Abstract
Metastasis of epithelial cancer cells to distant sites is a particularly critical stage of cancer progression that typically marks the incurability of the disease. It is governed by a complex series of events including invasion and intravasation of tumor cells into the stroma and blood, respectively. Epithelial‐to‐mesenchymal transition (EMT), a phenotypic change marked by the loss of epithelial characteristics and the acquisition of invasive mesenchymal properties, is implicated in the dissemination of tumor cells. Circulating tumor cells (CTCs), the precursors of metastasis, can be used to interrogate the contribution of EMT in metastasis and therapeutic responses. The analysis of these CTCs and in particular the presence of inter‐ and intrapatient heterogeneity for markers of EMT has provided new insights into the metastatic process. This review will focus on epithelial–mesenchymal plasticity in CTCs and its potential clinical implications.
Keywords: cancer, circulating tumor cells, epithelial to mesenchymal transition, metastasis
Abbreviations
- CTCs
circulating tumor cells
- EGFR
epidermal growth factor receptor
- EMT
epithelial‐to‐mesenchymal transition
- KRT
keratin
- MET
mesenchymal‐to‐epithelial transition
- RNA‐ISH
RNA in situ hybridization
1. Introduction
Epithelial‐to‐mesenchymal transition (EMT) is a tightly regulated lineage change occurring during gastrulation, neural crest delamination, and heart valve formation, developmental processes involved in embryonic patterning (Savagner, 2015). Hallmarks of EMT include the loss of epithelial markers and the acquisition of mesenchymal markers, disruption of cell–cell junctions, and apico‐basal polarity as well as remodeling of the cytoskeleton. These changes are associated with increased cell migration and resistance to anoikis (Savagner, 2015) and are triggered by several growth factors, cytokines, and numerous transcription factors. EMT is a reversible process that is dependent on the continued presence of the EMT‐inducing signal, the removal of which results in the reversion of the mesenchymal cell to an epithelial state [mesenchymal‐to‐epithelial transition (MET)].
In adult organisms, EMT is activated during physiologic and pathophysiologic responses including wound healing and fibrosis (Stone et al., 2016). EMT has been extensively implicated in cancer progression, specifically in promoting the early stages of metastasis, which involve invasion of tumor cells into the surrounding stroma and blood‐borne dissemination of cells to the lung, liver, bone, and brain (Nieto et al., 2016; Thiery et al., 2009). Once epithelial cancer cells, which have transitioned to a mesenchymal state, reach distant sites, it is postulated that they revert to the epithelial lineage through the process of MET (Gunasinghe et al., 2012). Tumor cells that are able to survive within the new microenvironment can acquire the ability to proliferate and establish metastatic tumors, which in most cancers marks a stage in which the cancer becomes incurable. In addition to a role in tumor cell invasion and dissemination, EMT of epithelial tumor cells is also linked to the acquisition of stem‐like characteristics and drug resistance (Aktas et al., 2009; Arumugam et al., 2009; Jolly et al., 2015; Marchini et al., 2013; Sequist et al., 2011; Witta et al., 2006). Interestingly, lineage tracing of epithelial and mesenchymal tumor cells within genetically engineered mice shows that EMT may not be a prerequisite for metastasis, but instead contributes to drug resistance, suggesting additional functions for EMT in cancer progression (Fischer et al., 2015; Maheswaran and Haber, 2015; Zheng et al., 2015).
Cancer cells co‐opting EMT to migrate and invade provides an attractive model to understand the critical steps involved in the initiation of metastasis, a highly complex process. However, identifying, within the primary tumor, epithelial tumor cells converting to a mesenchymal state has been complicated by the presence of stromal cells which express high levels of mesenchymal markers. As such, despite the dramatic invasive and tumorigenic phenotypes observed in mouse xenografts expressing master EMT‐inducing transcriptional regulators, Snail, Twist, and Slug among others (Ocana et al., 2012; Tran et al., 2014; Yang et al., 2004; Ye et al., 2015), the direct observation of EMT in the metastasis of human epithelial cancers has remained elusive. Increased invasion of tumor cells undergoing EMT suggests that mesenchymal characteristics would be prevalent in tumor cells entering the circulation. EMT of tumor cells in the blood has been evaluated by PCR analysis of peripheral blood mononuclear cells isolated from patients with cancer (Aktas et al., 2009; Lasa et al., 2013), but this approach has been unreliable due to the high rate of false signals detected in unpurified blood components. Analysis of circulating tumor cells (CTCs), precursors of metastasis which circulate in the blood as either individual tumor cells or as tumor cell clusters/tumor emboli, offers a rare opportunity to gain insight into the spread of cancer at a critical stage of metastasis (Maheswaran and Haber, 2010). Accumulating evidence from the analysis of these isolated CTCs has demonstrated significant heterogeneity of EMT markers supporting the concept of EMT as an important feature of invasive cancer cells (Fig. 1).
Figure 1.
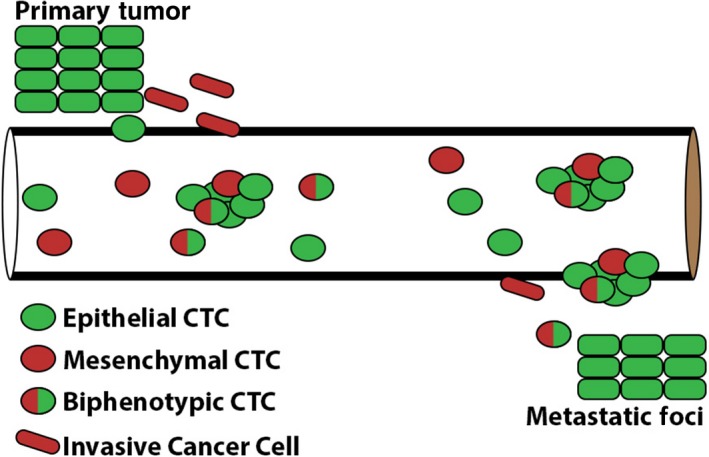
Epithelial‐to‐mesenchymal transition (EMT) in circulating tumor cells (CTCs): Invasive cells from the primary tumor exhibit epithelial plasticity including the loss of epithelial markers and gain of mesenchymal markers. Solitary and clusters of CTCs demonstrate heterogeneity of epithelial and mesenchymal markers.
Recent advances in technology have revolutionized our ability to isolate and characterize CTCs from the blood of patients with numerous forms of cancer (Ferreira et al., 2016). The development of these innovative CTC technologies has lead to the concept of using CTCs as a liquid biopsy that may be able to provide prognostic information upon initial diagnosis, track response to therapies, and identify early signs of resistance to treatment (Lianidou et al., 2015). Additionally, the analysis of CTCs that are derived from different sites of disease within a patient with metastatic disease has the potential to better detect the intrapatient heterogeneity of the cancer cells compared to a site‐directed biopsy (Fig. 2). The study of CTCs has revealed that markers of EMT are an important feature that can help define and characterize these cells and provide valuable and potentially actionable information to clinicians regarding disease prognosis and response to therapy for individual patients (Satelli et al., 2016; Yu et al., 2013; Zhao et al., 2017). This review will focus on the current state of knowledge regarding EMT in circulating tumor cells.
Figure 2.
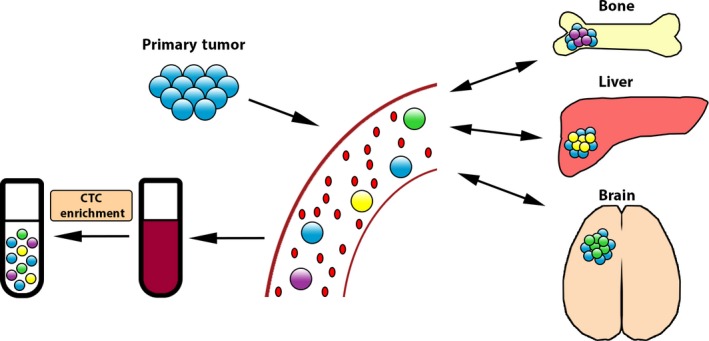
Heterogeneity of CTCs in metastatic disease: CTCs represent the intrapatient tumor heterogeneity of cancer cells residing at multiple metastatic sites.
2. Technologies to enrich epithelial and mesenchymal CTCs
2.1. Biophysical and functional property‐based enrichment of CTCs
CTCs were first observed in the blood of patients with cancer almost 150 years ago (Ashworth, 1869). They represent a rare population of cells, which are estimated to be present at a frequency of one CTC admixed with a billion blood cells (Maheswaran and Haber, 2010). Therefore, their isolation presents a significant technological challenge. Given their clinical utility as noninvasive blood‐based biomarkers, several platforms have been developed to enrich CTCs from the blood of patients with cancer. Some approaches rely on the functional properties of CTCs including the ability to breakdown collagen and invade or secrete detectable proteins (Deng et al., 2014; Lu et al., 2010). Other approaches use physical characteristics of the tumor cells including size, density, electric charge, and deformability to distinguish them from blood cells (Hou et al., 2013; Liu et al., 2015; Mitchell et al., 2015; Shaw Bagnall et al., 2015). Each method must be evaluated independently to determine the ability to enrich for tumor cells with either epithelial or mesenchymal features (Table 1). For example, there is little difference in the deformability between epithelial and mesenchymal tumor cells, suggesting that CTC enrichment relying on deformability might represent an unbiased approach to capture both CTC lineages (Shaw Bagnall et al., 2015). EMT, however, is associated with an increase in cell size (Lamouille and Derynck, 2007), suggesting that size‐based CTC separation methods might be biased toward the enrichment of mesenchymal CTC populations. In fact, size‐based CTC collection from patients with different types of metastatic cancers shows enrichment of CTCs with mesenchymal and stem‐like characteristics (Hou et al., 2013; Wu et al., 2015). This may reflect increased tumor aggressiveness, but needs to be considered with the caveat that size‐based CTC separation is biased toward enrichment of larger cells.
Table 1.
Epithelial and mesenchymal circulating tumor cell recovery from methods of CTC isolation
| Methods | Detection of Epithelial (E) versus Mesenchymal (M) CTCs | References |
|---|---|---|
| Size‐based filtration | E & M | Kim et al. (2017), Vona et al. (2000) |
| Density‐based centrifugation | E & M | Liu et al. (2015) |
| Size and deformability | M > E | Gogoi et al. (2016) |
| Cell surface charge | E & M | Gupta et al. (2012) |
| Negative selection | ||
| Microfluidic size based followed by negative selection for CD45 | E & M | Karabacak et al. (2014) |
| Density separation of tetrameric antibody complexes for CD45, CD66b, and glycophorin | E & M | Naume et al. (2004) |
| Positive selection | ||
| Cell surface vimentin | M | Satelli et al. (2015a) |
| High‐throughput microscopy for immunofluorescence or DNA/RNA FISH | E & M | Krivacic et al. (2004) |
| EpCAM‐based immunomagnetic separation | E | Allard et al. (2004) |
| Flow cytometry for surface epithelial markers | E | Gross et al. (1995), Magbanua et al. (2015) |
| Filtration using selective size amplification | E | Kim et al. (2012) |
2.2. Antibody‐based CTC isolation
The most common approach underlying CTC isolation technologies is dependent on immunoaffinity purification of tumor cells. In one form of this method, antibodies are directed against proteins that are exclusively expressed on the plasma membrane of the tumor cells and not on hematologic cells resulting in the positive selection of CTCs. Alternatively, antibodies against proteins that are expressed on the surface of the white blood cells can be used to mark and deplete the normal blood components—negative depletion—and the CTCs remaining in the flow‐through can be collected for further analysis.
2.3. Positive selection of CTCs
Numerous technologies relying on positive selection of CTCs utilize antibodies against EpCAM, a cell surface glycoprotein expressed on tumor cells of epithelial lineage, but not on leukocytes. CellSearch, the only FDA‐approved CTC isolation device, is based on immunomagnetic separation of CTCs expressing EpCAM and enumeration of the captured cells by staining with a cocktail of antibodies against the epithelial cytokeratins 8, 18, and/or 19 (Andree et al., 2016). CellSearch identifies CTCs in about 50% of patients with metastatic disease, which may represent the technical limitation of the technology, or the variable expression of the epithelial markers (Allard et al., 2004). Enumeration of CTCs using this technology is prognostic in patients presenting with many types of epithelial cancers, with the presence of ≥5 CTCs per 7.5 mL of blood being predictive of poor clinical outcome (de Bono et al., 2008; Hayes et al., 2006). CTC technologies including CellSearch, which rely on EpCAM‐mediated capture of CTCs, are limited by their inability to enrich mesenchymal CTCs which either express low levels or no EpCAM. CTCs that fail to be captured by EpCAM‐dependent methods are detectable with antibodies directed against mesenchymal as well as other tumor markers (Raimondi et al., 2011; Schneck et al., 2015), suggesting that CTCs are indeed heterogeneous for epithelial and mesenchymal markers. To overcome the limitation of EpCAM‐mediated CTC enrichment and to capture CTCs irrespective of their EMT status, antibody cocktails against EGFR and HER2 (tumor‐specific markers), CDH11 (a cadherin specifically overexpressed in mesenchymal tumor cells), as well as cell surface vimentin (expressed in cancer cells of mesenchymal lineage) (Armstrong et al., 2011; Satelli et al., 2015a; Yu et al., 2013), have been used in combination with EpCAM antibodies. Importantly, broadening the spectrum of detection antibodies beyond those solely against epithelial cytokeratins has improved the identification of mesenchymal CTCs in the blood (Armstrong et al., 2011; Satelli et al., 2015a; Yu et al., 2013).
2.4. Negative depletion of leukocytes
The efficiency of positive selection technologies is significantly impeded by the extensive heterogeneity exhibited by tumor cells in the blood. The negative depletion approach based on the removal of normal blood components circumvents this problem leading to antigen‐independent purification of CTCs. This minimizes the loss of CTC populations, whose heterogeneity can be attributed to various determinants including EMT. As highly specific antibodies with high affinity against mouse leukocyte populations are readily available, the negative selection approach can also be readily applied to isolate CTCs from mouse tumor models, negating the search for mouse‐specific antibodies required for positive selection of these CTCs (Ting et al., 2014). However, this approach may also enrich nonhematopoietic cells such as circulating endothelial cells as well as other circulating epithelial cells (Goon et al., 2006). Thus, CTC identification and enumeration following negative depletion requires carefully chosen epithelial and mesenchymal markers benchmarked against healthy donor blood, as well as molecular analyses that ensure their tumoral origin (Yu et al., 2013). Nevertheless, staining CTCs with epithelial and mesenchymal markers following negative depletion shows that CTCs in the blood are comprised of both epithelial and mesenchymal tumor cells (Yu et al., 2013).
3. Functional characteristics of epithelial and mesenchymal CTC populations
3.1. EMT of epithelial cancers
In addition to the significant advances in the ability to isolate CTCs, a second important advance in the study of CTCs is the development of analytical tools including single‐cell RNA‐Seq (Saliba et al., 2014; Tang et al., 2010), multicolor RNA in situ hybridization (Wu et al., 2016), and single‐cell DNA methylation analysis available to characterize CTCs (Pixberg et al., 2017). While still in the development stages, the potential rapid advancement of single‐cell proteomics will likely also prove to be a critical tool (Macaulay et al., 2017). The single or clustered nature of CTCs and their rarity necessitates the use of these technologies; however, the analysis of CTCs with these technologies has provided important insight into the biology of CTCs including their heterogeneity. Future technologic advancements will permit the multimodality analysis of the same single cell to give a richer view of CTC biology.
RNA in situ hybridization (RNA‐ISH) using multiple probes to mark epithelial and mesenchymal states has been applied to CTCs to increase detection sensitivity without compromising specificity (Yu et al., 2013). A dual colorimetric RNA‐ISH using two pools of probes, one against seven epithelial markers (keratins (KRT) 5, 7, 8, 18, and 19; EpCAM, and E‐cadherin) and the other against three mesenchymal markers [(FN1 (fibronectin 1), CDH2 (cadherin 2), and SERPINE1/PAI1 (serpin peptidase inhibitor, clade E)], was used to visualize breast CTCs isolated by positive selection using antibodies against HER2, EGFR, and EpCAM. Semiquantitative measurement of the punctate epithelial and mesenchymal RNA‐ISH signals can define the degree of epithelial and mesenchymal characteristics of each individual CTC. These results showed that CTCs exhibit varying degrees of epithelial and mesenchymal characteristics (e.g., epithelial (E), mesenchymal (M), E > M, E = M, and M > E), demonstrating that EMT is a continuous process. In fact, the biphenotypic tumor cells simultaneously expressing both epithelial and mesenchymal markers may represent the most plastic and potentially the cells most likely to contribute to metastatic outgrowth (Jolly et al., 2015). The EMT status of CTCs in patients with breast cancer is dependent on the breast cancer subtype: Mesenchymal CTCs are prevalent in triple‐negative and HER2‐positive breast cancer patients, whereas CTCs in lobular breast cancer patients are predominantly epithelial (Yu et al., 2013). Lobular breast carcinoma is marked by mutations or loss of E‐cadherin, a critical epithelial protein. Yet, tumor cells within the primary tumor or metastatic lesion do not exhibit complete conversion to a mesenchymal state although these tumors are marked by an invasive growth pattern (McCart Reed et al., 2016). Analysis of tumor cells invading through the blood is consistent with these findings. Further studies are needed to continue to define what elements of epithelial plasticity contribute to metastatic spread and potentially the cooperative interactions between cells with different phenotypes.
In addition to breast cancer, heterogeneity of EMT markers in CTCs has also been observed in other cancers. RNA‐ISH analysis of CTCs isolated from patients with liver, nasopharyngeal, breast, colon, gastric, and non‐small‐cell lung cancer showed the presence of epithelial, biphenotypic, and mesenchymal populations (Wu et al., 2015). The ratio of mesenchymal CTCs increased based on the TNM stage of the cancer for all of the cancers analyzed (Wu et al., 2015). A separate study of a mouse model of pancreatic cancer also identified varying degree of EMT in CTCs (Rhim et al., 2012). Interestingly, cells exhibiting mesenchymal markers are also seen in premalignant pancreatic intraepithelial neoplasms and in circulating pancreatic‐derived cells prior to the development of overt cancer. Therefore, EMT and malignant cell dissemination may occur early in the development of cancer. Additionally, while tumor cells displaying mesenchymal markers are not significantly enriched in the CTCs, functional analysis revealed that these cells had increased tumor‐initiating potential. Together, these results support the concept of epithelial and mesenchymal heterogeneity in CTCs and provide evidence that EMT may not only permit invasion and escape from the primary tumor, but may induce increased tumor‐initiating properties.
Analysis of epithelial and mesenchymal CTC populations in a cohort of patients with breast cancer showed a dynamic shift in the populations during treatment, such that increasing number of mesenchymal CTCs correlated with treatment failure and disease progression (Yu et al., 2013). Increase in the mesenchymal CTC population during relapse is associated with the appearance of tumor emboli or multicellular CTC clusters in the blood. The CTC clusters are highly mesenchymal and are not readily detectable with antibodies against epithelial cytokeratins. The mesenchymal state of the CTC clusters may be attributed to the coating of platelets, which are a rich source of TGF‐β, a potent inducer of EMT (Labelle et al., 2011). Indeed, transcriptome analysis of the breast CTC clusters shows the enrichment of TGF‐β and EMT signatures, suggesting that CTC clusters, which are oligoclonal and held together through cell–cell interactions, most likely arise through collective migration of tumor cells, a process dependent on partial EMT (Aceto et al., 2014; Friedl and Gilmour, 2009; Yu et al., 2013).
RNA sequencing of single CTCs has also enabled the comprehensive analysis of these cells and has been used to interrogate the EMT characteristics of CTCs in mouse tumor models as well as in patients. The microfluidic device, the iChip, which removes red blood cells and platelets through size‐based separation, enriches CTCs by magnetophoretic depletion of immunomagnetically tagged white blood cells (Karabacak et al., 2014). These CTCs collected in solution can be picked as single cells using a micromanipulator and analyzed with RNA‐Seq at single‐cell resolution (Miyamoto et al., 2015; Ting et al., 2014). This approach was applied to CTCs isolated from a genetically engineered mouse model of pancreatic cancer [(LSL‐KrasG12D, Trp53flox/flox or +, PDX‐Cre (KPC)], which closely mimics the progression of human pancreatic cancer. Single‐cell RNA‐Seq performed on individual CTCs identified three distinct populations: CTCs expressing epithelial markers (classical CTCs—CTC‐c), CTCs enriched for platelet markers (CTC‐plt) and CTCs with a robust proliferation signature (CTC‐Pro) (Ting et al., 2014). The classical CTCs, in comparison with tumor cells within the primary tumor, exhibited a universal loss of E‐cadherin, demonstrating their propensity to lose epithelial markers. This is consistent with the concept of EMT contributing to the invasion of these mouse pancreatic tumor cells into the stroma and blood (Rhim et al., 2012). However, the expression of mesenchymal genes in these CTCs is more complex and highly variable. When compared to the primary tumor, the expression of some EMT markers, CDH11 and vimentin, is elevated in CTCs, whereas others, S100A4, Itga5, Sdc1, are decreased. The heterogeneity exhibited by CTCs in these mice is consistent with the continuity and complexity associated with the EMT process and the presence of several intermediate EMT states in CTCs in human cancer patients. The transition of epithelial cancer cells to a mesenchymal state has been shown to be closely associated with the acquisition of stem‐like characteristics (Guo et al., 2012; Mani et al., 2008). Interestingly, the mouse pancreatic CTCs expressed high levels of Aldh1a1 and Aldh1a2 mRNA, but the expression of these stemness markers did not correlate with EMT, suggesting that these phenotypes are not linked in pancreatic CTCs. The absence of a correlation between the mesenchymal and stemness markers is also observed in CTCs isolated from castration‐resistant prostate cancer patients (Miyamoto et al., 2015).
A transgenic mouse model of prostate cancer tumorigenesis and metastasis was paired with a vimentin‐GFP reporter to track the expression of this mesenchymal marker in prostate tumor cells (Ruscetti et al., 2015). Analysis of the peripheral blood of these mice at different stages of tumorigenesis revealed an increase in mesenchymal (EpCAM−/GFP+) and biphenotypic (EpCAM+/GFP+) CTCs correlating with metastatic disease. Interestingly, while the mesenchymal and biphenotypic CTCs have increased tumor‐initiating ability in the prostate, only the biphenotypic and epithelial CTCs could form macrometastases. The mesenchymal CTCs persisted as micrometastatic foci. These results support the idea that epithelial plasticity is critical to metastasis. However, it is important to point out that in a study of prostate cancer CTCs derived from patients with metastatic disease, RNA‐Seq data of single cells did not identify downregulation of epithelial markers or upregulation of mesenchymal markers in subpopulations of the CTCs (Miyamoto et al., 2015), suggesting either that the heterogeneity can be lost in a moderately pretreated cohort or that epithelial and mesenchymal heterogeneity is not well represented in single‐cell RNA‐Seq analysis. Further studies are needed to investigate the presence of epithelial and mesenchymal marker heterogeneity in patient‐derived prostate samples.
3.2. EMT signatures in CTCs from nonepithelial tumors
Besides epithelial tumors, enrichment of EMT signatures is also observed in CTCs derived from nonepithelial tumor types, including melanoma and glioblastoma. Loss of E‐cadherin is observed in late‐stage malignant melanoma (Alexaki et al., 2010; Miller and Mihm, 2006) and the EMT‐inducing transcription factor network significantly changes during melanomagenesis and constitutes a risk factor for poor outcome in malignant melanoma (Caramel et al., 2013). The expression of EMT‐associated genes is a major determinant of melanoma metastasis (Alonso et al., 2007), suggesting that this pathway might be involved in promoting melanoma dissemination. Indeed, RNA‐Seq analysis of CTCs isolated from a B‐RAF/PTEN‐mutant mouse melanoma model, compared with primary tumor cells, shows the upregulation of EMT signatures coincident with genes implicated in tumor invasiveness (Luo et al., 2014). Similarly, glioblastoma CTCs isolated from patients as well as from a PDX mouse model, when compared with their matched parental tumor, show the upregulation of RNA encoding for the mesenchymal genes SERPINE1, TGFB1, TGFBR2, and vimentin. Overexpression of EMT regulators is observed in the mesenchymal subset of GBMs and is associated with therapeutic resistance and poor clinical outcome (Bhat et al., 2013; Colman et al., 2010; Phillips et al., 2006).
3.3. Potential genomic changes induced by EMT
The plasticity of EMT in epithelial cancer cells is generally attributed to epigenetic changes which are reversible (Bedi et al., 2014; McDonald et al., 2011). During development, proliferation is strictly restricted in gastrulating cells undergoing EMT; initiation of proliferation in these cells during EMT leads to severe developmental defects (Murakami et al., 2004; Seher and Leptin, 2000). The incompatibility between EMT and proliferation is also observed in cancer cells. TGF‐β and SNAIL‐induced EMT in proliferating mammary epithelial cells are associated with mitotic defects leading to binucleate cells and extensive chromosome missegregation (Comaills et al., 2016). These mitotic defects are mediated through TGF‐β and SNAIL‐mediated suppression of multiple nuclear envelope proteins including nuclear lamins, which maintain the nuclear architecture and orchestrate nuclear transport as well as mitotic processes. The mitotic aberrations induced during EMT are reversible; however, the resulting genomic instability leads to heritable changes. Single‐cell RNA‐Seq analysis of breast and prostate CTCs shows a correlation between TGF‐β and EMT signatures and aneuploidy. Furthermore, binucleated and micronuclei harboring cells were prevalent in CTCs of mesenchymal lineage compared to epithelial lineage, suggesting that EMT might be a mechanism leading to genomic instability. These findings that are consistent with developmental models (Murakami et al., 2004; Seher and Leptin, 2000) might explain the drug resistance associated with EMT (Maheswaran and Haber, 2015; Nieto et al., 2016). In fact, TGF‐β activation in the CD44+/CD24− stem‐like cells increases DNA copy number alterations and contributes to changes in drug responses (Pal et al., 2017). Taken together, these findings suggest that EMT in proliferating cancer cells could be a major contributor to the molecular evolution and heterogeneity of tumors during disease progression.
4. Clinical implications of epithelial‐to‐mesenchymal transition in circulating tumor cells
EMT provides a useful model to understand the mechanisms underlying tumor cell invasion and motility, and CTCs offer the possibility of capturing tumor cells during an intermediate step of cancer metastasis; therefore, the overlap of these two concepts will provide a more comprehensive understanding of metastasis. Importantly, though, the study of EMT in CTCs likely has clinical implications that are key to the development of novel cancer treatments and improved deployment of our current therapies. Markers of EMT in CTCs have the potential to provide both prognostic and predictive information. As discussed above, in breast cancer, the number of mesenchymal CTCs has been linked to disease progression and decreased overall survival (Polioudaki et al., 2015; Yu et al., 2013). In colorectal cancer, a study of CTCs in over a thousand patients found that biphenotypic and mesenchymal CTCs correlated with clinical stage, lymph node, and distant metastases (Zhao et al., 2017). Interestingly, a study of metastatic colorectal and prostate cancer patients observed that cell surface vimentin‐positive CTCs that expressed nuclear programmed death‐ligand 1 (PD‐L1) are associated with decreased survival, suggesting that there may exist subpopulations within the mesenchymal CTCs that correlate with clinical parameters (Satelli et al., 2016). In addition to potential prognostic markers of disease, markers of EMT in CTCs also have the potential to provide predictive information regarding a patient's response to therapy. Currently, this area of research is in its early stages. For example, in a recent clinical study of metastatic colorectal cancer, enumeration of CTCs with cell surface vimentin correlated with disease response to postsurgery chemotherapy, suggesting that markers of EMT in CTCs may also prove to be useful markers to predict response to therapy (Satelli et al., 2015b). In a preclinical model of breast cancer metastasis, FOXC2 has been identified as an inducer of EMT and an important regulator of metastasis (Werden et al., 2016). FOXC2 induction of EMT is dependent on MAP kinase activity, and in this study, an inhibitor of the p38 MAP kinase inhibited metastatic spread induced by FOXC2. Therefore, FOXC2 in CTCs may represent a potential marker predicting response to MAP kinase inhibitors. Future studies and investigation of additional markers will be needed.
5. Future directions
The study of EMT and the parallels between development and cancer have provided significant insight into the metastatic process. The recent advancements in the isolation and characterization of CTCs provide novel tools and information about how EMT contributes to cancer spread. However, one of the challenges of the current studies of both EMT and CTCs is the lack of standardization. For instance, the classification of epithelial, mesenchymal, and biphenotypic cells varies depending on the study even within the same cancer type. Similarly, the isolation of CTCs varies significantly based on the method and the markers used to purify these cells. It will be important to standardize the classifications and methods in order to increase the robustness of our current knowledge. With this in mind, it will be important to reliably establish the degree of heterogeneity of CTCs and clearly define their subpopulations. A consistent isolation and classification method will permit the accurate tracking of changes in CTCs and allow for testing of potential biomarkers of disease progression or response to therapy. Once biomarkers are identified and a reliable method of detection is established, prospective trials testing these biomarkers will be required to establish their clinical usefulness and validity. In addition to the focus on clinical tools based on EMT in CTCs, improved single‐cell technologies will continue to advance our understanding of CTCs. For example, it is likely that the expression of mesenchymal genes and misexpression of developmental EMT regulators are accompanied by epigenetic changes that activate these typically repressed genes. Epigenetic regulators are potential novel drug targets that offer a unique mechanism of action that could alter the cellular plasticity. Future investigations will need to further define the role of EMT in CTC generation, survival, extravasation, and colonization of distant organs with the hope of developing new therapies and using our current therapy more effectively.
Acknowledgements
This work was supported by National Institutes of Health (NIH) 2RO1CA129933, the Howard Hughes Medical Institute, the Breast Cancer Research Foundation, the National Foundation for Cancer Research (DAH), and T32‐CA071345‐20 (DSM). We thank Valentine Comaills for critical reading of the manuscript.
References
- Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H et al (2014) Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R and Kasimir‐Bauer S (2009) Stem cell and epithelial‐mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res 11, R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexaki VI, Javelaud D, Van Kempen LC, Mohammad KS, Dennler S, Luciani F, Hoek KS, Juarez P, Goydos JS, Fournier PJ et al (2010) GLI2‐mediated melanoma invasion and metastasis. J Natl Cancer Inst 102, 1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW and Terstappen LW (2004) Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 10, 6897–6904. [DOI] [PubMed] [Google Scholar]
- Alonso SR, Tracey L, Ortiz P, Perez‐Gomez B, Palacios J, Pollan M, Linares J, Serrano S, Saez‐Castillo AI, Sanchez L et al (2007) A high‐throughput study in melanoma identifies epithelial‐mesenchymal transition as a major determinant of metastasis. Cancer Res 67, 3450–3460. [DOI] [PubMed] [Google Scholar]
- Andree KC, van Dalum G and Terstappen LW (2016) Challenges in circulating tumor cell detection by the Cell Search system. Mol Oncol 10, 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, Herold CI, Marcom PK, George DJ and Garcia‐Blanco MA (2011) Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res 9, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam T, Ramachandran V, Fournier KF, Wang H, Marquis L, Abbruzzese JL, Gallick GE, Logsdon CD, McConkey DJ and Choi W (2009) Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res 69, 5820–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth TR (1869) A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Australian Med J 14, 146–147. [Google Scholar]
- Bedi U, Mishra VK, Wasilewski D, Scheel C and Johnsen SA (2014) Epigenetic plasticity: a central regulator of epithelial‐to‐mesenchymal transition in cancer. Oncotarget 5, 2016–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD et al (2013) Mesenchymal differentiation mediated by NF‐kappaB promotes radiation resistance in glioblastoma. Cancer Cell 24, 331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ and Raghavan D (2008) Circulating tumor cells predict survival benefit from treatment in metastatic castration‐resistant prostate cancer. Clin Cancer Res 14, 6302–6309. [DOI] [PubMed] [Google Scholar]
- Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A, Saldanha G, Osborne J, Hutchinson P, Tse G et al (2013) A switch in the expression of embryonic EMT‐inducers drives the development of malignant melanoma. Cancer Cell 24, 466–480. [DOI] [PubMed] [Google Scholar]
- Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG et al (2010) A multigene predictor of outcome in glioblastoma. Neuro Oncol 12, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comaills V, Kabeche L, Morris R, Buisson R, Yu M, Madden MW, LiCausi JA, Boukhali M, Tajima K, Pan S et al (2016) Genomic instability is induced by persistent proliferation of cells undergoing epithelial‐to‐mesenchymal transition. Cell Rep 17, 2632–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Zhang Y, Sun S, Wang Z, Wang M, Yu B, Czajkowsky DM, Liu B, Li Y, Wei W et al (2014) An integrated microfluidic chip system for single‐cell secretion profiling of rare circulating tumor cells. Sci Rep 4, 7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MM, Ramani VC and Jeffrey SS (2016) Circulating tumor cell technologies. Mol Oncol 10, 374–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J et al (2015) Epithelial‐to‐mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P and Gilmour D (2009) Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol 10, 445–457. [DOI] [PubMed] [Google Scholar]
- Gogoi P, Sepehri S, Zhou Y, Gorin MA, Paolillo C, Capoluongo E, Gleason K, Payne A, Boniface B, Cristofanilli M et al (2016) Development of an automated and sensitive microfluidic device for capturing and characterizing circulating tumor cells (CTCs) from clinical blood samples. PLoS One 11, e0147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goon PK, Lip GY, Boos CJ, Stonelake PS and Blann AD (2006) Circulating endothelial cells, endothelial progenitor cells, and endothelial microparticles in cancer. Neoplasia 8, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross HJ, Verwer B, Houck D, Hoffman RA and Recktenwald D (1995) Model study detecting breast cancer cells in peripheral blood mononuclear cells at frequencies as low as 10(‐7). Proc Natl Acad Sci U S A 92, 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasinghe NP, Wells A, Thompson EW and Hugo HJ (2012) Mesenchymal‐epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev 31, 469–478. [DOI] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer‐Hardi U, Bell G et al (2012) Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Jafferji I, Garza M, Melnikova VO, Hasegawa DK, Pethig R and Davis DW (2012) ApoStream(™), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 6, 24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, Matera J, Allard WJ, Doyle GV and Terstappen LW (2006) Circulating tumor cells at each follow‐up time point during therapy of metastatic breast cancer patients predict progression‐free and overall survival. Clin Cancer Res 12, 4218–4224. [DOI] [PubMed] [Google Scholar]
- Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS, Lim WT, Han J, Bhagat AA and Lim CT (2013) Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep 3, 1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly MK, Boareto M, Huang B, Jia D, Lu M, Ben‐Jacob E, Onuchic JN and Levine H (2015) Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol 5, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen PI et al (2014) Microfluidic, marker‐free isolation of circulating tumor cells from blood samples. Nat Protoc 9, 694–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Lim M, Park J, Oh JM, Kim H, Jeong H, Lee SJ, Park HC, Jung S, Kim BC et al (2017) FAST: Size‐selective, clog‐free isolation of rare cancer cells from whole blood at a liquid‐liquid interface. Anal Chem 89, 1155–1162. [DOI] [PubMed] [Google Scholar]
- Kim MS, Sim TS, Kim YJ, Kim SS, Jeong H, Park JM, Moon HS, Kim SI, Gurel O, Lee SS et al (2012) SSA‐MOA: a novel CTC isolation platform using selective size amplification (SSA) and a multi‐obstacle architecture (MOA) filter. Lab Chip 12, 2874–2880. [DOI] [PubMed] [Google Scholar]
- Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, Kepros JF, Barbera T, Ho MY, Chen LB et al (2004) A rare‐cell detector for cancer. Proc Natl Acad Sci U S A 101, 10501–10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labelle M, Begum S and Hynes RO (2011) Direct signaling between platelets and cancer cells induces an epithelial‐mesenchymal‐like transition and promotes metastasis. Cancer Cell 20, 576–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S and Derynck R (2007) Cell size and invasion in TGF‐β ‐induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol 178, 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa A, Garcia A, Alonso C, Millet P, Cornet M, Ramon y Cajal T, Baiget M and Barnadas A (2013) Molecular detection of peripheral blood breast cancer mRNA transcripts as a surrogate biomarker for circulating tumor cells. PLoS One 8, e74079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianidou ES, Markou A and Strati A (2015) The role of CTCs as tumor biomarkers. Adv Exp Med Biol 867, 341–367. [DOI] [PubMed] [Google Scholar]
- Liu HY, Qian HH, Zhang XF, Li J, Yang X, Sun B, Ma JY, Chen L and Yin ZF (2015) Improved method increases sensitivity for circulating hepatocellular carcinoma cells. World J Gastroenterol 21, 2918–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Fan T, Zhao Q, Zeng W, Zaslavsky E, Chen JJ, Frohman MA, Golightly MG, Madajewicz S and Chen WT (2010) Isolation of circulating epithelial and tumor progenitor cells with an invasive phenotype from breast cancer patients. Int J Cancer 126, 669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Mitra D, Sullivan RJ, Wittner BS, Kimura AM, Pan S, Hoang MP, Brannigan BW, Lawrence DP, Flaherty KT et al (2014) Isolation and molecular characterization of circulating melanoma cells. Cell Rep 7, 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Ponting CP and Voet T (2017) Single‐cell multiomics: multiple measurements from single cells. Trends Genet 33, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magbanua MJ, Carey LA, DeLuca A, Hwang J, Scott JH, Rimawi MF, Mayer EL, Marcom PK, Liu MC, Esteva FJ et al (2015) Circulating tumor cell analysis in metastatic triple‐negative breast cancers. Clin Cancer Res 21, 1098–1105. [DOI] [PubMed] [Google Scholar]
- Maheswaran S and Haber DA (2010) Circulating tumor cells: a window into cancer biology and metastasis. Curr Opin Genet Dev 20, 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswaran S and Haber DA (2015) Cell fate: transition loses its invasive edge. Nature 527, 452–453. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M et al (2008) The epithelial‐mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini S, Fruscio R, Clivio L, Beltrame L, Porcu L, Fuso Nerini I, Cavalieri D, Chiorino G, Cattoretti G, Mangioni C et al (2013) Resistance to platinum‐based chemotherapy is associated with epithelial to mesenchymal transition in epithelial ovarian cancer. Eur J Cancer 49, 520–530. [DOI] [PubMed] [Google Scholar]
- McCart Reed AE, Kutasovic JR, Vargas AC, Jayanthan J, Al‐Murrani A, Reid LE, Chambers R, Da Silva L, Melville L, Evans E et al (2016) An epithelial to mesenchymal transition programme does not usually drive the phenotype of invasive lobular carcinomas. J Pathol 238, 489–494. [DOI] [PubMed] [Google Scholar]
- McDonald OG, Wu H, Timp W, Doi A and Feinberg AP (2011) Genome‐scale epigenetic reprogramming during epithelial‐to‐mesenchymal transition. Nat Struct Mol Biol 18, 867–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ and Mihm MC Jr (2006) Melanoma. N Engl J Med 355, 51–65. [DOI] [PubMed] [Google Scholar]
- Mitchell MJ, Castellanos CA and King MR (2015) Surfactant functionalization induces robust, differential adhesion of tumor cells and blood cells to charged nanotube‐coated biomaterials under flow. Biomaterials 56, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J et al (2015) RNA‐Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349, 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami MS, Moody SA, Daar IO and Morrison DK (2004) Morphogenesis during Xenopus gastrulation requires Wee1‐mediated inhibition of cell proliferation. Development 131, 571–580. [DOI] [PubMed] [Google Scholar]
- Naume B, Borgen E, Tossvik S, Pavlak N, Oates D and Nesland JM (2004) Detection of isolated tumor cells in peripheral blood and in BM: evaluation of a new enrichment method. Cytotherapy 6, 244–252. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Huang RY, Jackson RA and Thiery JP (2016) Emt: 2016. Cell 166, 21–45. [DOI] [PubMed] [Google Scholar]
- Ocana OH, Corcoles R, Fabra A, Moreno‐Bueno G, Acloque H, Vega S, Barrallo‐Gimeno A, Cano A and Nieto MA (2012) Metastatic colonization requires the repression of the epithelial‐mesenchymal transition inducer Prrx1. Cancer Cell 22, 709–724. [DOI] [PubMed] [Google Scholar]
- Pal D, Pertot A, Shirole NH, Yao Z, Anaparthy N, Garvin T, Cox H, Chang K, Rollins F, Kendall J et al (2017) TGF‐β reduces DNA ds‐break repair mechanisms to heighten genetic diversity and adaptability of CD44+/CD24‐ cancer cells. Elife 6, e21615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L et al (2006) Molecular subclasses of high‐grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173. [DOI] [PubMed] [Google Scholar]
- Pixberg CF, Raba K, Muller F, Behrens B, Honisch E, Niederacher D, Neubauer H, Fehm T, Goering W, Schulz WA et al (2017) Analysis of DNA methylation in single circulating tumor cells. Oncogene. https://doi.org/10.1038/onc.2016.480. [DOI] [PubMed] [Google Scholar]
- Polioudaki H, Agelaki S, Chiotaki R, Politaki E, Mavroudis D, Matikas A, Georgoulias V and Theodoropoulos PA (2015) Variable expression levels of keratin and vimentin reveal differential EMT status of circulating tumor cells and correlation with clinical characteristics and outcome of patients with metastatic breast cancer. BMC Cancer 15, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi C, Gradilone A, Naso G, Vincenzi B, Petracca A, Nicolazzo C, Palazzo A, Saltarelli R, Spremberg F, Cortesi E et al (2011) Epithelial‐mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Cancer Res Treat 130, 449–455. [DOI] [PubMed] [Google Scholar]
- Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH et al (2012) EMT and dissemination precede pancreatic tumor formation. Cell 148, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscetti M, Quach B, Dadashian EL, Mulholland DJ and Wu H (2015) Tracking and functional characterization of epithelial‐mesenchymal transition and mesenchymal tumor cells during prostate cancer metastasis. Cancer Res 75, 2749–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba AE, Westermann AJ, Gorski SA and Vogel J (2014) Single‐cell RNA‐seq: advances and future challenges. Nucleic Acids Res 42, 8845–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satelli A, Batth IS, Brownlee Z, Rojas C, Meng QH, Kopetz S and Li S (2016) Potential role of nuclear PD‐L1 expression in cell‐surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. Sci Rep 6, 28910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satelli A, Brownlee Z, Mitra A, Meng QH and Li S (2015a) Circulating tumor cell enumeration with a combination of epithelial cell adhesion molecule‐ and cell‐surface vimentin‐based methods for monitoring breast cancer therapeutic response. Clin Chem 61, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satelli A, Mitra A, Brownlee Z, Xia X, Bellister S, Overman MJ, Kopetz S, Ellis LM, Meng QH and Li S (2015b) Epithelial‐mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin Cancer Res 21, 899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savagner P (2015) Epithelial‐mesenchymal transitions: from cell plasticity to concept elasticity. Curr Top Dev Biol 112, 273–300. [DOI] [PubMed] [Google Scholar]
- Schneck H, Gierke B, Uppenkamp F, Behrens B, Niederacher D, Stoecklein NH, Templin MF, Pawlak M, Fehm T, Neubauer H et al (2015) EpCAM‐independent enrichment of circulating tumor cells in metastatic breast cancer. PLoS One 10, e0144535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seher TC and Leptin M (2000) Tribbles, a cell‐cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation. Curr Biol 10, 623–629. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias‐Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK et al (2011) Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 3, 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw Bagnall J, Byun S, Begum S, Miyamoto DT, Hecht VC, Maheswaran S, Stott SL, Toner M, Hynes RO and Manalis SR (2015) Deformability of tumor cells versus blood cells. Sci Rep 5, 18542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone RC, Pastar I, Ojeh N, Chen V, Liu S, Garzon KI and Tomic‐Canic M (2016) Epithelial‐mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res 365, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K and Surani MA (2010) RNA‐Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc 5, 516–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY and Nieto MA (2009) Epithelial‐mesenchymal transitions in development and disease. Cell 139, 871–890. [DOI] [PubMed] [Google Scholar]
- Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW, Xega K et al (2014) Single‐cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep 8, 1905–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HD, Luitel K, Kim M, Zhang K, Longmore GD and Tran DD (2014) Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res 74, 6330–6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M et al (2000) Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol 156, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werden SJ, Sphyris N, Sarkar TR, Paranjape AN, LaBaff AM, Taube JH, Hollier BG, Ramirez‐Pena EQ, Soundararajan R, den Hollander P et al (2016) Phosphorylation of serine 367 of FOXC2 by p38 regulates ZEB1 and breast cancer metastasis, without impacting primary tumor growth. Oncogene 35, 5977–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, Helfrich B, Dziadziuszko R, Chan DC, Sugita M et al (2006) Restoring E‐cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res 66, 944–950. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu S, Liu Z, Huang J, Pu X, Li J, Yang D, Deng H, Yang N and Xu J (2015) Classification of circulating tumor cells by epithelial‐mesenchymal transition markers. PLoS One 10, e0123976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Park KJ, Deighan C, Amaya P, Miller B, Pan Q, Zborowski M, Lustberg M and Chalmers J (2016) Multiparameter evaluation of the heterogeneity of circulating tumor cells using integrated RNA in situ hybridization and immunocytochemical analysis. Front Oncol 6, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and Weinberg RA (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939. [DOI] [PubMed] [Google Scholar]
- Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E and Weinberg RA (2015) Distinct EMT programs control normal mammary stem cells and tumour‐initiating cells. Nature 525, 256–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM et al (2013) Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Cai Z, Li S, Cheng Y, Gao H, Liu F, Wu S, Liu S, Dong Y, Zheng L et al (2017) Expression and clinical relevance of epithelial and mesenchymal markers in circulating tumor cells from colorectal cancer. Oncotarget 8, 9293–9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS and Kalluri R (2015) Epithelial‐to‐mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]


