basel) 2014;2:785–796. [Google Scholar]
- 14.Ratto-Kim S. Heterologous prime-boost regimens using rAd35 and rMVA vectors elicit stronger cellular immune responses to HIV proteins than homologous regimens. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0045840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallengard D. A combination of intradermal jet-injection and electroporation overcomes in vivo dose restriction of DNA vaccines. Genet. Vaccines Ther. 2012;10(1):5. doi: 10.1186/1479-0556-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rerks-Ngarm S. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 17.Robb M.L. Risk behaviour and time as covariates for efficacy of the HIV vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAX B/E: a post-hoc analysis of the Thai phase 3 efficacy trial RV 144. Lancet Infect. Dis. 2012;12(7):531–537. doi: 10.1016/S1473-3099(12)70088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brave A. Induction of HIV-1-specific cellular and humoral immune responses following immunization with HIV-DNA adjuvanted with activated apoptotic lymphocytes. Vaccine. 2010;28(9):2080–2087. doi: 10.1016/j.vaccine.2009.12.040. [DOI] [PubMed] [Google Scholar]
- 19.Sandstrom E. Broad immunogenicity of a multigene: multiclade HIV-1 DNA vaccine boosted with heterologous HIV-1 recombinant modified vaccinia virus Ankara. J. Infect. Dis. 2008;198(10):1482–1490. doi: 10.1086/592507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsson C. Broad and potent cellular and humoral immune responses after a second late HIV-modified vaccinia virus ankara vaccination in HIV-DNA-primed and HIV-modified vaccinia virus Ankara-boosted Swedish vaccinees. AIDS Res. Hum. Retroviruses. 2014;30(3):299–311. doi: 10.1089/aid.2013.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brave A. Multigene/multisubtype HIV-1 vaccine induces potent cellular and humoral immune responses by needle-free intradermal delivery. Mol. Ther. 2005;12(6):1197–1205. doi: 10.1016/j.ymthe.2005.06.473. [DOI] [PubMed] [Google Scholar]
- 22.Chung A.W. Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell. 2015;163(4):988–998. doi: 10.1016/j.cell.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joachim A. Potent functional antibody responses elicited by HIV-I DNA priming and boosting with heterologous HIV-1 recombinant MVA in healthy Tanzanian adults. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0118486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjerrstrom A. Interactions of single and combined human immunodeficiency virus type 1 (HIV-1) DNA vaccines. Virology. 2001;284(1):46–61. doi: 10.1006/viro.2001.0905. [DOI] [PubMed] [Google Scholar]
- 25.Ljungberg K. Subtypes A, C, G, and recombinant HIV type 1 are circulating in Bangladesh. AIDS Res. Hum. Retroviruses. 2002;18(9):667–670. doi: 10.1089/088922202760019374. [DOI] [PubMed] [Google Scholar]
- 26.Joachim A. Three-Year Durability of Immune Responses Induced by HIV-DNA and HIV-Modified Vaccinia Virus Ankara and Effect of a Late HIV-Modified Vaccinia Virus Ankara Boost in Tanzanian Volunteers. AIDS Res. Hum. Retroviruses. 2017 doi: 10.1089/aid.2016.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakari M. Broad and potent immune responses to a low dose intradermal HIV-1 DNA boosted with HIV-1 recombinant MVA among healthy adults in Tanzania. Vaccine. 2011;29(46):8417–8428. doi: 10.1016/j.vaccine.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munseri P.J. Priming with a simplified intradermal HIV-1 DNA vaccine regimen followed by boosting with recombinant HIV-1 MVA vaccine is safe and immunogenic: a phase IIa randomized clinical trial. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0119629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brave A. Reduced cellular immune responses following immunization with a multi-gene HIV-1 vaccine. Vaccine. 2006;24(21):4524–4526. doi: 10.1016/j.vaccine.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Ljungberg K. Effective construction of DNA vaccines against variable influenza genes by homologous recombination. Virology. 2000;268(2):244–250. doi: 10.1006/viro.2000.0199. [DOI] [PubMed] [Google Scholar]
- 31.Ljungberg K. Enhanced immune responses after DNA vaccination with combined envelope genes from different HIV-1 subtypes. Virology. 2002;302(1):44–57. doi: 10.1006/viro.2002.1547. [DOI] [PubMed] [Google Scholar]
- 32.Yang X. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 2002;76(9):4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallengard D., Wahren B., Brave A. A truncated plasmid-encoded HIV-1 reverse transcriptase displays strong immunogenicity. Viral. Immunol. 2013;26(2):163–166. doi: 10.1089/vim.2012.0083. [DOI] [PubMed] [Google Scholar]
- 34.Earl P.L. Design and evaluation of multi-gene, multi-clade HIV-1 MVA vaccines. Vaccine. 2009;27(42):5885–5895. doi: 10.1016/j.vaccine.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su L. Characterization of a virtually full-length human immunodeficiency virus type 1 genome of a prevalent intersubtype (C/B') recombinant strain in China. J. Virol. 2000;74(23):11367–11376. doi: 10.1128/jvi.74.23.11367-11376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodenburg C.M. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res. Hum. Retroviruses. 2001;17(2):161–168. doi: 10.1089/08892220150217247. [DOI] [PubMed] [Google Scholar]
- 37.Cox J.H. Inclusion of a CRF01_AE HIV envelope protein boost with a DNA/MVA prime-boost vaccine: Impact on humoral and cellular immunogenicity and viral load reduction after SHIV-E challenge. Vaccine. 2012;30(10):1830–1840. doi: 10.1016/j.vaccine.2011.12.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roos A.K. Skin electroporation: effects on transgene expression, DNA persistence and local tissue environment. PLoS One. 2009;4(9) doi: 10.1371/journal.pone.0007226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kutzler M.A. Chemokine-adjuvanted electroporated DNA vaccine induces substantial protection from simian immunodeficiency virus vaginal challenge. Mucosal Immunol. 2016;9(1):13–23. doi: 10.1038/mi.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selby M. Enhancement of DNA vaccine potency by electroporation in vivo. J. Biotechnol. 2000;83(1-2):147–152. doi: 10.1016/s0168-1656(00)00308-4. [DOI] [PubMed] [Google Scholar]
- 41.Widera G. Increased DNA vaccine delivery and immunogenicity by electroporation in vivo. J. Immunol. 2000;164(9):4635–4640. doi: 10.4049/jimmunol.164.9.4635. [DOI] [PubMed] [Google Scholar]
- 42.Brave A. Vaccine delivery methods using viral vectors. Mol. Pharm. 2007;4(1):18–32. doi: 10.1021/mp060098+. [DOI] [PubMed] [Google Scholar]
- 43.Devito C. Cross-clade HIV-1-specific neutralizing IgA in mucosal and systemic compartments of HIV-1-exposed, persistently seronegative subjects. J. Acquir. Immune Defic. Syndr. 2002;30(4):413–420. doi: 10.1097/00042560-200208010-00007. [DOI] [PubMed] [Google Scholar]
- 44.Kopycinski J. Broad HIV epitope specificity and viral inhibition induced by multigenic HIV-1 adenovirus subtype 35 vector vaccine in healthy uninfected adults. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray G.E. Recombinant adenovirus type 5HIV gag/pol/nef vaccine in South Africa: unblinded, long-term follow-up of the phase 2b HVTN 503/Phambili study. Lancet Infect. Dis. 2014;14(5):388–396. doi: 10.1016/S1473-3099(14)70020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hulot S.L. Comparison of Immunogenicity in Rhesus Macaques of Transmitted-Founder, HIV-1 Group M Consensus, and Trivalent Mosaic Envelope Vaccines Formulated as a DNA Prime, NYVAC, and Envelope Protein Boost. J. Virol. 2015;89(12):6462–6480. doi: 10.1128/JVI.00383-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chamcha V. Strong, but Age-Dependent, Protection Elicited by a Deoxyribonucleic Acid/Modified Vaccinia Ankara Simian Immunodeficiency Virus Vaccine. Open Forum Infect. Dis. 2016 doi: 10.1093/ofid/ofw034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu H. Preferential infection of human Ad5-specific CD4 T cells by HIV in Ad5 naturally exposed and recombinant Ad5-HIV vaccinated individuals. Proc. Natl. Acad. Sci. USA. 2014;111(37):13439–13444. doi: 10.1073/pnas.1400446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen S.G. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science. 2013;340(6135) doi: 10.1126/science.1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen S.G. Broadly targeted CD8(+) T cell responses restricted by major histocompatibility complex E. Science. 2016;351(6274):714–720. doi: 10.1126/science.aac9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Currier J.R. Defining epitope coverage requirements for T cell-based HIV vaccines: theoretical considerations and practical applications. J. Transl. Med. 2011;9:212. doi: 10.1186/1479-5876-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rollman E. Multi-subtype gp160 DNA immunization induces broadly neutralizing anti-HIV antibodies. Gene Ther. 2004;11(14):1146–1154. doi: 10.1038/sj.gt.3302275. [DOI] [PubMed] [Google Scholar]
- 53.Wang S. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology. 2006;350(1):34–47. doi: 10.1016/j.virol.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 54.Liu L. Broad HIV-1 neutralizing antibody response induced by heterologous gp140/gp145 DNA prime-vaccinia boost immunization. Vaccine. 2012;30(28):4135–4143. doi: 10.1016/j.vaccine.2012.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muthumani K. HIV-1 Env DNA vaccine plus protein boost delivered by EP expands B- and T-cell responses and neutralizing phenotype in vivo. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0084234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine. 2008;26(31):3947–3957. doi: 10.1016/j.vaccine.2007.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joachim A. Boosting with Subtype C CN54rgp140 Protein Adjuvanted with Glucopyranosyl Lipid Adjuvant after Priming with HIV-DNA and HIV-MVA Is Safe and Enhances Immune Responses: A Phase I Trial. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0155702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hallengard D. Comparison of plasmid vaccine immunization schedules using intradermal in vivo electroporation. Clin. Vaccine Immunol. 2011;18(9):1577–1581. doi: 10.1128/CVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knudsen M.L. Kinetic and phenotypic analysis of CD8+ T cell responses after priming with alphavirus replicons and homologous or heterologous booster immunizations. J. Virol. 2014;88(21):12438–12451. doi: 10.1128/JVI.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mwau M. A human immunodeficiency virus 1 (HIV-1) clade A vaccine in clinical trials: stimulation of HIV-specific T-cell responses by DNA and recombinant modified vaccinia virus Ankara (MVA) vaccines in humans. J. Gen. Virol. 2004;85(Pt 4):911–919. doi: 10.1099/vir.0.19701-0. [DOI] [PubMed] [Google Scholar]
- 61.Vasan S. In vivo electroporation enhances the immunogenicity of an HIV-1 DNA vaccine candidate in healthy volunteers. PLoS One. 2011;6(5) doi: 10.1371/journal.pone.0019252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brave A. Biodistribution, persistence and lack of integration of a multigene HIV vaccine delivered by needle-free intradermal injection and electroporation. Vaccine. 2010;28(51):8203–8209. doi: 10.1016/j.vaccine.2010.08.108. [DOI] [PMC free article] [PubMed] [Google Scholar]