Abstract
Mitochondrial dysfunction is common in cancer and the mitochondrial electron transport chain is often affected in carcinogenesis. To date, little is known about the expression of the ATP synthase subunits in clear cell renal cell carcinoma (ccRCC). The NextBio database was used to determine an expression profile of the ATP synthase subunits based on published microarray studies. We observed down-regulation of 23 out of 29 subunits of the ATP synthase. Differential expression was validated exemplarily for 12 genes (ATP5A1, ATP5B, ATPAF1, ATP5C1, ATP5D, ATP5O, ATP5F1, ATP5G1, ATP5G2, ATP5G3, ATP5I, ATP5S; screening cohort ccRCC n = 18 and normal renal tissue n = 10) using real-time PCR. Additional eight genes (ATP5A1, ATP5B, ATPAF1, ATP5F1, ATP5G1, ATP5G2, ATP5G3, ATP5S) were internally validated within an enlarged cohort (ccRCC n = 74; normal renal tissue n = 36). Furthermore, down-regulation of ATP5A1, ATPAF1, ATP5G1/G2/G3 was confirmed on the protein level using Western Blot and immunohistochemistry. We observed that altered expression of ATPAF1 and ATP5G1/G2/G3 was correlated with overall survival in patients with ccRCC. In conclusion, down-regulation of many ATP Synthase subunits occurs in ccRCC and is the basis for the reduced activity of the mitochondrial electron chain. Alteration of the expression of ATP5A1, ATPAF1, and ATP5G1/G2/G3 is characteristic for ccRCC and may be prognostic for ccRCC patients' outcome.
Introduction
In 2012, renal tumors accounted for approximately 2% to 3% of all new cancer cases in the world, with cases occurring more frequently in developed regions [1]. The incidence of renal cell carcinoma (RCC) is increasing by 1.5% to 5.9% each year [1]. RCC with the predominant clear cell subtype is the most common renal tumor entity. Despite many efforts, there are no biomarkers available for patients with renal tumors; the identification of a suitable biomarker could aid the clinician in identifying patients needing treatment of a renal tumor.
In recent years, there has been an increasing interest in mitochondria because of their role as sensors and executioners of apoptosis [2], [3], [4] and their involvement in carcinogenesis [5], [6]. In 1924, Otto Warburg discovered that tumor cells receive energy from glycolysis rather than the mitochondrial oxidative phosphorylation. He established the hypothesis that faulty mitochondrial functions in tumor cells are major causes of carcinogenesis [7].
Usually, other metabolic pathways such as the mitochondrial electron transport chain are maintained along with the increased aerobic glycolysis [8]. However, some studies showed that, as opposed to other tumor types, most alternative pathways besides increased aerobic glycolysis are down-regulated in clear cell renal cell carcinomas (ccRCC) [9], [10], [11], [12]. Nevertheless, it is still not completely understood why ccRCC seems to knock down all other ways to earn energy except glycolysis.
The F1F0 ATP synthase represents the fifth complex of the mitochondrial electron transport chain and contains 29 subunits. It consists of two sub complexes, the membrane-spanning component (F0) and the water-soluble complex (F1). While the larger F1 particle, with its catalytic core, is composed by five different subunits (α, β, γ, δ and ε), the F0 particle embodies three main subunits (a, b and c) and, depending on the species, a various amount of additional ones (humans six: d, e, f, g, F6 and 8) [13].
The aim of this study was to develop a better understanding of the expression of ATP synthase in ccRCC and the determination of potential new ccRCC biomarkers.
Materials and Methods
NextBio Database
The NextBio database (Illumina, San Diego, CA, USA) was used to review published gene expression profiling studies (n = 17) [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29]. The last database query was performed on 20 August 2015. The retrieval for the comparison of normal renal and ccRCC tissue was restricted to the parameters of human, mRNA and fresh frozen tissue.
Patients
The prospective collection of tissue samples was performed within the framework of the Biobank at the CIO Cologne-Bonn. All patients underwent radical or partial nephrectomy at the Department of Urology at the University Hospital Bonn. Written informed consent for the collection of biomaterials was obtained from all patients and the study was approved by the local ethic committee (vote: 280/12).
The specimens were split for formalin fixation, paraffin embedding and fresh-frozen storage. Before use, the tissues were re-evaluated by an uropathologist and classified according the World Health Organization (WHO) classification from 2009. Fresh-frozen tissues were stored at −80 °C, and used for qPCR (92 ccRCC and 46 normal renal tissues) and Western Blot (corresponding 8 ccRCC and normal renal tissues) experiments. Formalin-fixed, paraffin embedded tissues were used for the construction of a tissue microarray with 191 RCC specimens [comprising 141 clear-cells, 29 papillary (pRCC), 10 chromophobe (chRCC) and 11 sarcomatoid (sRCC) RCC], 10 oncocytoma and 30 normal renal tissue samples were used for immunochemistry experiments [30]. The detailed clinical–pathological parameters are shown in Tables 1 and 2.
Table 1.
Clinical–Pathological Parameters of the Study Cohorts for PCR
| Screening Cohort |
Validation Cohort |
|||
|---|---|---|---|---|
| ccRCC |
Normal |
ccRCC |
Normal |
|
| n = 18 (%) | n = 10 (%) | n = 74 (%) | n = 36 (%) | |
| Sex | ||||
| Male | 11 (61.1) | 6 (60.0) | 53 (71.6) | 26 (72.2) |
| Female | 7 (38.8) | 4 (40.0) | 21 (28.4) | 10 (27.7) |
| Age | ||||
| Mean | 64.83 | 66.4 | 66.5 | 64.75 |
| Min–max | 43–83 | 46–89 | 38–89 | 43–86 |
| Pathological stage | ||||
| pT1 | 7 (38.8) | n.a. | 42 (56.8) | n.a. |
| pT2 | 2 (11.1) | n.a. | 7 (9.5) | n.a. |
| pT3 | 8 (44.4) | n.a. | 24 (32.4) | n.a. |
| pT4 | 1 (5.5) | n.a. | 1 (1.4) | n.a. |
| Vascular invasion | 7 (38.8) | n.a. | 24 (32.4) | n.a. |
| LN metastasis | 0 | n.a. | 2 (2.7) | n.a. |
| Distant metastasis | 1 (5.5) | n.a. | 14 (18.9) | n.a. |
| Grading | ||||
| Grade 1 | 1 (5.5) | n.a. | 9 (12.2) | n.a. |
| Grade 2 | 13 (72.2) | n.a. | 47 (63.5) | n.a. |
| Grade 3 | 3 (16.6) | n.a. | 15 (20.3) | n.a. |
| Grade 4 | 1 (5.5) | n.a. | 3 (4.1) | n.a. |
Table 2.
Clinical-Pathological Parameters of the Study Cohorts for Immunohistochemistry
| ccRCC |
pRCC |
chRCC |
sRCC |
RO |
normal |
|
|---|---|---|---|---|---|---|
| n = 141 (%) | n = 29 (%) | n = 10 (%) | n = 11 (%) | n = 10 (%) | n = 30 (%) | |
| Sex | ||||||
| Male | 88 (62.4) | 26 (89.6) | 6 (60.0) | 7 (63.6) | 0 | 21 (70.0) |
| Female | 53 (37.6) | 3 (10.3) | 4 (40.0) | 3 (27.3) | 10 (100) | 9 (30.0) |
| Age | ||||||
| Mean | 62,11 | 61,45 | 63,2 | 62,1 | 57,6 | 57,93 |
| Min–max | 26–85 | 35–82 | 27–85 | 51–75 | 26–73 | 28–80 |
| Pathological stage | ||||||
| pT1 | 59 (41.8) | 19 (65.5) | 6 (60.0) | 0 | n.a. | n.a. |
| pT2 | 31 (22.0) | 5 (17.2) | 4 (40.0) | 1 (9.1) | n.a. | n.a. |
| pT3 | 49 (34.8) | 5 (17.2) | 0 | 8 (72.7) | n.a. | n.a. |
| pT4 | 2 (1.4) | 0 | 0 | 1 (9.1) | n.a. | n.a. |
| Vascular invasion | 43 (30.5) | 0 | 0 | 8 (72.7) | n.a. | n.a. |
| LN metastasis | 8 (5.7) | 1 (3.4) | 0 | 5 (45.5) | n.a. | n.a. |
| Distant metastasis | 18 (12.8) | 3 (10.3) | 0 | 6 (54.5) | n.a. | n.a. |
| Grading | ||||||
| Grade 1 | 44 (31.2) | 11 (37.9) | 3 (30.0) | 0 | n.a. | n.a. |
| Grade 2 | 94 (66.7) | 16 (55.1) | 7 (70.0) | 1 (9.1) | n.a. | n.a. |
| Grade 3 | 3 (2.1) | 2 (6.9) | 0 | 7 (63.6) | n.a. | n.a. |
| Grade 4 | 0 | 0 | 0 | 2 (18.2) | n.a. | n.a. |
RO, renal oncocytoma; LN, Lymph node; n.a., not applicable.
Quantitative Real-Time PCR
RNA isolation was performed as described before [31]. In brief, total RNA was isolated with the mirVana miRNA Isolation Kit (Ambion, Foster City, CA, USA) and afterwards treated with DNase (DNA-free Kit, Ambion). RNA quantity was determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). RNA integrity was confirmed by evaluation of the 28S and 18S rRNA bands in a gel electrophoresis.
The expression of 12 differentially expressed genes (ATP5A1, ATP5B, ATPAF1, ATP5C1, ATP5D, ATP5O, ATP5F1, ATP5G1, ATP5G2, ATP5G3, ATP5I, ATP5S) was investigated in a screening cohort of 18 ccRCC and 10 normal renal tissue samples. Subsequently, the eight most down-regulated genes (ATP5A1, ATP5B, ATPAF1, ATP5F1, ATP5G1, ATP5G2, ATP5G3, ATP5S) were analyzed in a validation cohort (74 ccRCC and 36 normal renal tissues). cDNA was synthesized from 1 μg total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Saint-Germain-en Laye, France). For qPCR, we used 5 ng/μl cDNA templates with the 1× SYBR Premix Ex Taq II and ROX Plus with 10 pmol/μl forward/reverse primer (see Supplementary Table S1). PCR experiments were performed on an ABIPrism 7900 HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Data analysis was accomplished using QBase + (Biogazelle) with PPIA and ACTB [32], [33], earlier shown to be suitable reference genes, in the 2-ΔΔCT algorithm.
Western Blot
In order to validate gene expression at the protein level, we performed Western blot analyses for ATP5A1, ATPAF1, ATP5G1, ATP5G2 and ATP5G3; the experimental procedure was performed as described earlier [34]: Corresponding fresh-frozen tissues (50 mg ccRCC and normal renal tissue) from 8 patients (4× UICC stage I; 4× stage III) were homogenized in a Precellys 24 (Peqlab, Erlangen, Germany) with 400 μl Cell lysis Buffer (Cell Signaling, Cambridge, United Kingdom) including Complete Mini EDTA-free protease inhibitor (Roche, Basel, Switzerland). The protein concentration was determined (BCA Protein Assay Kit, Pierce Biotechnology, Rockford, IL, USA), 30 ng protein per well loaded into a NuPAGE 4–12% denaturating PAA Gel (Life Technologies, Carlsbad, CA, USA) and separated in a XCell4 SureLock electrophoresis system (Life Technologies). As molecular weight marker a biotinylated protein ladder (Cell Signaling Technology, Cambridge, UK) and PageRuler Prestained (Thermo Scientific, Waltham, MA, USA) was used. The specimens were transmitted on 0.2 μm nitrocellulose (XCell II, Life Technologies) and the proteins were blocked by 5% milk powder (Merck, Darmstadt, Germany). Subsequently, immunostaining was performed with antibodies against ATP5A1 1:1000 (#ab14748, Abcam, Cambridge, UK), ATPAF1 1:1000 (#HPA044950, Sigma-Aldrich, Munich, Germany), a combined antibody against ATP5G1, ATP5G2 and ATP5G3 1:5000 (#ab180149, Abcam), GAPDH (#2118, Cell Signaling Technology), and beta-actin (#A5316, Sigma-Aldrich). The detection was carried out with horseradish peroxidase conjugated to secondary antibodies (anti-rabbit-POD, #170–6515, Bio-Rad Laboratories, Munich, Germany; anti-mouse-POD, #170–6516, Bio-Rad; anti-biotin-POD, #7075, Cell Signaling Technology). The chemiluminescent signal was visualized using SuperSignal West Femto Kit (Thermo Scientific) and recorded by the LAS 3000 Image Reader (Fujifilm, Tokyo, Japan).
Immunohistochemistry
A tissue microarray was used to determine the expression of ATP5A1, ATPAF1, ATP5G1, ATP5G2, and ATP5G3 in RCC and benign renal tissue specimens. Paraffin sections were cut at 5 μm thickness, deparaffinized using xylene and rehydrated in graded ethanol. Slides were placed in citrate buffer (pH 6.0) for ATP5A1 and ATPAF1 and Tris/EDTA buffer (pH 9.0) for ATP5G1/G2/G3 and heated for 10 min at boiling temperature (microwave 600 W). After 30 min resting time and 15 min for cooling, the endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 10 minutes. The sections were washed with Tris-buffered saline and Tween 20 (Fa. Merck 8.22184). The primary antibodies diluted with Antibody diluent (Fa. Dako No. S3022) (anti-ATP5A1, dilution 1:1000; anti-ATPAF1, dilution 1:50; anti-ATP5G1/G2/G3, dilution 1:100) were applied according to the manufacturer's instructions. The slides were incubated at 4 °C overnight and signal detection was performed with Dako Envision + System-HRP Labeled Polymer (Dako, Hamburg, Germany) with secondary antibodies against rabbit and mouse. Finally, the slides were counterstained using hematoxylin and Bluing Reagent, dehydrated and mounted.
The staining was evaluated by three investigators and, in the case of disagreement, the scoring was discussed at a multi-headed microscope. The staining intensities were scored from 0 to 3, 0 being no staining to 3 being maximum staining. The expression of the target proteins was recorded for the sub-compartments (proximal/distal tubules, collecting duct, loop of Henle) in the normal tissues.
Statistical Analyses
The Mann–Whitney U test was used to correlate clinicopathological parameters and gene expression levels. Cox regression analyses and Kaplan Meier estimates were applied to correlate patient survival and gene expression. All statistical analyses were performed with SPSS Statistics v21 (IBM, Ehningen, Germany).
Results
Identification of Deregulated Subunits of Complex V
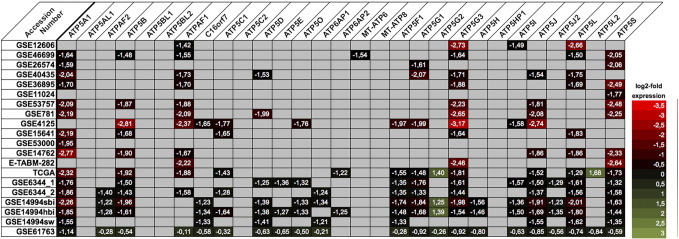
Using the NextBio database, we identified 16 microarray studies comparing the expression of normal and ccRCC tissue. Among the 29 analyzed subunits of the ATP synthase, 23 were significantly dysregulated in at least one microarray study: 21 were down- and 2 were up-regulated in ccRCC (see Figure 1).
Figure 1.
The expression profile of 29 subunits of the ATP synthase (complex V) was retrieved from NextBio database: 12 genes (ATP5A1, ATP5B, ATPAF1, ATP5C1, ATP5D, ATP5O, ATP5F1, ATP5G1, ATP5G2, ATP5G3, ATP5I, and ATP5S) had distinct expression differences of normal and clear cell renal cell carcinoma tissue in several microarray gene expression studies.
Validation of Gene Expression Profiling
Among the potentially dysregulated genes, we chose 12 ATP synthase subunits (ATP5A1, ATP5B, ATPAF1, ATP5C1, ATP5D, ATP5O, ATP5F1, ATP5G1, ATP5G2, ATP5G3, ATP5I, ATP5S) for validation within a cohort of ccRCC (n = 18) and normal (n = 10) renal tissue samples using qPCR. As expected, all studied mRNAs were expressed at lower levels in ccRCC compared to normal renal tissue (P ≤ .001; see Table 3 and Supplementary Figure S1).
Table 3.
Summary of Differentially Expressed mRNA in the Screening and Validation Cohort of Real-Time PCR
| Samples | Screening Cohort |
Validation Cohort |
|---|---|---|
| Fold Change | Fold Change | |
| ATP5A1 | −3.31 | −3.65 |
| ATP5B | −3.04 | −2.37 |
| ATPAF1 | −2.91 | −2.80 |
| ATP5C1 | −1.81 | n.d. |
| ATP5D | −2.12 | n.d. |
| ATP5O | −1.98 | n.d. |
| ATP5F1 | −1.93 | −1.62 |
| ATP5G1 | −8.09 | −5.88 |
| ATP5G2 | −2.69 | −2.11 |
| ATP5G3 | −3.10 | −2.37 |
| ATP5I | −1.72 | n.d. |
| ATP5S | −1.95 | −1.67 |
n.d., not determined.
In order to provide a substantiated validation, we further investigated 8 genes (ATP5A1, ATP5B, ATPAF1, ATP5F1, ATP5G1, ATP5G2, ATP5G3, ATP5S) in an enlarged independent cohort of 74 ccRCC and 36 normal renal tissues. Consistent with the microarray and qPCR expression results, we confirmed significant down-regulation of all Complex V subunits in ccRCC tissue (all P < .001; see Table 3 and Supplementary Figure S2).
ATP5S expression was correlated with presence of metastases (P = .013). There was no other correlation of mRNA expression levels with clinical-pathological parameters (i.e. pT-stage, metastasis or grading; all P > .05). ATP5G2 expression was correlated with overall survival following nephrectomy (log rank P = .024; see Supplementary Figure S3).
Western Blot: Validation on Protein Level
In order to confirm the findings on the protein level, we performed a Western blot for ATP5A1, ATPAF1, ATP5G1/G2/G3 with 8 corresponding normal renal and ccRCC tissues. Consistent with the mRNA expression studies, all studied proteins were down-regulated in ccRCC. The blots are summarized in Figure 2, and whole blot images are provided in Supplementary Figure S4.
Figure 2.
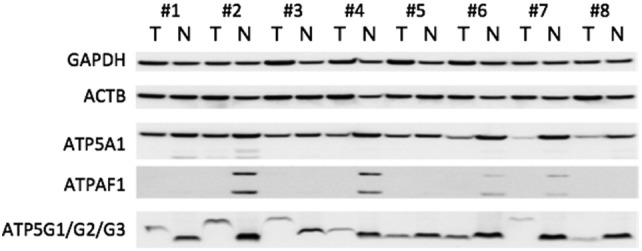
Western blot experiments were performed to determine the protein expression in 8 corresponding normal (N) and clear cell renal cell carcinoma (T) tissues (#1–#4 UICC grade 1; #5–#8 UICC grade 3). All protein levels were decreased in tumor compared to normal renal tissue.
Immunohistochemistry: Expression in RCC and Normal Renal Tissue
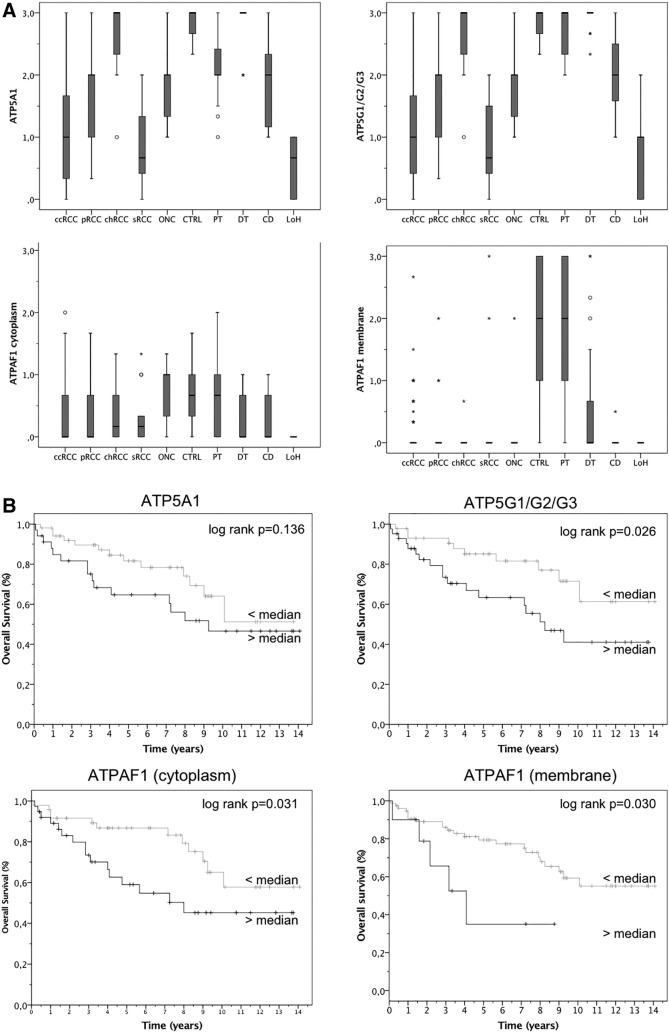
We next performed immunohistochemistry with a tissue microarray including ccRCC (n = 141), pRCC (n = 29), chRCC (n = 10), sRCC (n = 11), oncocytoma (n = 10) as well as normal renal tissues (n = 30). A representative staining of ATP5A1, ATPAF1, and ATP5G1/G2/G3 is shown in Supplementary Figure S5. Consistent with the western blot analysis, we observed decreased expression of ATP5A1, ATPAF1, and ATP5G1/G2/G3 in the cytoplasm of ccRCC compared to normal renal tissue (P < .001; see Figure 3). The more detailed analysis of normal tissue indicated that all proteins were expressed at a low level in the Loop of Henle and the collecting duct, whereas its expression was higher in the proximal and distal tubules. Interestingly, ATPAF1 also showed a membrane staining which was also decreased in ccRCC compared to normal renal tissue. ATPAF1 membrane staining was more intense than cytoplasmic staining in normal tissue. The comparison of ccRCC with other RCC subtypes showed that ATP5A1, and ATP5G1/G2/G3 expression was increased in pRCC (P < .001 and P = .002) and chRCC (both P < .001) compared to ccRCC. Furthermore, the expression of ATP5A1 (P = .001) and ATP5G1/G2/G3 (P = .008) as well as cytoplasmic ATPAF1 (P < .001) was higher in oncocytoma than in ccRCC tissue.
Figure 3.
(A) The expression of ATP5A1, ATP5G1/G2/G3 and ATPAF1 was determined using immunohistochemistry in a tissue microarray which included clear cell (ccRCC), papillary (pRCC) and sarcomatoid (sRCC) renal cell carcinoma as well as oncocytoma (ONC) and normal renal (proximal tubules, PT; distal tubules, DT; loop of Henle, LoH; collecting duct, CD) tissue. ccRCC tissues were characterized by lower levels of ATP5A1, ATP5G1/G2/G3, and ATPAF1 compared to other RCC subtypes and normal renal tissue. (B) Kaplan Meier estimates demonstrated significant shorter overall survival in ccRCC patients with high levels of ATPAF1 and ATP5G1/G2/G3.
We did not observe a correlation between the investigated proteins and clinical parameters (i.e. pT-stage, metastasis, grading; all >0.15) in patients with ccRCC. Follow-up information was available for 86 patients. Kaplan Meier estimates revealed a correlation of ATPAF1 (cytoplasmic and membrane staining) and ATP5G1/G2/G3 expression and overall survival of ccRCC patients (log rank P < .05; see Figure 3). The univariate Cox regression analysis also showed that an increased immunostaining of ATPAF1 and ATP5G1/G2/G3 was correlated with poor overall survival in a subset of 86 patients with available follow-up data (ATPAF1 cytoplasm: P = .036, hazard ratio (HR) = 2.23, 95% confidence interval (95% CI) 1.06–4.74; ATPAF1 membrane: P = .039, HR = 2.85, 95% CI 1.05–7.72; ATP5G1/G2/G3: P = .032, HR = 2.38, 95% CI 1.07–5.07). The prognostic value was lost in the multivariate model including TNM-stage and grading, but it should be noted that the number of cases (n = 86) limits the statistical power, and that the number of few patients with lymph node metastasis (n = 6; univariate analysis HR 10.22) limits the meaning of a multivariate analysis. See Table 4.
Table 4.
Uni- and Multivariate Cox Regression Analysis for the Prediction of Overall Survival in Patients with Clear Cell Renal Cell Carcinoma
| Univariate Analysis |
Multivariate Analysis |
|||||
|---|---|---|---|---|---|---|
| P value | HR | (95% CI) | P value | HR | (95% CI) | |
| pT-stage | 0.432 | 1.19 | (0.77–1.85) | |||
| LN-metastasis | <0.001 | 10.22 | (3.53–29.58) | 0.001 | 7.59 | (2.31–24.98) |
| Distant metastasis | 0.129 | 1.28 | (0.93–1.75) | |||
| Grading | 0.005 | 3.81 | (1.51–9.66) | 0.051 | 3.82 | (0.99–6.88) |
| ATPAF1 cytoplasm | 0.036 | 2.23 | (1.06–4.74) | 0.595 | 1.29 | (0.50–3.29) |
| ATPAF1 membrane | 0.039 | 2.85 | (1.05–7.72) | 0.751 | 1.21 | (0.37–3.94) |
| ATP5G1/G2/G3 | 0.032 | 2.38 | (1.07–5.07) | 0.238 | 1.78 | (0.67–4.29) |
| ATP5A1 | 0.143 | 1.75 | (0.83–3.69) | |||
HR, hazard ratio; 95% CI, 95% confidence interval.
Discussion
Renal cell carcinoma is termed as metabolic disease that is characterized by: dysregulation of metabolic pathways involved in oxygen sensing (VHL/HIF pathway alterations); energy sensing (Warburg shift augmented lipogenesis; and reduced AMPK and Krebs cycle activity) and/or nutrient sensing cascade (deregulation of AMPK-TSC1/2-mTOR and PI3K-Akt–mTOR pathways) [35]. Otto Warburg has already described that cancer cells unlike normal cells metabolize glucose mostly via glycolysis even in the presence of sufficient oxygen [36]. It is assumed that impaired VHL function is the initiating genetic event for many of the dysregulated pathways in ccRCC [29]. In brief, the VHL protein binds under conditions of normal oxygen tension to the hypoxia-inducible factor subunit α (HIF-α) and directs it to proteosomal degradation. Thus, in the absence of functional VHL, even under normoxic conditions, HIF-alpha is not degraded and translocates to the nucleus to form a dimer with HIF-β to generate HIF-1. HIF-1, as a transcription factor, is involved in the regulation of many biological processes such as glucose uptake and energy metabolism, angiogenesis, erythropoiesis, cell proliferation and apoptosis, cell–cell and cell-matrix interactions, and barrier function [37], [38]. In RCC, the increased levels of HIF and its target gene activation accounts for much of its unique attributes such as ADRP, neutral lipid accumulation and clear cell histology; VEGF and other angiokines, vascularity; erythropoietin, paraneoplastic polycythemia [39].
So far, little information is available about the detailed expression of the subunits of the electron transport chain proteins. We earlier demonstrated down-regulation of UQCRC1, UQCRFS1, NDUFS1 and NDUFA4 [34], [40], [41] and up-regulation of its paralogue NDUFA4L2 [42]. We subsequently reviewed the expression profile of the mitochondrial ATP synthase (Complex V) subunits in ccRCC by retrieval of published microarray expression profiling studies. 80% of the 29 subunits showed dysregulation (mostly down-regulation) in at least one microarray study, and several genes were dysregulated in several studies. We were able to validate the former microarray gene expression profiling studies on the mRNA (ATP5A1, ATP5B, ATPAF1, ATP5C1, ATP5D, ATP5O, ATP5F1, ATP5G1, ATP5G2, ATP5G3, ATP5I, ATP5S) and protein (ATP5A1, ATPAF1, ATP5G1/G2/G3) level. These findings support the idea that the activity of the mitochondrial electron chain is reduced by down-regulation of its members, and that this leads to reduced oxidative phosphorylation.
Biomarkers are available for many malignancies, but unfortunately this is not the case for RCC. Our study revealed several - so far unknown biomarker candidates - in ccRCC: among the in detail validated ATP-synthase subunits, we observed 6-fold down-regulation of ATP5G1 on the mRNA level, and also the ATP5G1/G2/G3 protein was highly down-regulated in ccRCC tissue. It should be noted that the antibody for ATP5G1 was not highly specific and ATP5G2 and ATP5G3 subunits are also recognized according to the manufacturer's information, but a more specific antibody was not available for immunohistochemistry and Western blot analysis. To the best of our knowledge, this is the first study showing dysregulation of ATP5G1 in cancer. Earlier studies demonstrated that siRNA knockdown of ATP5G1 resulted in reduced ATP levels as a consequence of reduced oxidative phosphorylation activity, and increased the amount of reactive oxygen species (ROS) [43].
We also highlighted a role for ATP5A1 in the pathogenesis of ccRCC: its mRNA was down-regulated 3.5-fold in ccRCC compared to normal renal tissue, and also protein levels were significantly decreased in ccRCC tissue. The subunit ATP5A1 was linked to cancer by other researchers: ATP5A1 expression facilitated the development of colorectal tumors with microsatellite instability [44], and its expression was up-regulated in glioblastoma and endothelial cells of tumor microenvironment [45].
Another distinctly down-regulated gene in ccRCC was ATPAF1. Interestingly, ATPAF1 was not only detected in the cytoplasm of normal renal tissue, but we also observed regular immunohistochemical membrane staining of the proximal tubules. The relevance of ATPAF1 localization remains unknown. The functional consequence of altered ATPAF1 expression remains largely unknown, but it was shown that RNAi-mediated knockdown of ATPAF1 reduces cell growth of prostate cancer cells in androgen-deficient conditions [46].
This study enhanced our understanding of the dysregulation in RCC. Specific genes involved in the tumor-forced glycolysis may represent potential therapeutic targets of agents that specifically interact with the key factors of tumor phenotype. Furthermore our findings could serve as a basis for future studies in search for biomarkers in RCC. To substantiate our findings further analysis like reactive-oxygen-species analysis (ROS) or extracellular flux analysis (ECAR) in vitro are warranted to demonstrate the functional relevance of altered expression of the complex V subunits.
The following are the supplementary data related to this article.
Supplementary material. (Supplementary Table S1; Supplementary Figures S1-S4).
Funding Source
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of Interest
None.
Acknowledgements
The tissue samples were collected within the framework of the Biobank of the Center for Integrated Oncology Cologne Bonn at the University Hospital Bonn.
References
- 1.McLaughlin J, Lipworth L, Tarone R. Epidemiologic Aspects of Renal Cell Carcinoma. Semin Oncol. 2006;33:527–533. doi: 10.1053/j.seminoncol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Green DR, Reed JC. Mitochondria and Apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 3.Ferri KF, Kroemer G. Organelle-specific initiation of cell death pathways. Nat Cell Biol. 2001;3:E255–E263. doi: 10.1038/ncb1101-e255. [DOI] [PubMed] [Google Scholar]
- 4.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 5.Thompson C. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 6.Reed JC. Mechanisms of apoptosis avoidance in cancer. Curr Opin Oncol. 1999;11 doi: 10.1097/00001622-199901000-00014. [DOI] [PubMed] [Google Scholar]
- 7.Warburg O. On the Origin of Cancer Cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 8.Ward PS, Thompson CB. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatto F, Nookaew I, Nielsen J. Chromosome 3p loss of heterozygosity is associated with a unique metabolic network in clear cell renal carcinoma. Proc Natl Acad Sci U S A. 2014;111:E866–E875. doi: 10.1073/pnas.1319196111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unwin RD, Craven RA, Harnden P, Hanrahan S, Totty N, Knowles M, Eardley I, Selby PJ, Banks RE. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics. 2003;3:1620–1632. doi: 10.1002/pmic.200300464. [DOI] [PubMed] [Google Scholar]
- 11.Perroud B, Lee J, Valkova N, Dhirapong A, Lin P, Fiehn O, Kültz D, Weiss RH. Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol Cancer. 2006;5:64. doi: 10.1186/1476-4598-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meierhofer D. Decrease of mitochondrial DNA content and energy metabolism in renal cell carcinoma. Carcinogenesis. 2004;25:1005–1010. doi: 10.1093/carcin/bgh104. [DOI] [PubMed] [Google Scholar]
- 13.Bonora M, Bononi A, Marchi E de, Giorgi C, Lebiedzinska M, Marchi S, Patergnani S, Rimessi A, Suski JM, Wojtala A. Role of the c subunit of the F O ATP synthase in mitochondrial permeability transition. Cell Cycle. 2014;12:674–683. doi: 10.4161/cc.23599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stickel JS, Weinzierl AO, Hillen N, Drews O, Schuler MM, Hennenlotter J, Wernet D, Muller CA, Stenzl A, Rammensee H. HLA ligand profiles of primary renal cell carcinoma maintained in metastases. Cancer Immunol Immunother. 2009;58:1407–1417. doi: 10.1007/s00262-008-0655-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckel-Passow JE, Serie DJ, Bot BM, Joseph RW, Hart SN, Cheville JC, Parker AS. Somatic expression of ENRAGE is associated with obesity status among patients with clear cell renal cell carcinoma. Carcinogenesis. 2014;35:822–827. doi: 10.1093/carcin/bgt485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ooi A, Wong J, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, Min BWH, Tan M, Zhang Z, Yang XJ. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20:511–523. doi: 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 17.Wozniak MB, Le Calvez-Kelm F, Abedi-Ardekani B, Byrnes G, Durand G, Carreira C, Michelon J, Janout V, Holcatova I, Foretova L. Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United States. PLoS One. 2013;8:e57886. doi: 10.1371/journal.pone.0057886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 19.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, Yamasaki T, Zhrebker L, Sivanand S, Spence P. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kort EJ, Farber L, Tretiakova M, Petillo D, Furge KA, Yang XJ, Cornelius A, Teh BT. The E2F3-Oncomir-1 axis is activated in Wilms' tumor. Cancer Res. 2008;68:4034–4038. doi: 10.1158/0008-5472.CAN-08-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roemeling CA von, Radisky DC, Marlow LA, Cooper SJ, Grebe SK, Anastasiadis PZ, Tun HW, Copland JA. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res. 2014;74:4796–4810. doi: 10.1158/0008-5472.CAN-14-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beroukhim R, Brunet J, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins JPT, Shinghal R, Gill H, Reese JH, Terris M, Cohen RJ, Fero M, Pollack JR, van de Rijn M, Brooks JD. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol. 2003;162:925–932. doi: 10.1016/S0002-9440(10)63887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 26.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Roche O, Yan MS, Finak G, Evans AJ, Metcalf JL, Hast BE, Hanna SC, Wondergem B, Furge KA. Regulation of endocytosis via the oxygen-sensing pathway. Nat Med. 2009;15:319–324. doi: 10.1038/nm.1922. [DOI] [PubMed] [Google Scholar]
- 28.Deng M, Blondeau JJ, Schmidt D, Perner S, Müller SC, Ellinger J. Identification of novel differentially expressed lncRNA and mRNA transcripts in clear cell renal cell carcinoma by expression profiling. Genom Data. 2015;5:173–175. doi: 10.1016/j.gdata.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comprehensive molecular characterization of clear cell renal cell carcinomaNature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellinger J, Kahl P, Mertens C, Rogenhofer S, Hauser S, Hartmann W, Bastian PJ, Büttner R, Müller SC, Ruecker A von. Prognostic relevance of global histone H3 lysine 4 (H3K4) methylation in renal cell carcinoma. Int J Cancer. 2010;127:2360–2366. doi: 10.1002/ijc.25250. [DOI] [PubMed] [Google Scholar]
- 31.Blondeau J, Deng M, Syring I, Schrödter S, Schmidt D, Perner S, Müller SC, Ellinger J. Identification of novel long non-coding RNAs in clear cell renal cell carcinoma. Clin Epigenetics. 2015;7:10. doi: 10.1186/s13148-015-0047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung M, Ramankulov A, Roigas J, Johannsen M, Ringsdorf M, Kristiansen G, Jung K. In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR. BMC Mol Biol. 2007;8:47. doi: 10.1186/1471-2199-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dupasquier S, Delmarcelle A, Marbaix E, Cosyns J, Courtoy PJ, Pierreux CE. Validation of housekeeping gene and impact on normalized gene expression in clear cell renal cell carcinoma: critical reassessment of YBX3/ZONAB/CSDA expression. BMC Mol Biol. 2014;15:9. doi: 10.1186/1471-2199-15-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrödter S, Braun M, Syring I, Klümper N, Deng M, Schmidt D, Perner S, Müller SC, Ellinger J. Identification of the dopamine transporter SLC6A3 as a biomarker for patients with renal cell carcinoma. Mol Cancer. 2016;15:1–10. doi: 10.1186/s12943-016-0495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massari F, Ciccarese C, Santoni M, Brunelli M, Piva F, Modena A, Bimbatti D, Fantinel E, Santini D, Cheng L. Metabolic alterations in renal cell carcinoma. Cancer Treat Rev. 2015;41:767–776. doi: 10.1016/j.ctrv.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 37.Haase VH. The VHL/HIF oxygen-sensing pathway and its relevance to kidney disease. Kidney Int. 2006;69:1302–1307. doi: 10.1038/sj.ki.5000221. [DOI] [PubMed] [Google Scholar]
- 38.Pinthus JH, Whelan KF, Gallino D, Lu J, Rothschild N. Metabolic features of clear-cell renal cell carcinoma: mechanisms and clinical implications. Can Urol Assoc J. 2011;5:274–282. doi: 10.5489/cuaj.10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rathmell WK, Chen S. VHL inactivation in renal cell carcinoma: implications for diagnosis, prognosis and treatment. Expert Rev Anticancer Ther. 2008;8:63–73. doi: 10.1586/14737140.8.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellinger J, Poss M, Brüggemann M, Gromes A, Schmidt D, Ellinger N, Tolkach Y, Dietrich D, Kristiansen G, Müller SC. Systematic Expression Analysis of Mitochondrial Complex I Identifies NDUFS1 as a Biomarker in Clear-Cell Renal-Cell Carcinoma. Clin Genitourin Cancer. 2016 doi: 10.1016/j.clgc.2016.11.010. [1558–7673(16)30346–9] [DOI] [PubMed] [Google Scholar]
- 41.Ellinger J, Gromes A, Poss M, Brüggemann M, Schmidt D, Ellinger N, Tolkach Y, Dietrich D, Kristiansen G, Müller SC. Systematic expression analysis of the mitochondrial complex III subunits identifies UQCRC1 as biomarker in clear cell renal cell carcinoma. Oncotarget. 2016;7:86490–86499. doi: 10.18632/oncotarget.13275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller FE, Braun M, Syring I, Klümper N, Schmidt D, Perner S, Hauser S, Müller SC, Ellinger J. NDUFA4 expression in clear cell renal cell carcinoma is predictive for cancer-specific survival. Am J Cancer Res. 2015;5:2816–2822. [PMC free article] [PubMed] [Google Scholar]
- 43.Natera-Naranjo O, Kar AN, Aschrafi A, Gervasi NM, Macgibeny MA, Gioio AE, Kaplan BB. Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol Cell Neurosci. 2012;49:263–270. doi: 10.1016/j.mcn.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 44.Seth R, Keeley J, Abu-Ali G, Crook S, Jackson D, Ilyas M. The putative tumour modifier gene ATP5A1 is not mutated in human colorectal cancer cell lines but expression levels correlate with TP53 mutations and chromosomal instability. J Clin Pathol. 2009;62:598–603. doi: 10.1136/jcp.2009.064436. [DOI] [PubMed] [Google Scholar]
- 45.Xu G, Li JY. ATP5A1 and ATP5B are highly expressed in glioblastoma tumor cells and endothelial cells of microvascular proliferation. J Neurooncol. 2016;126:405–413. doi: 10.1007/s11060-015-1984-x. [DOI] [PubMed] [Google Scholar]
- 46.Schinke EN, Bii V, Nalla A, Rae DT, Tedrick L, Meadows GG, Trobridge GD. A novel approach to identify driver genes involved in androgen-independent prostate cancer. Mol Cancer. 2014;13:120. doi: 10.1186/1476-4598-13-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material. (Supplementary Table S1; Supplementary Figures S1-S4).