Abstract
Epithelial‐mesenchymal transition (EMT) is an important process in embryonic development, fibrosis, and cancer metastasis. During cancer progression, the activation of EMT permits cancer cells to acquire migratory, invasive, and stem‐like properties. A growing body of evidence supports the critical link between EMT and cancer stemness. However, contradictory results have indicated that the inhibition of EMT also promotes cancer stemness, and that mesenchymal‐epithelial transition, the reverse process of EMT, is associated with the tumor‐initiating ability required for metastatic colonization. The concept of ‘intermediate‐state EMT’ provides a possible explanation for this conflicting evidence. In addition, recent studies have indicated that the appearance of ‘hybrid’ epithelial‐mesenchymal cells is favorable for the establishment of metastasis. In summary, dynamic changes or plasticity between the epithelial and the mesenchymal states rather than a fixed phenotype is more likely to occur in tumors in the clinical setting. Further studies aimed at validating and consolidating the concept of intermediate‐state EMT and hybrid tumors are needed for the establishment of a comprehensive profile of cancer metastasis.
Keywords: epithelial‐mesenchymal transition, metastasis, plasticity, stemness
Abbreviations
- CBP
CREB binding protein
- CSCs
cancer stem cells
- CtBP1
C‐terminal binding protein 1
- EMT
epithelial‐mesenchymal transition
- EMT‐TF
epithelial‐mesenchymal transition transcription factor
- ERCC1
ERCC excision repair 1, endonuclease noncatalytic subunit
- HDAC
histone deacetylase
- HIF‐1α
hypoxia‐inducible factor 1 alpha
- Id1
inhibitor of differentiation 1
- IL‐8
interleukin‐8
- LSD1
lysine‐specific demethylase
- MET
mesenchymal‐epithelial transition
- OVOL2
ovo‐like zinc finger 2
- PRC2
polycomb repressive complex 2
- Snail1
Snail family zinc finger 1
- TGF‐β
transforming growth factor beta
- Twist1
twist family bHLH transcription factor 1
- ZEB1
zinc finger E‐box binding homeobox 1
- ZEB2
zinc finger E‐box binding homeobox 2
1. General overview of EMT
During embryonic development, epithelial cells lose their polarity and are converted into a mesenchymal phenotype. This process is referred to as epithelial‐mesenchymal transition (EMT) (Nieto et al., 2016). The classic view of EMT is that epithelial cells transform into mesenchymal cells. Morphological changes in cells have been considered the characteristic feature of EMT (Hay, 1995; Nieto, 2013). EMT presents certain features that are considered as its hallmarks, including disruption of intercellular junctions, loss of cell polarity, reorganization of the cytoskeleton, and increased cell motility. Therefore, in most experimental models, epithelial (E‐cadherin) and mesenchymal (N‐cadherin and vimentin) markers and morphological changes are examined as indicators to confirm the occurrence of EMT. In cancers, EMT is triggered by diverse signaling pathways through the regulation of EMT transcription factors (EMT‐TFs) and/or microRNAs (miRNAs) (Nieto et al., 2016). EMT not only enhances cancer motility and dissemination through the disruption of intercellular junctions but also allows cells to acquire stem‐like properties (Nieto et al., 2016). However, the reverse process of EMT, that is, mesenchymal‐epithelial transition (MET), is an important process for cancer cell re‐differentiation and metastatic colonization (Bonnomet et al., 2012). Therefore, the association between EMT‐MET and stemness is controversial and debated. The major factors and signaling pathways that trigger the changes in EMT/MET are summarized in Fig. 1. In this review, we summarize and discuss the connection between epithelial and mesenchymal states and the acquisition of stemness in cancer cells.
Figure 1.
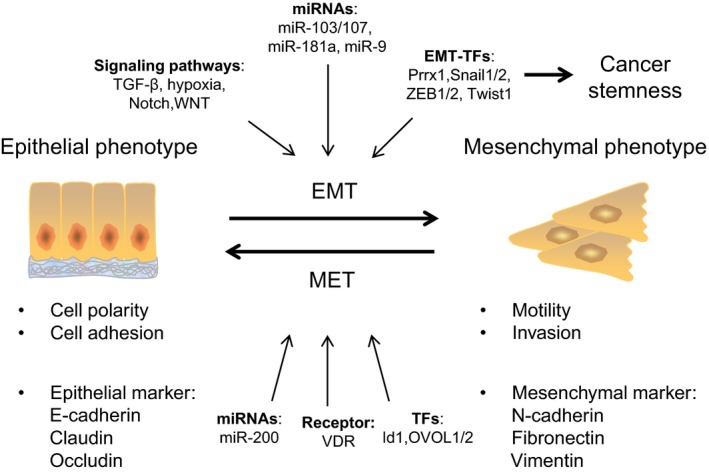
The dynamic change between the epithelial and the mesenchymal phenotype in cancer cells during metastasis. In response to EMT‐triggering events, such as the activation of signaling pathways (e.g., TGF‐β, hypoxia, Notch, WNT) or the expression of EMT‐TFs (e.g., Snail1/2, Twist1, ZEB1/2, Prrx1) and miRNAs (e.g., miR‐103/107, miR‐181a, miR‐9), cancer cells transition from an epithelial phenotype to a mesenchymal phenotype, with the suppression of epithelial markers and expression of mesenchymal markers. Activation of an EMT program results in the acquisition of migration and invasion abilities for facilitating cancer dissemination. Furthermore, EMT‐TFs promote cancer cells to acquire the stem‐like features. After the mesenchymal‐type cancer cells reaching the metastatic sites, the cancer cells reverse back to the epithelial type through MET, which is critical for cancer colonization. The effectors of MET include the activation of certain transcriptional factors (e.g., Id1, OVOL1/2), miRNAs (e.g., miR‐200), and receptor (VDR).
1.1. EMT transcription factors
One of the major events contributing to EMT is the activation of EMT‐TFs, such as Snail1, Twist1, ZEB1, and ZEB2. These EMT‐TFs often control the expression of each other and cooperate with other TFs to regulate the expression of target genes, and EMT‐TFs often function as repressors for epithelial genes and activators for mesenchymal genes (De Craene and Berx, 2013; Peinado et al., 2007).
1.1.1. Snail1
Snail1 (also known as Snail) functions as a suppressor by binding to the E‐box in the promoters of the junction proteins E‐cadherin, claudin, and occludin and recruiting histone modifiers, including SIN3A‐histone deacetylase 1 and 2 (HDAC1 and HDAC2) complex, polycomb repressive complex 2 (PRC2), and lysine‐specific demethylase 1, to repress the transcription of target genes (Batlle et al., 2000; Cano et al., 2000; Herranz et al., 2008; Ikenouchi et al., 2003; Lin et al., 2010a,b; Peinado et al., 2004). However, Snail1 also acts as an activator that increases the expression of mesenchymal genes such as fibronectin 1, an extracellular matrix protein (Stanisavljevic et al., 2011); excision repair 1 endonuclease noncatalytic subunit (ERCC1), an endonuclease noncatalytic subunit that is required for the repair of DNA lesions (Hsu et al., 2010); and interleukin‐8 (Hwang et al., 2011) to contribute to the mesenchymal phenotype. Moreover, Snail1 acts as an activator by interacting with CREB binding protein, which prevents repressor complex formation and remodels the tumor microenvironment (Hsu et al., 2014).
1.1.2. Slug
Slug (also known as Snail2) belongs to the Snail superfamily of zinc finger transcriptional factors (Nieto, 2002). Slug interacts with the corepressor nuclear receptor coreceptor and recruits C‐terminal binding protein 1 (CtBP1) for repressing E‐cadherin and triggering EMT (Hajra et al., 2002; Molina‐Ortiz et al., 2012; Nieto, 2002). Slug also binds to E2‐box sequence of the target genes promoter (BRCA2 and VDR) and recruits CtBP1 and HDAC1 to suppress the gene expression (Hemavathy et al., 2000; Molina‐Ortiz et al., 2012; Tripathi et al., 2005). Overexpression of VDR upregulates E‐cadherin, downregulates SNAI1, TWIST1, and MMP9, and reduces the ability to form mammospheres, an attribute of breast normal and cancer stem cells (CSCs; Larriba et al., 2016; Pervin et al., 2013). Degradation of Slug consequently enhances E‐cadherin expression and represses cancer cell invasion (Mittal et al., 2008; Shih and Yang, 2011; Wang et al., 2009).
1.1.3. ZEB1
Zinc finger E‐box binding homeobox 1 (ZEB1) binds to E‐boxes and represses the expression of E‐cadherin to induce EMT (Eger et al., 2005; Spoelstra et al., 2006; Witta et al., 2006). ZEB1 can function as an activator by interacting with Smads, signaling mediators of the transforming growth factor beta (TGF‐β) pathway, and the transcriptional coactivator p300 (Pena et al., 2006; Postigo et al., 2003). The EMT‐inhibiting transcription factor ovo‐like zinc finger 2 restricts EMT by directly inhibiting EMT‐inducing factor ZEB1 and induces MET (Hong et al., 2015; Kitazawa et al., 2016; Roca et al., 2013; Watanabe et al., 2014). ZEB1 is indicated as a key factor for pancreatic cancer progression. Depletion of ZEB1 suppresses stemness and colonization capacity of tumor cells in Pdx1‐cre‐mediated activation of mutant Kras and p53 (KPC) model of pancreatic cancer. In this model, EMT‐TFs Snail1 and Twist1 had no such effect (Krebs et al., 2017; Zheng et al., 2015). Krebs et al. (2017) also suggested that there are considerable functional variabilities and tissue specificities among different EMT‐TFs. With regard to the interplay between ZEB1 and other EMT‐TFs, Snail1 acts cooperatively with Twist1 to control the expression of ZEB1 (Dave et al., 2011).
1.1.4. ZEB2
Zinc finger E‐box binding homeobox 2 (ZEB2) acts as a transcriptional repressor and regulates downstream targets either dependent or independent of the CtBP1 corepressor complex (van Grunsven et al., 2003; Shi et al., 2003). ZEB2 induces EMT by binding to the E‐cadherin promoter and repressing the transcription of E‐cadherin (Comijn et al., 2001). Moreover, ZEB2 has been shown to repress the expression of several genes encoding junctional proteins, including desmosomal proteins desmoplakin and plakophilin 2 and tight junction protein claudin 4 (Vandewalle et al., 2005). ZEB2 is regulated by sumoylation, which attenuates gene repression by the disruption of CtBP1 recruitment (Long et al., 2005).
1.1.5. Twist1
Twist1, a basic helix‐loop‐helix transcriptional factor, is a master regulator of gastrulation and mesoderm specification (Castanon and Baylies, 2002; Furlong et al., 2001) and is recently demonstrated to be essential to mediate cancer metastasis (Yang et al., 2004). Ectopic expression of Twist1 upregulates mesenchymal cell markers (fibronectin, vimentin, smooth muscle actin, and N‐cadherin) and a loss of epithelial markers (E‐cadherin, and α‐ and γ‐catenin), and induces EMT (Kang and Massague, 2004; Yang et al., 2004). Twist1 has been shown to play a vital role in the intravasation step of metastasis, angiogenesis, and chromosomal instability (Mironchik et al., 2005; Yang et al., 2004). Under hypoxic condition, a principal feature of malignancies, HIF‐1α promotes EMT through the induction of Twist1 (Yang et al., 2008). Twist1 in turn activates Bmi1, and both of them are essential for promoting EMT and tumor‐initiating capacity (Yang et al., 2008, 2010). A report by Tsai et al. (2012) also indicated that turning off Twist1 reversed the EMT process, leading to the subsequent occurrence of MET for colonization and the formation of metastases, indicating that Twist1 is an important regulator of epithelial plasticity during cancer metastasis.
1.2. Signaling pathways for EMT induction
EMT transcription factors can be activated through different pathways, which strongly suggest the convergence of diverse pathways on common targets during EMT (Lamouille et al., 2014). TGF‐β deposited in the surrounding stroma or secreted from tumor cells induces the expression of both ZEB1 and Snail1, thereby triggering EMT to promote tumor progression and metastasis (Korpal et al., 2008; Zavadil and Bottinger, 2005). Notch signaling pathway plays an important role in physiological and pathologic conditions through the induction of EMT (Niessen et al., 2008; Timmerman et al., 2004; Wang et al., 2010; Zavadil et al., 2004). WNT family proteins and growth factors that act through receptor tyrosine kinases have also been shown to induce EMT (Lamouille et al., 2014). Hypoxia induces the expression of Twist1 or Snail to promote EMT during cancer progression (Peinado and Cano, 2008).
1.3. miRNAs for regulation of EMT
miRNA that selectively target mRNA for the degradation of mRNA or translational repression also participate in the regulation of the EMT process (Ambros, 2004; Lamouille et al., 2013). For example, the miR‐200 family miRNAs have been shown to repress the expression of ZEB1 and ZEB2, thereby maintaining cancer cells in the epithelial phenotype (Gregory et al., 2008; Korpal et al., 2008; Park et al., 2008). ZEB1/2 and miR‐200 family members have a double‐negative feedback loop that controls the balance between epithelial and mesenchymal states (Bracken et al., 2008; Gregory et al., 2011). miR‐103/107 induces EMT by targeting Dicer, a key component of the miRNA processing machinery, to downregulate the level of miR‐200 in breast cancer cells (Martello et al., 2010). Inhibition of the let‐7d causes EMT (Huleihel et al., 2014; Pandit et al., 2010). miR‐181a mediates TGF‐β‐induced EMT (Brockhausen et al., 2015). miR‐9 directly targets the E‐cadherin‐encoding mRNA CDH1, leading to an EMT‐like conversion (Ma et al., 2010). In summary, signaling within the microenvironment triggers the activation of EMT‐TFs, resulting in the occurrence of EMT in cancer cells. miRNAs also function as major mediators of EMT by regulating the expression of EMT‐TFs.
2. EMT and cancer stemness
In the past decade, accumulating evidence has shown that EMT permits cancer cells to acquire stem cell properties for metastasis and dissemination. Here, we will focus on the association between EMT and cancer stemness.
2.1. Cancer stem cells
Intratumoral heterogeneity contributes to therapeutic resistance and results in disease recurrence (Hanahan and Weinberg, 2011). CSCs are a small population of cancer cells with the characteristics of self‐renewal, tumor initiation, and chemotherapy resistance (O'Brien et al., 2007; Ricci‐Vitiani et al., 2007; Todaro et al., 2007, 2014). The existence of CSCs was initially intensively debated; however, the concept of CSCs has been strongly supported by the application of spontaneous tumor mouse models and genetic tracing (Chen et al., 2012; Driessens et al., 2012; Schepers et al., 2012). Moreover, the term ‘stemness’, which was initially used to describe the properties of normal stem cells, has been expanded to illustrate the feature of CSCs with reference to the molecular signatures that control and maintain the stem cell state. In experimental models, stemness is generally defined as an increase in cancer type‐specific stem cell markers. The reported markers for CSCs in different types of cancers are illustrated in Table 1. Furthermore, serial replating of tumorspheres and in vivo serial repopulation assays have been applied as the standard procedures for testing the self‐renewal ability of cancer cells.
Table 1.
CSC markers for different tumor types
| Cancer types | CSC markers | Features/Reference |
|---|---|---|
| Breast | ALDH1 | Tumor initiation in xenograft, poor prognostic factor, metastasis (Ginestier et al., 2007) |
| CD44 | Mammosphere formation, tumor initiation in xenograft, poor prognostic factor, metastasis (Al‐Hajj et al., 2003; Leth‐Larsen et al., 2012; Ponti et al., 2005) | |
| Sox2 | Mammosphere formation, tumor initiation in xenograft (Leis et al., 2012) | |
| Colon | LGR5 | Increase pluripotency and self‐renewal (lineage tracing); induces clonogenicity and tumorigenicity (Barker et al., 2007; Kemper et al., 2012) |
| CD24 | Increase carcinogenesis; express in spheroid cultures (Sagiv et al., 2006; Vermeulen et al., 2008) | |
| CD29 | Increase colony formation; express in spheroid cultures (Fujimoto et al., 2002; Vermeulen et al., 2008) | |
| CD44 | Tumor initiation in xenograft, colony formation; poor prognostic factor, lymph node infiltration (Dalerba et al., 2007; Du et al., 2008; Huh et al., 2009) | |
| CD133 | Tumor initiation in xenograft, sphere formation (Ricci‐Vitiani et al., 2007) | |
| Head and neck | Oct4 | Sphere formation, chemoresistance, invasion, migration, tumor initiation in xenograft, poor prognostic factor (Koo et al., 2015; Liao et al., 2016) |
| CD44 | Tumor initiation in xenograft, colony formation, sphere formation (Krishnamurthy et al., 2010; Prince et al., 2007) | |
| ALDH1 | Tumor initiation in xenograft, colony formation, sphere formation, radioresistance (Krishnamurthy et al., 2010; Major et al., 2013) | |
| Liver | CD133 | Tumor initiation in xenograft, clonogenicity (Yin et al., 2007) |
| SALL4 | Poor prognostic factor, tumor proliferation, chemoresistance, tumor initiation in xenograft (Oikawa et al., 2013) | |
| ALDH1 | Tumor initiation in xenograft, proliferation, sphere formation (Ma et al., 2008) | |
| Pancreas | CD24/CD44/EpCAM | Tumor initiation in xenograft (Li et al., 2009) |
| CD133 | Metastasis, poor prognostic factor (Hermann et al., 2007; Li et al., 2015) | |
| CXCR4 | Metastasis, poor prognostic factor (Hermann et al., 2007; Marechal et al., 2009; Wang et al., 2015) | |
| Prostate | CD133 | Proliferation, invasion, clonogenicity, glandular regeneration (Collins et al., 2005; Vander Griend et al., 2008) |
| CD44 | Tumor initiation in xenograft, proliferation, clonogenicity, metastasis, poor prognostic factor (Hurt et al., 2008; Li et al., 2007; Patrawala et al., 2006) | |
| EpCAM | Tumor initiation in xenograft, metastasis (Deng et al., 2015; Li et al., 2007) |
2.2. Correlation between EMT and stemness
Exposing human mammary epithelial cells to TGF‐β or the ectopic expression of Snail1/Twist1 induces a cell population with stem cell characteristics, including enhanced expression of CD44 (CD44high) and low expression of CD24 (CD24low) and the ability to form mammospheres (Mani et al., 2008). Prostate cancer cells with the mesenchymal phenotype display stem‐like properties, including increased expression of the pluripotency genes Sox2, Nanog, and Oct4, enhanced clonogenic and sphere‐forming ability, and tumorigenicity in vivo (Kong et al., 2010). In pancreatic cancer, ZEB1 is the critical link between the activation of EMT and the acquisition of stem‐like properties and functions by suppressing miR‐200 family members, which are strong inducers of epithelial differentiation. Activation of ZEB1 promotes EMT and the expression of stem cell factors such as Sox2 and Klf4 (Wellner et al., 2009). Bmi1, a polycomb‐group protein that maintains self‐renewal, is directly regulated by Twist1, which links EMT to tumor‐initiating ability (Wu and Yang, 2011; Wu et al., 2012; Yang et al., 2010). The EMT process can also confer resistance to senescence. Twist1/2 and ZEB1/2 override oncogene‐induced premature senescence by inhibiting p53‐ and Rb‐dependent pathways (Ansieau et al., 2008; Morel et al., 2012; Ohashi et al., 2010). Furthermore, Twist1 acts together with Bmi1 to suppress the expression of let‐7, a microRNA expressed during stem cell differentiation, leading to cancer stemness (Yang et al., 2012). Downregulation of let‐7 activates the chromatin modifier ARID3B to promote expression of stemness genes through histone modification (Liao et al., 2016). In colon CSCs, Snail1 mediates the switch from asymmetric to symmetric cell division, indicating a role for EMT in increasing the size of the CSC pool (Hwang et al., 2014). Slug‐driven EMT program is important for inducing the entrance into adult stem cell state; however, it is not sufficient to induce this change in ‘differentiated’ luminal cells. Instead, activation of an additional genetic program through expression of Sox9 is required to work in concert with the EMT program to induce stem cells (Guo et al., 2012).
Intriguingly, EMT has also been shown to inhibit the development of stem‐like traits in certain studies (Celia‐Terrassa et al., 2012; Korpal et al., 2011; Sarrio et al., 2012), a finding that contradicts the concept of EMT‐induced stemness. Further evidence has shown that, in human breast cancer cells, knockdown of paired‐related homeobox transcription factor 1 (Prrx1), a recently identified EMT inducer, increased mammosphere formation, self‐renewal capacity, and the proportion of enhanced expression of CD44 (CD44high) and low expression of CD24 (CD24low) CSCs (Ocana et al., 2012). Moreover, another study showed that Twist1 is essential for the acquisition of CSC properties; however, cancer stemness is independent of EMT or tumor invasion, implying that EMT and stemness are regulated separately (Beck et al., 2015). Transient activation of Twist1 promotes cancer stemness, even when EMT has not been induced (Schmidt et al., 2015). Taken together, this indicates that EMT is closely associated with but is not necessary for cancer stemness. EMT‐TFs are the critical mediators that link EMT to stemness, but the mechanisms are different, including epigenetic and miRNA regulation; in other words, the regulation of EMT and stemness are an independent function of the same EMT‐TFs. This correlation between EMT and cancer stemness is more complicated than expected and deserves intensive investigation in the future.
3. Cell plasticity and cancer stemness
Studies in induced pluripotent stem cells (iPSCs) showed that MET, the reverse process of EMT, is a prerequisite for the reprogramming of fibroblasts to iPSCs (Li et al., 2010; Samavarchi‐Tehrani et al., 2010). During the reprogramming process, Oct4/Sox2 represses the expression of Snail1, c‐Myc reduces the expression of TGF‐β1 and TGF‐β receptor П, and Klf4 activates the expression of E‐cadherin. All these events result in MET (Li et al., 2010). During tumor progression, MET is considered an essential process for metastatic colonization (Nieto, 2013). Evidence of EMT in clinical specimen is the fact that the histology of metastatic tumors exhibits the epithelial phenotype rather than the mesenchymal‐like phenotype, suggesting that the reversion of EMT occurs during metastatic colonization (Yao et al., 2011). Moreover, miR‐200 family miRNAs were shown to promote MET, which was also found to increase metastatic colonization (Dykxhoorn et al., 2009; Perdigao‐Henriques et al., 2016). In addition to metastatic colonization, MET has also been noted to promote the stemness of cancer cells. For example, inhibitor of differentiation 1 (Id1) induces MET and the stem‐like phenotype by antagonizing Twist1 (Stankic et al., 2013). Connective tissue growth factor has been noted to enhance stem‐like properties and trigger MET in head and neck cancer cells (Chang et al., 2013). Furthermore, transient expression of Twist1 induces long‐term invasiveness and colonization capability by promoting the coexistence of the features of epithelial and mesenchymal cells (Schmidt et al., 2015). This result suggests that an ‘intermediate state’ of cancer cells may be more flexible in terms of cell invasion and the regulation of stem‐like properties.
A concern of previous studies is that most instances of EMT or MET were achieved by the forced expression of certain factors, which fixed cells in a terminal epithelial or mesenchymal state and may not reflect the dynamic process of transition between epithelial and mesenchymal status in vivo. For example, circulating tumor cells (CTCs) have been shown to express both epithelial and mesenchymal markers (Bonnomet et al., 2012; Lecharpentier et al., 2011; Paterlini‐Brechot and Benali, 2007; Raimondi et al., 2011; Yu et al., 2013). In patients with advanced metastatic cancer, a high frequency of ‘hybrid’ CTC populations expresses CSC markers (Armstrong et al., 2011; Theodoropoulos et al., 2010). A recent study that used intravital microscopy to observe epithelial‐mesenchymal plasticity without artificially modifying the expression of EMT regulators showed that epithelial‐mesenchymal plasticity occurs during the migration process but not when cells enter the circulation. This study also observed that mesenchymal cells adopt the epithelial state after several rounds of cell division upon reaching metastatic sites (Beerling et al., 2016). Furthermore, the hybrid epithelial/mesenchymal (E/M) cells in primary ovarian cancer cells and prostate cancer cells showed higher self‐renewal and tumor‐initiating ability (Ruscetti et al., 2015; Strauss et al., 2011). The concept of hybrid E/M cells in metastatic colonization is shown in Fig. 2. Therefore, stemness properties are no longer a feature of a fixed state, but follow the changes in the cells as a flexible feature. Further studies are necessary to clarify the mechanism and significance of epithelial plasticity and stemness in tumor cells.
Figure 2.
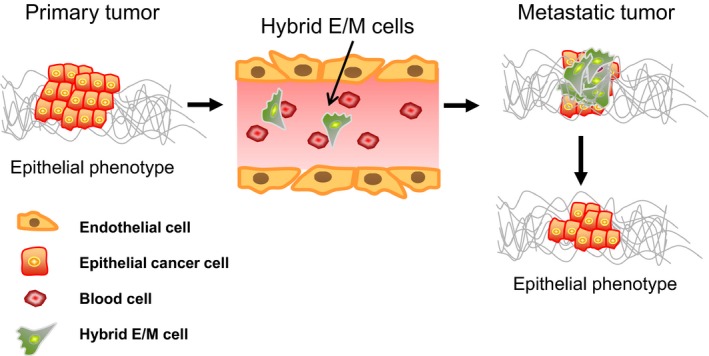
A model for depicting cellular plasticity for cancer metastasis. In primary tumors, most cancer cells have an epithelial type. In metastatic cancer, hybrid epithelial/mesenchymal (E/M) cells or partial EMT is favorable for cancer dissemination. When the hybrid E/M cells reach the metastatic site, they will revert back to epithelial cells to form metastatic colonies, possibly via rapid kinetics. Therefore, the epithelial/mesenchymal features and stem‐like properties are no longer a fixed state. A dynamic or a flexible feature of E/M phenotype is a better description for the plasticity of cancer cells.
4. Conclusions
Experimental models of EMT have been used for decades and have established a foundation for us to elucidate the mechanisms underlying EMT, metastasis, and tumor initiation. However, this dichotomy between the epithelial and the mesenchymal states may be oversimplified and may not precisely reflect the situation in vivo. The concept of an ‘intermediate‐state’, or so‐called partial EMT, provides a possible explanation for this controversy. The phenomenon of partial EMT has been found to occur during the process of embryo development and in wound healing, and a growing body of evidence indicates the existence of partial EMT in cancer biology. Hence, the development of an in vivo model will be important for providing a research tool for us to use in elucidating the dynamic changes in the epithelial‐mesenchymal phenotype and the regulation of stemness properties in pathophysiological microenvironments. Considering a process of plastic change between the epithelial and the mesenchymal states is more useful than considering the process of a fixed transition for our understanding of cancer progression and metastasis.
Acknowledgements
This work was supported by grants from the Ministry of Science and Technology (104‐2321‐B‐010‐005, 104‐0210‐01‐09‐02, and 103‐2633‐H‐010‐001 to M‐HY); Taipei Veterans General Hospital (V106C‐090 and VTA106‐V1‐3‐3 to M‐HY); the Ministry of Education, Aim for the Top University Plan (to M‐HY); and the Ministry of Health and Welfare, Center of Excellence for Cancer Research (MOHW106‐TDU‐B‐211‐144‐003 to M‐HY).
References
- Al‐Hajj M, Wicha MS, Benito‐Hernandez A, Morrison SJ and Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100, 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V (2004) The functions of animal microRNAs. Nature 431, 350–355. [DOI] [PubMed] [Google Scholar]
- Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S et al (2008) Induction of EMT by twist proteins as a collateral effect of tumor‐promoting inactivation of premature senescence. Cancer Cell 14, 79–89. [DOI] [PubMed] [Google Scholar]
- Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, Herold CI, Marcom PK, George DJ and Garcia‐Blanco MA (2011) Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res 9, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ et al (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J and Garcia De Herreros A (2000) The transcription factor snail is a repressor of E‐cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2, 84–89. [DOI] [PubMed] [Google Scholar]
- Beck B, Lapouge G, Rorive S, Drogat B, Desaedelaere K, Delafaille S, Dubois C, Salmon I, Willekens K, Marine JC et al (2015) Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell 16, 67–79. [DOI] [PubMed] [Google Scholar]
- Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, Schafer R, van Diest P, Voest E, van Oudenaarden A et al (2016) Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis‐enhancing stem cell capacity. Cell Rep 14, 2281–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnomet A, Syne L, Brysse A, Feyereisen E, Thompson EW, Noel A, Foidart JM, Birembaut P, Polette M and Gilles C (2012) A dynamic in vivo model of epithelial‐to‐mesenchymal transitions in circulating tumor cells and metastases of breast cancer. Oncogene 31, 3741–3753. [DOI] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF and Goodall GJ (2008) A double‐negative feedback loop between ZEB1‐SIP1 and the microRNA‐200 family regulates epithelial‐mesenchymal transition. Cancer Res 68, 7846–7854. [DOI] [PubMed] [Google Scholar]
- Brockhausen J, Tay SS, Grzelak CA, Bertolino P, Bowen DG, d'Avigdor WM, Teoh N, Pok S, Shackel N, Gamble JR et al (2015) miR‐181a mediates TGF‐beta‐induced hepatocyte EMT and is dysregulated in cirrhosis and hepatocellular cancer. Liver Int 35, 240–253. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez‐Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA (2000) The transcription factor snail controls epithelial‐mesenchymal transitions by repressing E‐cadherin expression. Nat Cell Biol 2, 76–83. [DOI] [PubMed] [Google Scholar]
- Castanon I and Baylies MK (2002) A Twist in fate: evolutionary comparison of Twist structure and function. Gene 287, 11–22. [DOI] [PubMed] [Google Scholar]
- Celia‐Terrassa T, Meca‐Cortes O, Mateo F, Martinez de Paz A, Rubio N, Arnal‐Estape A, Ell BJ, Bermudo R, Diaz A, Guerra‐Rebollo M et al (2012) Epithelial‐mesenchymal transition can suppress major attributes of human epithelial tumor‐initiating cells. J Clin Invest 122, 1849–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Hsu WH, Wang CC, Chou CH, Kuo MY, Lin BR, Chen ST, Tai SK, Kuo ML and Yang MH (2013) Connective tissue growth factor activates pluripotency genes and mesenchymal‐epithelial transition in head and neck cancer cells. Cancer Res 73, 4147–4157. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG and Parada LF (2012) A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488, 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AT, Berry PA, Hyde C, Stower MJ and Maitland NJ (2005) Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 65, 10946–10951. [DOI] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D and van Roy F (2001) The two‐handed E box binding zinc finger protein SIP1 downregulates E‐cadherin and induces invasion. Mol Cell 7, 1267–1278. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM et al (2007) Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A 104, 10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave N, Guaita‐Esteruelas S, Gutarra S, Frias A, Beltran M, Peiro S and de Herreros AG (2011) Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J Biol Chem 286, 12024–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Wu Y, Ma W, Zhang S and Zhang YQ (2015) Adoptive T‐cell therapy of prostate cancer targeting the cancer stem cell antigen EpCAM. BMC Immunol 16, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B and Berx G (2013) Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 13, 97–110. [DOI] [PubMed] [Google Scholar]
- Driessens G, Beck B, Caauwe A, Simons BD and Blanpain C (2012) Defining the mode of tumour growth by clonal analysis. Nature 488, 527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Wang H, He L, Zhang J, Ni B, Wang X, Jin H, Cahuzac N, Mehrpour M, Lu Y et al (2008) CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res 14, 6751–6760. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B and Lieberman J (2009) miR‐200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One 4, e7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H and Foisner R (2005) DeltaEF1 is a transcriptional repressor of E‐cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 24, 2375–2385. [DOI] [PubMed] [Google Scholar]
- Fujimoto K, Beauchamp RD and Whitehead RH (2002) Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology 123, 1941–1948. [DOI] [PubMed] [Google Scholar]
- Furlong EE, Andersen EC, Null B, White KP and Scott MP (2001) Patterns of gene expression during Drosophila mesoderm development. Science 293, 1629–1633. [DOI] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe‐Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S et al (2007) ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew‐Goodall Y and Goodall GJ (2008) The miR‐200 family and miR‐205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10, 593–601. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, Morris M, Wyatt L, Farshid G, Lim YY et al (2011) An autocrine TGF‐beta/ZEB/miR‐200 signaling network regulates establishment and maintenance of epithelial‐mesenchymal transition. Mol Biol Cell 22, 1686–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Grunsven LA, Michiels C, Van de Putte T, Nelles L, Wuytens G, Verschueren K and Huylebroeck D (2003) Interaction between Smad‐interacting protein‐1 and the corepressor C‐terminal binding protein is dispensable for transcriptional repression of E‐cadherin. J Biol Chem 278, 26135–26145. [DOI] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer‐Hardi U, Bell G et al (2012) Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajra KM, Chen DY and Fearon ER (2002) The SLUG zinc‐finger protein represses E‐cadherin in breast cancer. Cancer Res 62, 1613–1618. [PubMed] [Google Scholar]
- Hanahan D and Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Hay ED (1995) An overview of epithelio‐mesenchymal transformation. Acta Anat 154, 8–20. [DOI] [PubMed] [Google Scholar]
- Hemavathy K, Guru SC, Harris J, Chen JD and Ip YT (2000) Human Slug is a repressor that localizes to sites of active transcription. Mol Cell Biol 20, 5087–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ and Heeschen C (2007) Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 1, 313–323. [DOI] [PubMed] [Google Scholar]
- Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez‐Munoz I, Di Croce L, Helin K et al (2008) Polycomb complex 2 is required for E‐cadherin repression by the Snail1 transcription factor. Mol Cell Biol 28, 4772–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong T, Watanabe K, Ta CH, Villarreal‐Ponce A, Nie Q and Dai X (2015) An Ovol2‐Zeb1 mutual inhibitory circuit governs bidirectional and multi‐step transition between epithelial and mesenchymal states. PLoS Comput Biol 11, e1004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu DS, Lan HY, Huang CH, Tai SK, Chang SY, Tsai TL, Chang CC, Tzeng CH, Wu KJ, Kao JY et al (2010) Regulation of excision repair cross‐complementation group 1 by Snail contributes to cisplatin resistance in head and neck cancer. Clin Cancer Res 16, 4561–4571. [DOI] [PubMed] [Google Scholar]
- Hsu DS, Wang HJ, Tai SK, Chou CH, Hsieh CH, Chiu PH, Chen NJ and Yang MH (2014) Acetylation of snail modulates the cytokinome of cancer cells to enhance the recruitment of macrophages. Cancer Cell 26, 534–548. [DOI] [PubMed] [Google Scholar]
- Huh JW, Kim HR, Kim YJ, Lee JH, Park YS, Cho SH and Joo JK (2009) Expression of standard CD44 in human colorectal carcinoma: association with prognosis. Pathol Int 59, 241–246. [DOI] [PubMed] [Google Scholar]
- Huleihel L, Ben‐Yehudah A, Milosevic J, Yu G, Pandit K, Sakamoto K, Yousef H, LeJeune M, Coon TA, Redinger CJ et al (2014) Let‐7d microRNA affects mesenchymal phenotypic properties of lung fibroblasts. Am J Physiol Lung Cell Mol Physiol 306, L534–L542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt EM, Kawasaki BT, Klarmann GJ, Thomas SB and Farrar WL (2008) CD44+ CD24(‐) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br J Cancer 98, 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WL, Yang MH, Tsai ML, Lan HY, Su SH, Chang SC, Teng HW, Yang SH, Lan YT, Chiou SH et al (2011) SNAIL regulates interleukin‐8 expression, stem cell‐like activity, and tumorigenicity of human colorectal carcinoma cells. Gastroenterology 141, 279–291, 291.e271–275. [DOI] [PubMed] [Google Scholar]
- Hwang WL, Jiang JK, Yang SH, Huang TS, Lan HY, Teng HW, Yang CY, Tsai YP, Lin CH, Wang HW et al (2014) MicroRNA‐146a directs the symmetric division of Snail‐dominant colorectal cancer stem cells. Nat Cell Biol 16, 268–280. [DOI] [PubMed] [Google Scholar]
- Ikenouchi J, Matsuda M, Furuse M and Tsukita S (2003) Regulation of tight junctions during the epithelium‐mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci 116, 1959–1967. [DOI] [PubMed] [Google Scholar]
- Kang Y and Massague J (2004) Epithelial‐mesenchymal transitions: twist in development and metastasis. Cell 118, 277–279. [DOI] [PubMed] [Google Scholar]
- Kemper K, Prasetyanti PR, De Lau W, Rodermond H, Clevers H and Medema JP (2012) Monoclonal antibodies against Lgr5 identify human colorectal cancer stem cells. Stem Cells 30, 2378–2386. [DOI] [PubMed] [Google Scholar]
- Kitazawa K, Hikichi T, Nakamura T, Mitsunaga K, Tanaka A, Nakamura M, Yamakawa T, Furukawa S, Takasaka M, Goshima N et al (2016) OVOL2 maintains the transcriptional program of human corneal epithelium by suppressing epithelial‐to‐mesenchymal transition. Cell Rep 15, 1359–1368. [DOI] [PubMed] [Google Scholar]
- Kong D, Banerjee S, Ahmad A, Li Y, Wang Z, Sethi S and Sarkar FH (2010) Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS One 5, e12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BS, Lee SH, Kim JM, Huang S, Kim SH, Rho YS, Bae WJ, Kang HJ, Kim YS, Moon JH et al (2015) Oct4 is a critical regulator of stemness in head and neck squamous carcinoma cells. Oncogene 34, 2317–2324. [DOI] [PubMed] [Google Scholar]
- Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia‐Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y et al (2011) Direct targeting of Sec23a by miR‐200s influences cancer cell secretome and promotes metastatic colonization. Nat Med 17, 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G and Kang Y (2008) The miR‐200 family inhibits epithelial‐mesenchymal transition and cancer cell migration by direct targeting of E‐cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283, 14910–14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D, Reichardt W, Bronsert P et al (2017) The EMT‐activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol 19, 518–529. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, Dong Z, Vodopyanov D, Imai A, Helman JI, Prince ME, Wicha MS and Nor JE (2010) Endothelial cell‐initiated signaling promotes the survival and self‐renewal of cancer stem cells. Cancer Res 70, 9969–9978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Subramanyam D, Blelloch R and Derynck R (2013) Regulation of epithelial‐mesenchymal and mesenchymal‐epithelial transitions by microRNAs. Curr Opin Cell Biol 25, 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Xu J and Derynck R (2014) Molecular mechanisms of epithelial‐mesenchymal transition. Nat Rev Mol Cell Biol 15, 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larriba MJ, Garcia de Herreros A and Munoz A (2016) Vitamin D and the epithelial to mesenchymal transition. Stem Cells Int 2016, 6213872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecharpentier A, Vielh P, Perez‐Moreno P, Planchard D, Soria JC and Farace F (2011) Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non‐small cell lung cancer. Br J Cancer 105, 1338–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis O, Eguiara A, Lopez‐Arribillaga E, Alberdi MJ, Hernandez‐Garcia S, Elorriaga K, Pandiella A, Rezola R and Martin AG (2012) Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 31, 1354–1365. [DOI] [PubMed] [Google Scholar]
- Leth‐Larsen R, Terp MG, Christensen AG, Elias D, Kuhlwein T, Jensen ON, Petersen OW and Ditzel HJ (2012) Functional heterogeneity within the CD44 high human breast cancer stem cell‐like compartment reveals a gene signature predictive of distant metastasis. Mol Med 18, 1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF and Simeone DM (2007) Identification of pancreatic cancer stem cells. Cancer Res 67, 1030–1037. [DOI] [PubMed] [Google Scholar]
- Li C, Lee CJ and Simeone DM (2009) Identification of human pancreatic cancer stem cells. Methods Mol Biol 568, 161–173. [DOI] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q et al (2010) A mesenchymal‐to‐epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63. [DOI] [PubMed] [Google Scholar]
- Li X, Zhao H, Gu J and Zheng L (2015) Prognostic value of cancer stem cell marker CD133 expression in pancreatic ductal adenocarcinoma (PDAC): a systematic review and meta‐analysis. Int J Clin Exp Pathol 8, 12084–12092. [PMC free article] [PubMed] [Google Scholar]
- Liao TT, Hsu WH, Ho CH, Hwang WL, Lan HY, Lo T, Chang CC, Tai SK and Yang MH (2016) let‐7 modulates chromatin configuration and target gene repression through regulation of the ARID3B complex. Cell Rep 14, 520–533. [DOI] [PubMed] [Google Scholar]
- Lin T, Ponn A, Hu X, Law BK and Lu J (2010a) Requirement of the histone demethylase LSD1 in Snai1‐mediated transcriptional repression during epithelial‐mesenchymal transition. Oncogene 29, 4896–4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM and Zhou BP (2010b) The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine‐specific demethylase 1. EMBO J 29, 1803–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Zuo D and Park M (2005) Pc2‐mediated sumoylation of Smad‐interacting protein 1 attenuates transcriptional repression of E‐cadherin. J Biol Chem 280, 35477–35489. [DOI] [PubMed] [Google Scholar]
- Ma S, Chan KW, Lee TK, Tang KH, Wo JY, Zheng BJ and Guan XY (2008) Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res 6, 1146–1153. [DOI] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya‐Feldstein J, Reinhardt F, Onder TT, Valastyan S et al (2010) miR‐9, a MYC/MYCN‐activated microRNA, regulates E‐cadherin and cancer metastasis. Nat Cell Biol 12, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major AG, Pitty LP and Farah CS (2013) Cancer stem cell markers in head and neck squamous cell carcinoma. Stem Cells Int 2013, 319489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M et al (2008) The epithelial‐mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal R, Demetter P, Nagy N, Berton A, Decaestecker C, Polus M, Closset J, Deviere J, Salmon I and Van Laethem JL (2009) High expression of CXCR4 may predict poor survival in resected pancreatic adenocarcinoma. Br J Cancer 100, 1444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T et al (2010) A MicroRNA targeting dicer for metastasis control. Cell 141, 1195–1207. [DOI] [PubMed] [Google Scholar]
- Mironchik Y, Winnard PT Jr, Vesuna F, Kato Y, Wildes F, Pathak AP, Kominsky S, Artemov D, Bhujwalla Z, Van Diest P et al (2005) Twist overexpression induces in vivo angiogenesis and correlates with chromosomal instability in breast cancer. Cancer Res 65, 10801–10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal MK, Myers JN, Misra S, Bailey CK and Chaudhuri G (2008) In vivo binding to and functional repression of the VDR gene promoter by SLUG in human breast cells. Biochem Biophys Res Comm 372, 30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina‐Ortiz P, Villarejo A, MacPherson M, Santos V, Montes A, Souchelnytskyi S, Portillo F and Cano A (2012) Characterization of the SNAG and SLUG domains of Snail2 in the repression of E‐cadherin and EMT induction: modulation by serine 4 phosphorylation. PLoS One 7, e36132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel AP, Hinkal GW, Thomas C, Fauvet F, Courtois‐Cox S, Wierinckx A, Devouassoux‐Shisheboran M, Treilleux I, Tissier A, Gras B et al (2012) EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin‐low tumors in transgenic mice. PLoS Genet 8, e1002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen K, Fu Y, Chang L, Hoodless PA, McFadden D and Karsan A (2008) Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J Cell Biol 182, 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA (2002) The snail superfamily of zinc‐finger transcription factors. Nat Rev Mol Cell Biol 3, 155–166. [DOI] [PubMed] [Google Scholar]
- Nieto MA (2013) Epithelial plasticity: a common theme in embryonic and cancer cells. Science 342, 1234850. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Huang RY, Jackson RA and Thiery JP (2016) Emt: 2016. Cell 166, 21–45. [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S and Dick JE (2007) A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445, 106–110. [DOI] [PubMed] [Google Scholar]
- Ocana OH, Corcoles R, Fabra A, Moreno‐Bueno G, Acloque H, Vega S, Barrallo‐Gimeno A, Cano A and Nieto MA (2012) Metastatic colonization requires the repression of the epithelial‐mesenchymal transition inducer Prrx1. Cancer Cell 22, 709–724. [DOI] [PubMed] [Google Scholar]
- Ohashi S, Natsuizaka M, Wong GS, Michaylira CZ, Grugan KD, Stairs DB, Kalabis J, Vega ME, Kalman RA, Nakagawa M et al (2010) Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial‐to‐mesenchymal transition through ZEB transcription factors. Cancer Res 70, 4174–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y, Wauthier E, Tajiri H, Miller LD, Wang XW et al (2013) Sal‐like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology 57, 1469–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D et al (2010) Inhibition and role of let‐7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 182, 220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E and Peter ME (2008) The miR‐200 family determines the epithelial phenotype of cancer cells by targeting the E‐cadherin repressors ZEB1 and ZEB2. Genes Dev 22, 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterlini‐Brechot P and Benali NL (2007) Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett 253, 180–204. [DOI] [PubMed] [Google Scholar]
- Patrawala L, Calhoun T, Schneider‐Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K et al (2006) Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene 25, 1696–1708. [DOI] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M and Cano A (2004) Snail mediates E‐cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 24, 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D and Cano A (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7, 415–428. [DOI] [PubMed] [Google Scholar]
- Peinado H and Cano A (2008) A hypoxic twist in metastasis. Nat Cell Biol 10, 253–254. [DOI] [PubMed] [Google Scholar]
- Pena C, Garcia JM, Garcia V, Silva J, Dominguez G, Rodriguez R, Maximiano C, Garcia de Herreros A, Munoz A and Bonilla F (2006) The expression levels of the transcriptional regulators p300 and CtBP modulate the correlations between SNAIL, ZEB1, E‐cadherin and vitamin D receptor in human colon carcinomas. Int J Cancer 119, 2098–2104. [DOI] [PubMed] [Google Scholar]
- Perdigao‐Henriques R, Petrocca F, Altschuler G, Thomas MP, Le MT, Tan SM, Hide W and Lieberman J (2016) miR‐200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene 35, 158–172. [DOI] [PubMed] [Google Scholar]
- Pervin S, Hewison M, Braga M, Tran L, Chun R, Karam A, Chaudhuri G, Norris K and Singh R (2013) Down‐regulation of vitamin D receptor in mammospheres: implications for vitamin D resistance in breast cancer and potential for combination therapy. PLoS One 8, e53287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA and Daidone MG (2005) Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 65, 5506–5511. [DOI] [PubMed] [Google Scholar]
- Postigo AA, Depp JL, Taylor JJ and Kroll KL (2003) Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J 22, 2453–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF and Ailles LE (2007) Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A 104, 973–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi C, Gradilone A, Naso G, Vincenzi B, Petracca A, Nicolazzo C, Palazzo A, Saltarelli R, Spremberg F, Cortesi E et al (2011) Epithelial‐mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Cancer Res Treat 130, 449–455. [DOI] [PubMed] [Google Scholar]
- Ricci‐Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C and De Maria R (2007) Identification and expansion of human colon‐cancer‐initiating cells. Nature 445, 111–115. [DOI] [PubMed] [Google Scholar]
- Roca H, Hernandez J, Weidner S, McEachin RC, Fuller D, Sud S, Schumann T, Wilkinson JE, Zaslavsky A, Li H et al (2013) Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS One 8, e76773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscetti M, Quach B, Dadashian EL, Mulholland DJ and Wu H (2015) Tracking and functional characterization of epithelial‐mesenchymal transition and mesenchymal tumor cells during prostate cancer metastasis. Cancer Res 75, 2749–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagiv E, Memeo L, Karin A, Kazanov D, Jacob‐Hirsch J, Mansukhani M, Rechavi G, Hibshoosh H and Arber N (2006) CD24 is a new oncogene, early at the multistep process of colorectal cancer carcinogenesis. Gastroenterology 131, 630–639. [DOI] [PubMed] [Google Scholar]
- Samavarchi‐Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A and Wrana JL (2010) Functional genomics reveals a BMP‐driven mesenchymal‐to‐epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77. [DOI] [PubMed] [Google Scholar]
- Sarrio D, Franklin CK, Mackay A, Reis‐Filho JS and Isacke CM (2012) Epithelial and mesenchymal subpopulations within normal basal breast cell lines exhibit distinct stem cell/progenitor properties. Stem Cells 30, 292–303. [DOI] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M and Clevers H (2012) Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337, 730–735. [DOI] [PubMed] [Google Scholar]
- Schmidt JM, Panzilius E, Bartsch HS, Irmler M, Beckers J, Kari V, Linnemann JR, Dragoi D, Hirschi B, Kloos UJ et al (2015) Stem‐cell‐like properties and epithelial plasticity arise as stable traits after transient Twist1 activation. Cell Rep 10, 131–139. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, el Affar B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y and Shi Y (2003) Coordinated histone modifications mediated by a CtBP co‐repressor complex. Nature 422, 735–738. [DOI] [PubMed] [Google Scholar]
- Shih JY and Yang PC (2011) The EMT regulator slug and lung carcinogenesis. Carcinogenesis 32, 1299–1304. [DOI] [PubMed] [Google Scholar]
- Spoelstra NS, Manning NG, Higashi Y, Darling D, Singh M, Shroyer KR, Broaddus RR, Horwitz KB and Richer JK (2006) The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res 66, 3893–3902. [DOI] [PubMed] [Google Scholar]
- Stanisavljevic J, Porta‐de‐la‐Riva M, Batlle R, de Herreros AG and Baulida J (2011) The p65 subunit of NF‐kappaB and PARP1 assist Snail1 in activating fibronectin transcription. J Cell Sci 124, 4161–4171. [DOI] [PubMed] [Google Scholar]
- Stankic M, Pavlovic S, Chin Y, Brogi E, Padua D, Norton L, Massague J and Benezra R (2013) TGF‐beta‐Id1 signaling opposes Twist1 and promotes metastatic colonization via a mesenchymal‐to‐epithelial transition. Cell Rep 5, 1228–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss R, Li ZY, Liu Y, Beyer I, Persson J, Sova P, Moller T, Pesonen S, Hemminki A, Hamerlik P et al (2011) Analysis of epithelial and mesenchymal markers in ovarian cancer reveals phenotypic heterogeneity and plasticity. PLoS One 6, e16186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoropoulos PA, Polioudaki H, Agelaki S, Kallergi G, Saridaki Z, Mavroudis D and Georgoulias V (2010) Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett 288, 99–106. [DOI] [PubMed] [Google Scholar]
- Timmerman LA, Grego‐Bessa J, Raya A, Bertran E, Perez‐Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua‐Belmonte JC et al (2004) Notch promotes epithelial‐mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev 18, 99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G, Medema JP et al (2007) Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin‐4. Cell Stem Cell 1, 389–402. [DOI] [PubMed] [Google Scholar]
- Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S, Cocorullo G et al (2014) CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 14, 342–356. [DOI] [PubMed] [Google Scholar]
- Tripathi MK, Misra S, Khedkar SV, Hamilton N, Irvin‐Wilson C, Sharan C, Sealy L and Chaudhuri G (2005) Regulation of BRCA2 gene expression by the SLUG repressor protein in human breast cells. J Biol Chem 280, 17163–17171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JH, Donaher JL, Murphy DA, Chau S and Yang J (2012) Spatiotemporal regulation of epithelial‐mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Griend DJ, Karthaus WL, Dalrymple S, Meeker A, DeMarzo AM and Isaacs JT (2008) The role of CD133 in normal human prostate stem cells and malignant cancer‐initiating cells. Cancer Res 68, 9703–9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van Roy F and Berx G (2005) SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell‐cell junctions. Nucleic Acids Res 33, 6566–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G and Medema JP (2008) Single‐cell cloning of colon cancer stem cells reveals a multi‐lineage differentiation capacity. Proc Natl Acad Sci U S A 105, 13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ding X, Nan L, Wang Y, Wang J, Yan Z, Zhang W, Sun J, Zhu W, Ni B et al (2015) Investigation of the roles of exosomes in colorectal cancer liver metastasis. Oncol Rep 33, 2445–2453. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li Y, Kong D and Sarkar FH (2010) The role of Notch signaling pathway in epithelial‐mesenchymal transition (EMT) during development and tumor aggressiveness. Curr Drug Targets 11, 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, Yuan A, Lin CW, Yang SC, Chan WK et al (2009) p53 controls cancer cell invasion by inducing the MDM2‐mediated degradation of Slug. Nat Cell Biol 11, 694–704. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Villarreal‐Ponce A, Sun P, Salmans ML, Fallahi M, Andersen B and Dai X (2014) Mammary morphogenesis and regeneration require the inhibition of EMT at terminal end buds by Ovol2 transcriptional repressor. Dev Cell 29, 59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A et al (2009) The EMT‐activator ZEB1 promotes tumorigenicity by repressing stemness‐inhibiting microRNAs. Nat Cell Biol 11, 1487–1495. [DOI] [PubMed] [Google Scholar]
- Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, Helfrich B, Dziadziuszko R, Chan DC, Sugita M et al (2006) Restoring E‐cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res 66, 944–950. [DOI] [PubMed] [Google Scholar]
- Wu CY, Hung JJ and Wu KJ (2012) Linkage between Twist1 and Bmi1: molecular mechanism of cancer metastasis/stemness and clinical implications. Clin Exp Pharmacol Physiol 39, 668–673. [DOI] [PubMed] [Google Scholar]
- Wu KJ and Yang MH (2011) Epithelial‐mesenchymal transition and cancer stemness: the Twist1‐Bmi1 connection. Biosci Rep 31, 449–455. [DOI] [PubMed] [Google Scholar]
- Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK et al (2010) Bmi1 is essential in Twist1‐induced epithelial‐mesenchymal transition. Nat Cell Biol 12, 982–992. [DOI] [PubMed] [Google Scholar]
- Yang WH, Lan HY, Huang CH, Tai SK, Tzeng CH, Kao SY, Wu KJ, Hung MC and Yang MH (2012) RAC1 activation mediates Twist1‐induced cancer cell migration. Nat Cell Biol 14, 366–374. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and Weinberg RA (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939. [DOI] [PubMed] [Google Scholar]
- Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC and Wu KJ (2008) Direct regulation of TWIST by HIF‐1alpha promotes metastasis. Nat Cell Biol 10, 295–305. [DOI] [PubMed] [Google Scholar]
- Yao D, Dai C and Peng S (2011) Mechanism of the mesenchymal‐epithelial transition and its relationship with metastatic tumor formation. Mol Cancer Res 9, 1608–1620. [DOI] [PubMed] [Google Scholar]
- Yin S, Li J, Hu C, Chen X, Yao M, Yan M, Jiang G, Ge C, Xie H, Wan D et al (2007) CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int J Cancer 120, 1444–1450. [DOI] [PubMed] [Google Scholar]
- Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM et al (2013) Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J and Bottinger EP (2005) TGF‐beta and epithelial‐to‐mesenchymal transitions. Oncogene 24, 5764–5774. [DOI] [PubMed] [Google Scholar]
- Zavadil J, Cermak L, Soto‐Nieves N and Bottinger EP (2004) Integration of TGF‐beta/Smad and Jagged1/Notch signalling in epithelial‐to‐mesenchymal transition. EMBO J 23, 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS and Kalluri R (2015) Epithelial‐to‐mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]


