Rab7 promotes fusion of autophagosomes and late endosomes with lysosomes. Lőrincz et al. show that Rab2 is critical for the delivery of autophagic and endocytic cargo to lysosomes and for their degradation, and that it promotes autophagosome–lysosome fusion. The results suggest Rab2 and Rab7 coordinately promote autophagic and endosomal degradation and lysosome function.
Abstract
Rab7 promotes fusion of autophagosomes and late endosomes with lysosomes in yeast and metazoan cells, acting together with its effector, the tethering complex HOPS. Here we show that another small GTPase, Rab2, is also required for autophagosome and endosome maturation and proper lysosome function in Drosophila melanogaster. We demonstrate that Rab2 binds to HOPS, and that its active, GTP-locked form associates with autolysosomes. Importantly, expression of active Rab2 promotes autolysosomal fusions unlike that of GTP-locked Rab7, suggesting that its amount is normally rate limiting. We also demonstrate that RAB2A is required for autophagosome clearance in human breast cancer cells. In conclusion, we identify Rab2 as a key factor for autophagic and endocytic cargo delivery to and degradation in lysosomes.
Introduction
The two main pathways of lysosomal degradation are endocytosis and autophagy. Double-membrane autophagosomes (generated in the main pathway of autophagy) and endosomes can fuse with each other to generate amphisomes, and mature into degradative endo- and autolysosomes, respectively, by ultimately fusing with lysosomes (Mizushima et al., 2008). One of the main regulators of intracellular trafficking and vesicle fusions are Rab small GTPases. Active, GTP-bound Rab proteins recruit various effectors including tethers and molecular motors (Zhen and Stenmark, 2015), of which Rab7 is the only known direct regulator of both autophagosome–lysosome and endosome–lysosome fusions.
The tethering complex homotypic fusion and vacuole protein sorting (HOPS) was identified in yeast, and it simultaneously binds two yeast Rab7 (Ypt7) molecules on its opposing ends. In animal cells, Rab7 binds to RILP, ORPL1, FYCO1, and PLEKHM1 to recruit dyneins and HOPS and ensure the fusion of late endosomes and autophagosomes with lysosomes (Pankiv et al., 2010; van der Kant et al., 2013; McEwan et al., 2015). This way, HOPS could cross-link two Rab7-positive membranes to prompt tethering and fusion (Balderhaar and Ungermann, 2013; Solinger and Spang, 2013). Rab7 is present on lysosomes, autophagosomes, and endosomes (Hegedűs et al., 2016), but it is not clear whether another Rab is involved in degradative auto- and endolysosome formation, which also requires transport of hydrolases from the Golgi (Saftig and Klumperman, 2009).
Rab2 is known to control anterograde and retrograde traffic between the ER and Golgi (Saraste, 2016). A recent biochemical screen identified Rab2 as a direct binding partner of HOPS, and active Rab2 was found to localize to Rab7-positive vacuoles in cultured Drosophila melanogaster cells (Gillingham et al., 2014). Here we propose an updated model in which Rab7 and Rab2 coordinately promote the HOPS-dependent degradation of autophagosomes and endosomes via fusion of these as well as biosynthetic vesicles with lysosomes.
Results and discussion
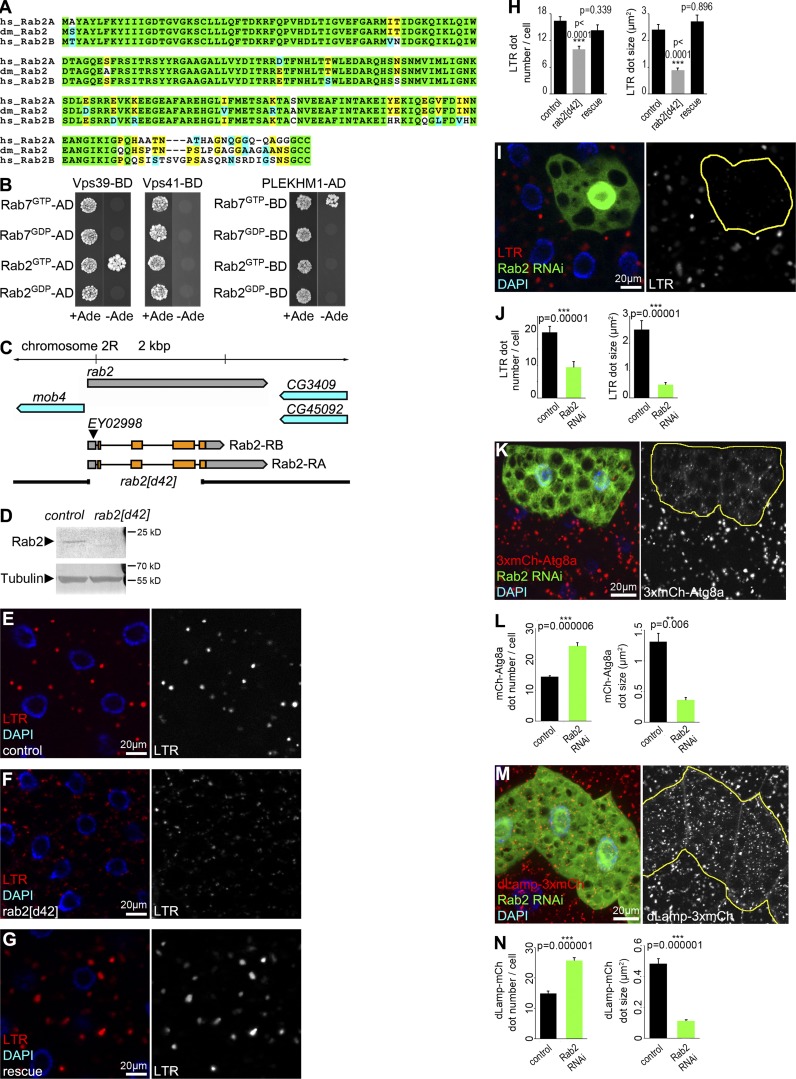
Rab2 is highly conserved among higher eukaryotes, including Drosophila melanogaster and humans (Fig. 1 A). The HOPS subunits Vps39 and Vps41 directly bind to Ypt7/Rab7 in yeast, whereas their interaction may be indirect in mammalian cells (van der Kant et al., 2013; McEwan et al., 2015; Wijdeven et al., 2016). No binding was detected between Drosophila Rab7 and Vps39 or Vps41, whereas GTP-locked Rab7 bound to its known effector PLEKHM1 in yeast two-hybrid (Y2H) experiments (Fig. 1 B). Vps39 directly bound Rab2GTP in both Y2H and recombinant protein pull-down experiments (Figs. 1 B and S1 A; Gillingham et al., 2014), and Rab2GTP immunoprecipitated endogenous Vps16A (another HOPS subunit) from fly lysates (Fig. S1 B). Consistently, we have reported that recombinant mammalian RAB2A pulls down Vps39 but not Vps41 from cell lysates (Kajiho et al., 2016), and human HOPS subunits did not show Rab7 binding in Y2H experiments (Caplan et al., 2001; Khatter et al., 2015).
Figure 1.
Rab2 is required for proper autolysosome formation in starved fat cells. (A) Alignment of Drosophila (dm) Rab2 with human (hs) Rab2A and Rab2B proteins. Identical (green/yellow) and similar (blue) amino acids are indicated. (B) Y2H assays reveal that GTP-locked Drosophila Rab2 binds to Vps39 and Rab7GTP binds to PLEKHM1. (C) Genomic map of rab2, showing the size of d42 deletion that arose from imprecise P element EY02998 excision. (D) Rab2 protein is absent from homozygous mutant larvae. (E–J) LTR staining reveals that starvation-induced autolysosome formation seen in control (E) and rescued (G) fat cells is impaired in rab2 mutants (F). Quantification of LTR data in E–G, n = 20 cells (H). RNAi knockdown of Rab2 in a GFP+ fat cell impairs punctate LTR staining compared with neighboring non-GFP control cells (I), quantified in (J), n = 10 cells. (K and L) Rab2 knockdown in GFP+ cells impairs proper formation of 3xmCherry-Atg8a+ autophagic vesicles (K). Red dots are bigger and brighter in control cells compared with the many smaller and fainter dots in RNAi cells, quantified in L, n = 10 cells. (M and N) Rab2 silencing in GFP-marked cells decreases the size and increases the number of dLamp-3xmCherry+ lysosomes (M), quantified in N, n = 10 cells. Error bars mark ± SEM in H, J, L, and N. Red channels are shown in grayscale in E–G, I, K, and M, and RNAi cells are encircled in I, K, and M.
To address whether Rab2 functions in autophagy and endocytosis, we knocked out rab2 by imprecise excision of a transposon from the 5′ UTR. The resulting rab2d42 allele carries a 2,047-bp deletion, which removes most of the protein coding sequences of both predicted Rab2 isoforms and eliminates protein expression (Fig. 1, C and D). Rab2 mutant animals die as L2/L3-stage larvae, and their viability is fully rescued by expression of YFP-Rab2.
Larval fat cells are widely used for autophagy analyses because of their massive autophagic potential. Numerous Lysotracker Red (LTR)-positive vesicles appear upon starvation, which represent newly formed autolysosomes with likely increased v-ATPase–mediated acidification in these cells (Mauvezin et al., 2014; Nagy et al., 2015). LTR dot number and size (and signal intensity as a likely consequence) decreased in rab2-null cells compared with controls, which was rescued by expression of YFP-Rab2 (Fig. 1, E–H; and Fig. S1 C). RNAi knockdown of Rab2 in GFP-marked fat cell clones also impaired starvation-induced punctate LTR staining compared with surrounding GFP-negative cells (Fig. 1, I and J).
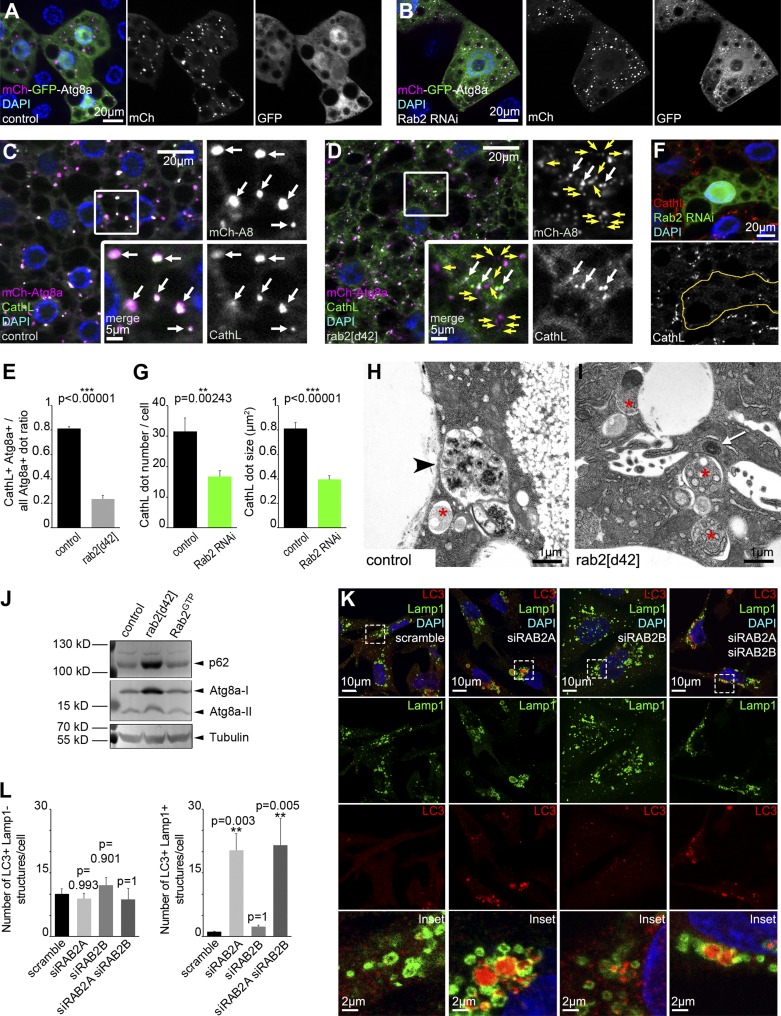
A 3xmCherry-Atg8a reporter that labels all autophagic structures via retained fluorescence of mCherry inside autolysosomes revealed increased number and decreased size of such vesicles in both starved rab2 RNAi and mutant fat cells (Fig. 1, K and L; and Fig. S1, D and E). A dLamp-3xmCherry reporter of late endosomes and lysosomes showed similar changes in rab2 RNAi or mutant fat cells of starved animals (Fig. 1, M and N; and Fig. S1, F and G). Tandem tagged mCherry-GFP-Atg8a reporters are commonly used to follow autophagic flux, because GFP is quenched in lysosomes, whereas mCherry signal persists (Mauvezin et al., 2014; Nagy et al., 2015). Knockdown of rab2 prevented the quenching of GFP that is seen in starved control fat cells: dots positive for both GFP and mCherry accumulated (Fig. 2, A and B), raising the possibility that Rab2 promotes autophagosome–lysosome fusion, similar to HOPS. We thus looked at colocalization of 3xmCherry-Atg8a with the lysosomal hydrolase cathepsin L (CathL). The overlap of these markers of autophagic and lysosomal structures strongly decreased in rab2 mutant fat cells compared with controls, and rab2 RNAi also impaired endogenous CathL-positive vesicle formation (Fig. 2, C–G), suggesting that formation of degradative autolysosomes requires Rab2.
Figure 2.
Rab2 in Drosophila fat and RAB2A in human cells are required for autophagosome clearance. (A and B) Tandem mCherry-GFP-Atg8a shows that autophagic flux proceeds normally in starved control cells, based on quenching of GFP (A). GFP remains fluorescent and colocalizes with mCherry in Rab2 RNAi cells (B). (C–E) Most 3xmCherry-Atg8a autophagic structures contain the lysosomal hydrolase CathL in starved control fat cells (C), whereas their overlap is reduced in rab2 mutants (D), quantified in E, n = 30–45 cells, indicating that the convergence of autophagic and lysosomal compartments is blocked in rab2 mutants. White arrows, overlapping signal; yellow arrows, mCherry-Atg8a–only vesicles in C and D. (F and G) Rab2 knockdown in GFP+ cells reduces the number and size of CathL vesicles (F), quantified in G, n = 9 cells. (H and I) Ultrastructural analysis of starved fat cells. Degrading autolysosomes (arrowhead) form in control cells (H), unlike in rab2 mutants (I) that accumulate double-membrane autophagosomes (asterisks) containing nondegraded cytoplasm and dense structures likely representing amphisomes (arrow). (J) Western blots reveal accumulation of p62 and both forms of Atg8a in starved rab2-null mutants, and similar protein levels in control and Rab2GTP-expressing larvae. (K and L) Confocal analysis of human MDA-MB-231 cells transfected with scramble oligo or siRNA against RAB2A, RAB2B, or both (K). Bottom, magnification of boxed areas. Knockdown of RAB2A, but not RAB2B, causes accumulation of endogenous LC3+ autophagic structures, which colocalize with the late endosomal-lysosomal marker Lamp1. Quantification of LC3+ Lamp1− autophagosomes and LC3+ Lamp1+ amphisomes (L), n = 13–23 cells. Error bars mark ± SEM in E, G, and L. Indicated channels are shown in grayscale in A–D and F.
These phenotypes resembled the autophagosome–lysosome fusion defect of mutants for the autophagosomal SNARE syntaxin 17, HOPS, and Rab7 (Takáts et al., 2013, 2014; Hegedűs et al., 2016). Accordingly, ultrastructural analysis of starved fat cells revealed accumulation of double-membrane autophagosomes and small dense structures likely representing amphisomes (Fig. 2, H and I), similar to HOPS mutants (Takáts et al., 2014). Recently, rab2 RNAi was reported to cause accumulation of autophagosomes in Drosophila muscles and enlarged amphisomes in fat cells (Fujita et al., 2017). Autophagosome accumulation in our rab2-null mutant fat cells is likely caused by a complete loss-of-function condition.
Western blots detected increased levels of the selective autophagy cargo p62/Ref2p (Nezis et al., 2008; Pircs et al., 2012), along with both free and lipidated autophagosome-associated forms of Atg8a in starved rab2 mutants (Figs. 2 J and S1 H). Basal autophagic degradation was also impaired in rab2 mutants, based on increased numbers of endogenous Atg8a and p62 dots in well-fed conditions (Fig. S1, I–K).
We confirmed the importance of Rab2 for autophagic degradation in human cells. Knockdown of RAB2B had no effect on endogenous LC3 structures in breast cancer cells, whereas RAB2A or combined siRNA treatment caused accumulation of autophagic vesicles (Fig. S1, L–Q). LC3 accumulated within Lamp1-positive structures upon RAB2A knockdown, which likely represent amphisomes unable to mature into autolysosomes in these cells (Fig. 2, K and L), consistent with the recently reported role of Rab2 homologs for degradation of autophagic cargo in mouse embryonic fibroblasts (Fujita et al., 2017).
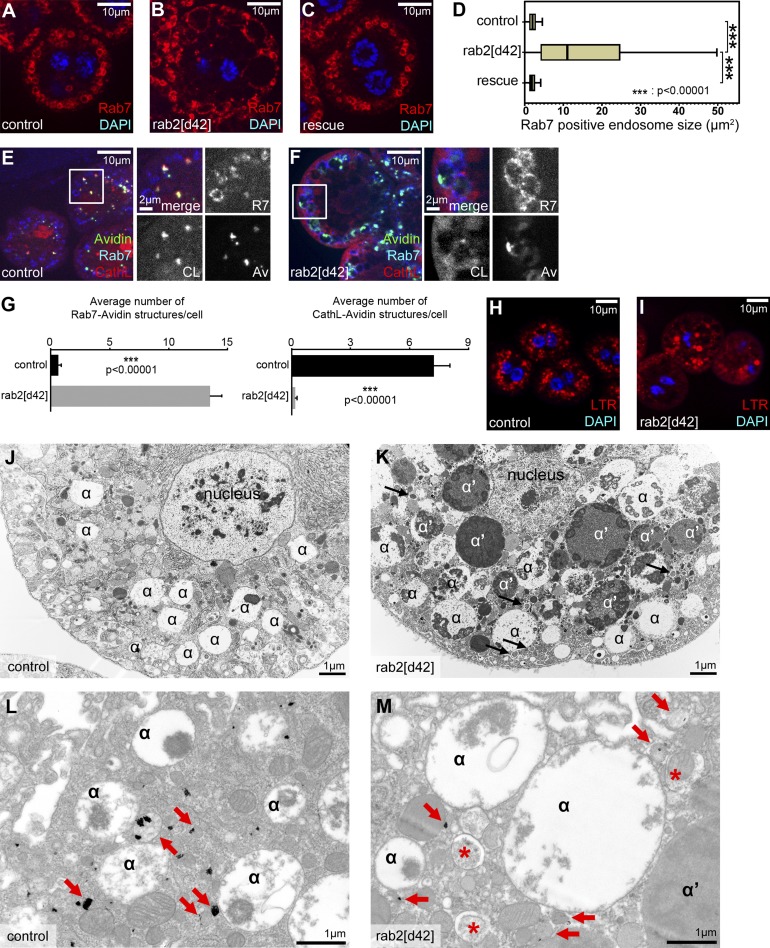
To analyze the possible involvement of Drosophila Rab2 in endosomal degradation, we incubated dissected nephrocytes with fluorescent avidin for 5 min. Trafficking of this endocytic tracer was clearly perturbed in rab2 mutant cells, similar to vps41/lt and rab7 mutants (Fig. S2, A–E). Loss of HOPS leads to enlargement of late endosomes (Lőrincz et al., 2016). Similarly, Rab7 endosomes are enlarged in rab2 mutant nephrocytes compared with control or rescued cells (Fig. 3, A–D). Importantly, fluorescent avidin was trapped in Rab7 endosomes and failed to reach CathL-positive lysosomes after a 30-min chase in rab2 mutants (Fig. 3, E–G). LTR staining showed the presence of acidic vacuoles in rab2 mutant nephrocytes (Fig. 3, H and I), which probably include the enlarged late endosomes in rab2 mutant nephrocytes, based on ultrastructural analysis (Fig. 3, J and K). Aberrant late endosomes accumulated in mutant cells, which were apparently unable to fuse with neighboring acid phosphatase–positive lysosomes (Fig. 3, L and M). Of note, the number of acid phosphatase–positive lysosomes also decreased in mutant nephrocytes (Fig. 3, L and M; and Fig. S2 F), suggesting that Rab2 promotes both endosome–lysosome fusion and biosynthetic transport to lysosomes.
Figure 3.
Rab2 is required for endosome–lysosome fusion in Drosophila nephrocytes. (A–D) Rab7+ late endosomes are enlarged in rab2 mutant nephrocytes (B) compared with control (A) and genetically rescued mutant (C) cells. Quantification of data in A–C (D), n = 10 cells. Box plots show the data ranging between upper and lower quartiles; medians are indicated within boxes. (E–G) Uptake assays reveal that FITC-avidin reaches CathL+ lysosomes in control cells (E), whereas it is trapped in Rab7 endosomes in rab2 mutant nephrocytes (F). Quantification of triple colocalization data in E and F (G), n = 12–13 cells; error bars mark ± SEM. (H and I) LTR+ acidic vacuoles are present in both control (H) and rab2 mutant (I) cells. (J and K) Ultrastructure of late endosomes (α-vacuoles) in control (J) and rab2 mutant nephrocytes (K). Late endosomes (α) are enlarged in rab2 mutant cells, and many abnormal vacuoles (α′) containing dense but still nondegraded endocytic cargo as well as autophagosomes (arrows) are seen only in mutants. (L and M) Acid phosphatase activity (black precipitate, arrows) is detected in endolysosomes of control cells (L), whereas in rab2 mutants (M), only smaller lysosomes are seen next to the enlarged late endosomes (α) and autophagosomes (asterisks).
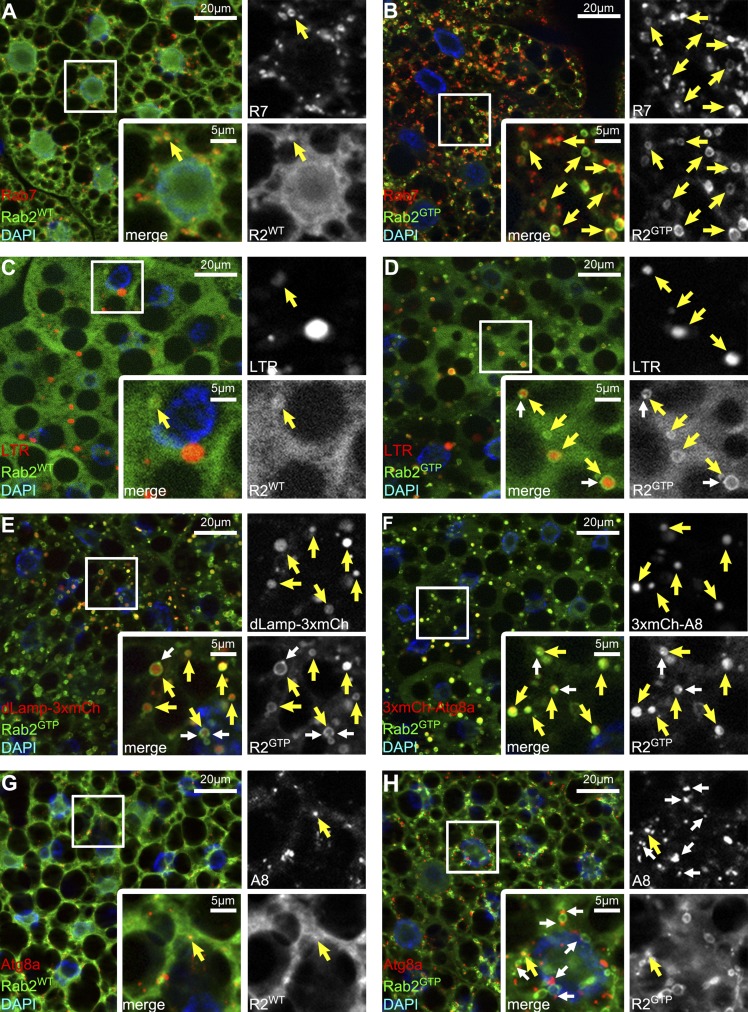
GTP-locked, constitutively active Rab2GTP redistributes from the Golgi onto Rab7 vacuoles in cultured Drosophila cells (Gillingham et al., 2014). Similarly, Rab2GTP colocalized with endogenous Rab7 in starved fat cells, unlike wild-type Rab2 (Fig. 4, A and B; and Fig. S3 A). Rab2GTP appeared as large pronounced rings around LTR-positive autolysosomes in starved fat cells, unlike wild-type Rab2 (Fig. 4, C and D; and Fig. S3 A). Similarly, Rab2GTP formed rings around lysosomes and autophagic structures marked by dLamp-3xmCherry and 3xmCherry-Atg8a, respectively (Fig. 4, E and F; and Fig. S3 A). Of note, small Rab2GTP dots often closely associated with large Rab2GTP rings in these experiments (Fig. 4, D–F; and Fig. S3 A), raising the possibility that Rab2 vesicles fuse with autolysosomes. Finally, wild-type Rab2 or Rab2GTP modestly overlapped with autophagosomes marked by endogenous Atg8a (Fig. 4, G and H; and Fig. S3 A).
Figure 4.
Active Rab2 localizes to autolysosomes in starved fat cells. (A) Wild-type (WT) YFP-Rab2WT rarely overlaps with endogenous Rab7. (B) Active YFP-Rab2GTP shows extensive colocalization with Rab7. (C) YFP-Rab2WT rarely associates with LTR+ autolysosomes. (D–F) LTR vesicles are often surrounded by YFP-Rab2GTP rings (D), similar to lysosomal (dLamp-3xmCherry+; E) and autophagic (3xmCherry-Atg8a+; F) structures. Note that small Rab2 dots (white arrows in D–F) associate with the rings. (G and H) Rab2WT (G) and Rab2GTP (H) show modest overlap with autophagosomes marked by endogenous Atg8a. Note that Atg8a vesicles dock to Rab2GTP rings (white arrows in H). Boxed areas are enlarged, with yellow arrows indicating colocalization and green/red channels shown in grayscale in all panels.
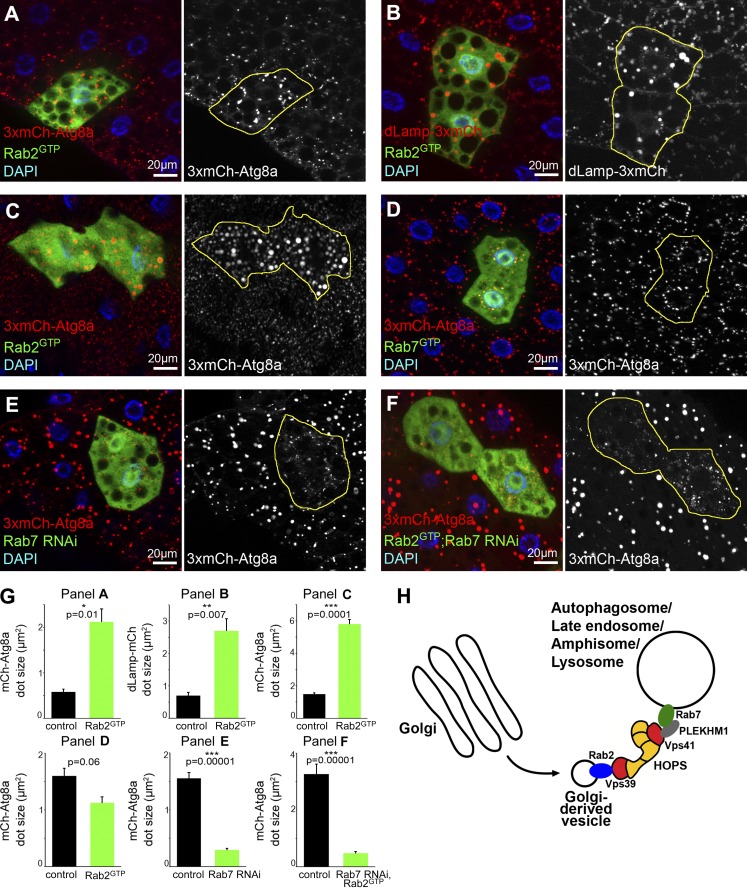
These localization and loss-of-function data pointed to Rab2 as a positive regulator of autolysosome formation. Indeed, fat and midgut cells expressing Rab2GTP contained enlarged and brighter 3xmCherry-Atg8a autophagic structures and dLamp-3xmCherry lysosomes compared with surrounding control cells (Fig. 5, A–C and G), suggesting that Rab2 controls autolysosome size. Increased lysosomal input or a block of degradation can cause enlargement of autolysosomes. Systemic expression of Rab2GTP did not impair the viability of animals, and Western blots of starved L3 larval lysates revealed no changes in p62 and Atg8a levels (Fig. 2 J), suggesting that autophagic degradation proceeds normally in cells expressing Rab2GTP. Thus, Rab2GTP may increase autolysosome size by accelerating fusions with other vesicles. Importantly, expression of GTP-locked, active Rab7 did not increase the size of autophagic structures (Fig. 5, D and G). Rab7 is required for autophagosome–lysosome fusion, and its knockdown prevents the formation of large, bright 3xmCherry-Atg8a-positive autolysosomes: these cells contain only small, faint autophagosomes (Fig. 5, E and G; Hegedűs et al., 2016). Similarly, only small, faint 3xmCherry-Atg8a dots appeared in Rab2GTP-expressing fat cells undergoing Rab7 RNAi (Fig. 5, F and G), indicating that Rab2-dependent fusions also require Rab7 and there is no functional redundancy between them.
Figure 5.
Expression of Rab2GTP increases autolysosome size in starved Drosophila larvae. (A–C) Expression of active YFP-Rab2GTP leads to a striking increase in the size of 3xmCherry-Atg8a (A, fat cells; C, midgut cells) and dLamp-3xmCherry structures (B, fat cells), compared with surrounding control cells. Note that Rab-expressing cells coexpress free GFP to visualize cell outlines in these and all subsequent panels. (D) YFP-Rab7GTP expression does not affect 3xmCherry-Atg8a vesicle size. (E and F) Knockdown of Rab7 (E) in GFP+ cells causes accumulation of small faint autophagosomes and lack of bigger, brighter autolysosomes seen in neighboring control cells, based on 3xmCherry-Atg8a. Rab7 RNAi also prevents degradative autolysosome formation in cells expressing YFP-Rab2GTP (F). Quantification of data in A–F (G), n = 10 cells, error bars mark ± SEM. (H) A model of lysosomal fusions. We hypothesize that Rab2 is transported in Golgi-derived vesicles to fuse with Rab7-positive autophagosomes, late endosomes, amphisomes, and auto/endolysosomes. Rab2 promotes fusions until its release from these vesicles via GTP hydrolysis. In this scenario, Rab2 may directly bind to the Vps39 end of HOPS, whereas Rab7 may interact with Vps41 via adaptors such as PLEKHM1.
Eye pigment granules are lysosome-related organelles. Changes in lysosomal transport often lead to eye discoloration caused by pigment granule alterations, such as in HOPS mutants (Lloyd et al., 1998; Sevrioukov et al., 1999). Rab2GTP expression led to a slight darkening of eyes and appearance of enlarged pigment granules (Fig. S3, B–D), consistent with the role of Rab2 in promoting lysosomal fusions.
Several homo- and heterotypic fusions occur during endosome and autophagosome maturation into degradative lysosomes. Known metazoan factors acting at lysosomal fusions include HOPS and EPG5 tethers and Rab7 together with its effectors (Pols et al., 2013; van der Kant et al., 2013; Jiang et al., 2014; Takáts et al., 2014; McEwan et al., 2015; Wang et al., 2016). Because biosynthetic transport to lysosomes also requires input from Golgi, the role of Golgi-associated Rab2 in various lysosomal fusions fits well into this picture. Consistently, Rab2 promotes breakdown of phagocytosed apoptotic bodies and lysosome-related acrosome biogenesis (Mountjoy et al., 2008; Guo et al., 2010).
Accumulation of unfused autophagosomes and enlarged late endosomes in rab2 mutants resembles the fusion defect of rab7 mutant cells (Hegedűs et al., 2016). The decreased function of lysosomes in rab2 mutants is unlikely to account for these fusion defects, because we have shown that autophagosome–lysosome fusion proceeds and gives rise to enlarged, nondegrading autolysosomes in fat cells with perturbed acidification or biosynthetic transport to lysosomes (Maruzs et al., 2015; Mauvezin et al., 2015).
The role of Rab2 in the fusion of lysosomes with other vesicles is also supported by the autolysosomal localization of its active form and by its binding to the Vps39-containing end of HOPS, the tethering complex required for autophagosomal, endosomal, and biosynthetic transport to lysosomes. Consistently, Rab2 recruits HOPS to Rab7-positive vesicles in cultured Drosophila cells (Gillingham et al., 2014). Expression of Rab2GTP increases degradative autolysosome and pigment granule size, suggesting that it is rate limiting during these fusion reactions, unlike Rab7. This is supported by low levels of wild-type Rab2 on these organelles, unlike wild-type Rab7 that is abundant on autophagosomes, late endosomes, and lysosomes (Cherry et al., 2013; Hegedűs et al., 2016). Consistent with this, we have recently shown that expression of RAB2AGTP also increases Rab7 vesicle size in human cells (Kajiho et al., 2016). Based on binding of Rab2 to one end of HOPS, we propose an updated model of lysosomal fusions in animal cells (Fig. 5 H). We hypothesize that GTP-loaded Rab2 is transported on Golgi-derived carrier vesicles toward Rab7 positive vesicles, and its interaction with Vps39 promotes fusions. Vps41 located on the other end of HOPS may bind Rab7 vesicles via adaptors such as PLEKHM1. These interactions help the tethering and fusion of autophagic, endocytic, and lysosomal vesicles to generate degrading compartments. Lysosomal membranes may contain active Rab2 for only a short period of time, and it likely dissociates upon GTP hydrolysis to limit organelle size. Rab asymmetry is also observed during homotypic vacuole fusion in yeast: GTP-bound Ypt7/Rab7 is necessary on only one of the vesicles, and its nucleotide status is irrelevant on the opposing membrane (Zick and Wickner, 2016). Importantly, Rab7 directly interacts with both ends of HOPS in the absence of a Rab2 homolog in yeast. This difference may explain why yeast cells contain one large vacuole instead of the many smaller lysosomes seen in animal cells. Collectively, these data indicate that Rab2 and Rab7 coordinately promote autophagic and endosomal degradation and lysosome function.
Materials and methods
Fly work and treatments
Flies were raised at 25°C on regular food. Rab2[d42]-null allele was generated by imprecise excision of the transposable element Rab2[EY02998] (FlyBase ID: FBst0019993; Bloomington Drosophila Stock Center). The mutant was identified by PCR screening and sequencing using primers 5′-ACGTCTGTGCCTACGCCTTGATG-3′ and 5′-CACGCACGACATTCACGTACACA-3′.
Df(2R)ED1612 (FlyBase ID: FBst0008045), GTP-locked Rab protein expressing UAS-YFP-Rab7[Q67L] (Flybase ID: FBst0042707) and UAS-YFP-Rab2[Q65L] (FlyBase ID: FBst0009761), GDP-locked Rab2 expressing UAS-YFP-Rab2[S20N] (FlyBase ID: FBst0023640), UAS-YFP-Rab2 (FlyBase ID: FBst0023246), cg-Gal4 (FlyBase ID: FBst0007011), da-Gal4 (FlyBase ID: FBst0051669), Rab2-Gal4 (FlyBase ID: FBst0051581), and GMR-Gal4 (FlyBase ID: FBst0001104) came from Bloomington Drosophila Stock Center. UAS-Rab2[GD34767] (FlyBase ID: FBst0460794) and UAS-Rab7[GD40337] (FlyBase ID: FBst0463506) RNAi flies were purchased from the Vienna Drosophila Resource Center. For rescue experiments, Rab2-Gal4 was used to drive YFP-Rab2 expression in a homozygous rab2[d42] background. Da-Gal4 was used to drive the ubiquitous expression of GTP-locked, YFP-Rab2GTP Q65L mutant protein. Cg-Gal4 was used for localization of YFP-Rab2WT or YFP-Rab2GTP in fat cells.
We generated Gal4-expressing fat cell clones using hs-Flp; dLamp-3xmCherry, UAS-GFP; Act>CD2>Gal4, UAS-Dcr2, or hs-Flp; 3xmCherry-Atg8a, UAS-GFP; Act>CD2>Gal4, UAS-Dcr2 (Hegedűs et al., 2016). Starvations were performed by floating larvae in 20% sucrose solution for 4 h at RT. We estimated autophagic flux by the tandem mCherry-GFP-Atg8a reporter as earlier (Mauvezin et al., 2014; Nagy et al., 2015), and we used 95- to 100-h-old animals in these experiments. Other mutant lines used in this study were lt[LL07138] (FlyBase ID: FBst0328511; Lőrincz et al., 2014) and rab7[1] (Flybase ID: FBal0325096; Hegedűs et al., 2016).
Cell culture and siRNA experiments
Mycoplasma-free MDA-MB-231 cells were obtained from ATCC and grown in L15 Medium (Gibco) supplemented with 10% South American serum (EuroClone). Cells were grown at 37°C in 0% CO2. siRNA delivery was achieved by mixing 10 nM specific siRNAs (Silencer Select siRNA; Thermo Fisher Scientific) with Optimem and Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific) in one round of transfection according to the manufacturer’s instructions. Target sequences were as follows: hRAB2A #1: 5′-GAAGGAGUCUUUGACAUUAtt-3′ and hRAB2B #1: 5′-GAAUCCUUCCGUUCUAUCAtt-3′. For each RNA interference experiment, Silencer Select Negative Control No. 2 siRNA (Thermo Fisher Scientific) was used as scrambled siRNA. Knockdown efficiency was tested by quantitative PCR (Fig. S1 Q). All experiments were repeated on a different day, with similar results.
Quantitative PCR
Gene expression was analyzed using TaqMan Gene expression assay (Applied Biosystems). Real-time PCR was performed on the 14 ABI/Prism 7700 Sequence Detector System (PerkinElmer/Applied Biosystems), using a pre-PCR step of 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Specificity of the amplified products was confirmed by melting curve analysis (Dissociation Curve TM; PerkinElmer/Applied Biosystems). Samples were amplified with primers for each gene (GADPH, Hs99999905_m1; RAB2A, Hs00234094_m1; RAB2B, Hs00375685_m1; Thermo Fisher Scientific). The Ct values were normalized to the GAPDH curve. Results were quantified using the 2−ΔΔCT method. PCR experiments were performed in triplicate.
Y2H assay
PLEKHM1 was amplified from the EST SD27034 using primers 5′-ATGAGCTCCCTGTTCCGCAG-3′ and 5′-TTAGGCAACCTCATTCTTCTGTTT-3′, Vps41/Lt from LD33620 using 5′-ATGGCTAAAGCGTTGCCGCTC-3′ and 5′-CTATTTCCCCACGGTTAACTTCCAAA-3′, and Vps39 from GH10703 using 5′-ATGCACCAGGCCTACAGTGTTCACTCG-3′ and 5′-TTATTGCTGAGCAGCCGCCCT-3′ (all ESTs were from Drosophila Genomics Resource Center). GTP (QL mutant) and GDP (SN mutant) locked versions of Rab2 and Rab7 were amplified from genomic DNA of transgenic flies using primers 5′-ATGTCCTACGCGTACTTGTTCAA-3′ and 5′-CTAGCAGCAGCCACTGTTTGC-3′ and primers 5′-ATGTCCGGACGTAAGAAATCC-3′ and 5′-TTAGCACTGACAGTTGTCAGGA-3′, respectively. Appropriate 5′ overhangs were added to primers for cloning into pGADT7 AD (Gal4 DNA activation domain) and pGBKT7 BD (Gal4 DNA binding domain) vectors (Takara Bio Inc.), followed by cotransfection into the yeast strain PJ69-4A using Frozen-EZ Yeast Transformation II kit (Zymo Research). We selected transformants by their ability to grow in Trp−, Leu− medium, and interactions were assessed by growth on Trp−, Leu−, Ade− plates, with empty vectors serving as negative controls (Lőrincz et al., 2016).
GST pull-downs and coimmunoprecipitations
Full-length Vps39 was amplified from EST GH10703 (Drosophila Genomics Resource Center) with primers 5′-GATGGATCCATGCACCAGGCCTACAGTGTT-3′ and 5′-ATCCTCGAGTTGCTGAGCAGCCGCCCTGGCGAGCCGA-3′ and cloned as a BamHI–XhoI fragment into pETMBP vector (Glatz et al., 2013) encoding an N-terminal Maltose Binding Protein/MBP and C-terminal hexahistidine tag. GST-Rab2-GTP and -GDP locked constructs were gifts from S. Munro (Medical Research Council Laboratory of Molecular Biology, Cambridge, England, UK) (Gillingham et al., 2014). GST-Rab2 and MBP-Vps39 expression was performed overnight at 18°C in Escherichia coli Rosetta (DE3) pLysS strain (EMD Millipore). Expression was induced by 0.1 mM IPTG at OD 0.6–0.7. Recombinant Vps39 was purified with sequential nickel and MBP affinity chromatography according to the manufacturer’s recommendations (resins from GE Healthcare). GST-Rab2–expressing cells were collected and resuspended in lysis buffer (20 mM Tris-HCl, pH 8.0, 110 mM KCl, 5 mM MgCl2, 1% CHAPS, 5 mM β-mercaptoethanol, protease inhibitors, and 200 µM GDP or GTPγS). Lysates were centrifuged at 12,000 g for 30 min. Glutathione resin (GE Healthcare) was added to the supernatant and incubated for 30 min at 4°C. Beads were washed with lysis buffer (10-column volume) before pull-down assays. For GST pull-down assays, first the glutathione resin (GE Healthcare) with immobilized GST-Rab2 (or GST alone) was equilibrated with binding buffer (20 mM Tris, 50 mM NaCl, 0.1% Triton X-100, 2 mM β-mercaptoethanol, and 200 µM GDP or GTPγS). 20 µl resin saturated with baits was incubated with 1.5 µM prey (MBP-Vps39 or MBP alone; this concentration was chosen to prevent MBP-Vps39 precipitation) in binding buffer (total volume 200 µl) for 30 min at 4°C. Glutathione beads were pelleted (2,000 g, 2 min) and washed three times with wash buffer (20 mM Tris, 300 mM NaCl, 0.1% Triton X-100, 2 mM 2-mercaptoethanol, and 200 µM GTPγS or GDP). Proteins were eluted by boiling in 30 µl Laemmli buffer. Samples were subjected to SDS-PAGE, followed by staining with Coomassie Brilliant Blue (Sigma-Aldrich) to visualize GST-fused proteins and Western blots to detect MBP-Vps39 or MBP. GST pull-down experiments were repeated using independently purified proteins, with similar results.
Lysates from adult flies expressing YFP-Rab2[Q65L], YFP-Rab2[S20N], or GFP (driven by da-Gal4) were prepared as described (Takáts et al., 2013). In brief, 100 mg adult flies (starved for 2 h) were homogenized for 2× 10 s on ice in 1 ml lysis buffer containing 1% Triton X-100, using an Ultra-Turret T10 (IKA) with S10N-5G disperser (IKA). Lysates were cleared by centrifugation at 30,130 g for 2× 10 min at 4°C. Immunoprecipitation was performed using GFP-Trap nanobodies coupled to magnetic agarose beads (ChromoTek) according to the manufacturer’s instructions. Beads were boiled in 30 µl Laemmli buffer, followed by Western blot analysis.
Western blots and immunohistochemistry
Immunofluorescence analyses and Western blots for Drosophila samples were performed as described (Takáts et al., 2013; Lőrincz et al., 2016). In brief, protein samples of L3 larvae were separated by SDS-PAGE and transferred to Immobilon-P PVDF membrane, 0.45 µm (EMD Millipore). After blocking with 0.5% casein in TBS containing 0.1% Tween-20 (pH 7.6) for 1 h at RT, membranes were incubated with the appropriate primary antibodies diluted in blocking solution (1 h, RT). Membranes were washed 3× 10 min, followed by incubation with secondary antibody in blocking solution (1 h, RT). After 3× 10-min final washes, signal was detected with nitroblue tetrazolium–5-bromo-4-chloro-3-indolyl phosphate (Sigma) for AP-conjugated antibodies, and membranes were scanned dry by transilluminating in a 4990 photo scanner (Epson). For HRP-conjugated antibodies, chemiluminescence was developed using Immobilon ECL kit (EMD Millipore), and signal was detected on a ChemiDoc MP System (Bio-Rad Laboratories).
For immunostaining of Drosophila tissues, larvae were dissected in PBS and fixed with 4% formaldehyde in PBS (45 min at RT). Samples were washed (3× 10 min at RT) and permeabilized in PBS plus 0.1% Triton X-100 (PBTX) for 15 min at RT, followed by incubation in blocking solution (5.0% FCS in PBTX, 30 min at RT). Samples were incubated with primary antibodies diluted in the blocking solution overnight at 4°C. Samples were rinsed 3× and washed 3× 15 min in PBTX at RT, then incubated in blocking solution for 30 min at RT, followed by incubation with secondary antibodies in blocking solution for 3 h at RT. Washing steps were repeated, nuclei were counterstained with DAPI, and samples were mounted in Vectashield (Vector Laboratories). All experiments were repeated on a different day, with similar results.
The following antibodies were used for Drosophila experiments: mouse anti-Rab7 (1:10, DSHB; Riedel et al., 2016), rabbit anti-CathL (1:100; ab58991; Abcam), rat anti-Atg8a (1:300; Takáts et al., 2013), rabbit anti-Atg8a (Western blot, 1:5,000; Takáts et al., 2013), rabbit anti-p62/Ref2p (1:2,000; Pircs et al., 2012), rabbit anti-Rab2 (1:200; FL-212-sc:28567; Santa Cruz Biotechnology, Inc.) mouse anti-tubulin (1:2,000; AA4.3-s; DSHB), rat anti-GFP (1:3,000; Pircs et al., 2012), and rabbit anti-Vps16A (1:2,000; Pulipparacharuvil et al., 2005). Secondary antibodies were Alexa Fluor 350 goat anti–mouse, Alexa Fluor 568 goat anti–rabbit, Alexa Fluor 568 goat anti–rat, Alexa Fluor 568 goat anti–rabbit, Alexa Fluor 488 goat anti–rabbit (all 1:1,000; Invitrogen) for immunofluorescence and AP-conjugated goat anti–rat, anti–rabbit, and anti–mouse (all 1:5,000; EMD Millipore) for Western blots, with the exception of GST pulldowns, for which interactions were probed using HRP-conjugated monoclonal mouse anti-MBP (1:5,000; New England Biolabs, Inc.).
Immunofluorescence labeling of human cells was performed as follows. Cells were plated on glass coverslips (preincubated with 0.5% gelatin in PBS at 37°C for 30 min). After 72 h, cells were fixed in methanol at −20°C for 10 min, washed with PBS, and incubated in PBS/0.02% saponin for 10 min at RT. Cells were then incubated with PBS supplemented with 1% BSA and 0.02% saponin for 10 min. The coverslips were then gently deposited, face down, on 50 µl primary antibody diluted in PBS, 0.02% saponin, and 1% BSA on Parafilm. After 40-min incubation at RT, coverslips were transferred to 12-well plates and washed 3× with PBS/0.02% saponin. Cells were then incubated for 40 min at RT with the appropriate secondary antibody in PBS, 0.02% saponin, and 1% BSA. After three washes in PBS/0.02% saponin, coverslips were transferred to 12-well plates and incubated in PBS containing DAPI (1:3,000) for 5 min at RT. Coverslips were washed 3× in PBS and mounted in 20% Mowiol (Sigma-Aldrich), 5% glycerol, 2.5% DABCO (Molecular Probes), and 0.02% NaN3 in PBS. The following primary antibodies were used: mouse anti-LAMP1 (1:100, APC Mouse Anti-Human CD107a; BD) and rabbit anti-LC3A/B (1:100, D3U4C; Cell Signaling Technology). The following secondary antibodies were used: Alexa Fluor 488 donkey anti–mouse (Invitrogen) and Cy3 donkey anti–rabbit (Invitrogen). All experiments were repeated on a different day, with similar results.
Uptake assays and LTR staining
For uptake assays without chase, L3 larval proventriculi with garland nephrocytes were dissected in cold M3 medium (Sigma-Aldrich) and incubated in M3 supplemented with Texas red–conjugated Avidin D (Vector Laboratories) in 0.1 mg/ml for 5 min at RT, rinsed 3×, and fixed with 4% formaldehyde in PBS (30 min at RT). Samples were washed 3× 10 min in PBS, counterstained with DAPI, and processed for microscopy. For uptake assays with chase, samples were incubated in M3 supplemented with FITC-conjugated avidin (1:100; Invitrogen) for 5 min at RT, rinsed 3×, then incubated in tracer-free M3 for 30 min at RT. Samples were fixed and processed for immunofluorescence as described earlier.
For LysoTracker experiments, dissected fat bodies or late L3-stage larval proventriculi with the loosely attached garland nephrocytes were dissected in cold PBS and incubated in LTR (1:1,000 in PBS; Thermo Fisher Scientific) for 2 min at RT. Samples were rinsed three times, mounted in 80% glycerol in PBS, and photographed immediately. All experiments were repeated on a different day, with similar results.
Fluorescent imaging
Fluorescent images of Drosophila fat cells or garland nephrocytes were obtained at RT with an AxioImager.M2 microscope (ZEISS) with an ApoTome2 grid confocal unit (ZEISS) using EC Plan-Neofluar 40×/0.75-NA Air (ZEISS) or Plan-Apochromat 40×/0.95-NA Air (ZEISS) objectives for fat cells, and Plan-Apochromat 63×/1.40-NA Oil (ZEISS) objective for nephrocytes, an Orca Flash 4.0 LT sCMOS camera (Hamamatsu Photonics), and Efficient Navigation 2 software (ZEISS). Immersol 518F (ZEISS) immersion oil was used. To enhance focus depths in Fig. 1 (K and M), Fig. 3 (A–C, H, and I), Fig. 5, and Fig. S1 (D–G, I, and J), images from five consecutive focal planes (section thickness 0.24 µm for nephrocytes and 0.55 µm for fat cells) were projected onto one single image. Single focal planes are shown in other images, including all colocalization experiments. Microscope and imaging settings were identical for all experiments of the same kind. Primary images of Drosophila experiments were processed in Efficient Navigation 2 and Photoshop CS4 (Adobe Systems) to produce final figures.
Immunostained MDA-MB-231 cells were examined at RT by fluorescent microscopy (Fig. S1, L–O) on an upright AX70 microscope (Olympus) equipped with CoolSnap EZ camera (Photometrics) using a UPlanSapo 60×/1.35-NA Oil objective (Olympus), or by confocal microscopy (Fig. 2 K) on a DM IRE2 inverted microscope (Leica Biosystems) with TCS SP2 AOBS confocal scanner unit (Leica Biosystems) and 405-, 488-, and 561-nm excitation laser lines using a HCX PL Apochromat 63×/1.4-NA oil (Leica Biosystems) objective. The following immersion oils were used: UM3082 (Carlo Erba) and type F (Leica Biosystems), respectively. Imaging was performed using MetaMorph (Olympus) orconfocal software (Leica Biosystems), respectively. Image acquisition conditions were set to remove channel cross-talk, optimizing spectral detection bands and scanning modalities. Images were analyzed using ImageJ. We quantified the area of LC3 signal above background (using the adjust threshold function of ImageJ). Values were normalized over the scramble siRNA-treated control cells and shown as fold increase in Fig. S1 P. For analyzing colocalization of LC3 with Lamp1 (Fig. 2 K), ImageJ coloc2 plugin was used. Final images were prepared in Photoshop CS4, by adjusting brightness and contrast.
Statistics
Fluorescence structures from original, unmodified single focal planes were quantified using ImageJ. The signal threshold for the relevant fluorescent channel was set by the same person when quantifying one type of experiment. For clonal experiments, a GFP-positive fat cell from one cell clone was randomly selected, and one of its immediate neighbor GFP-negative control cells was also randomly selected for quantification. Please note that fat cell clones are spontaneously and randomly generated independent of each other in mosaic animals. Cells were randomly selected for counting from pictures of mutant or control fat cells or nephrocytes. In all cases, only cells with their nuclei in the focal plane were selected to make sure that both perinuclear and peripheral regions are included in quantifications. Rab2 colocalization was manually quantified by the same skilled researcher. Both double-positive, overlapping dots and YFP-Rab2 rings around the red markers were counted as colocalization.
The quantified data were evaluated by performing the appropriate statistical tests as described previously (Takáts et al., 2013, 2014). We used SPSS17 (IBM) for data analysis. t tests were used for comparing two and analysis of variance for comparing multiple samples that all showed normal distribution, and U tests for comparing two and Kruskal–Wallis tests for comparing multiple samples that contained at least one variable showing non-Gaussian data distribution. Analysis of variance was used in Fig. 1 H (dot number; n = 20 cells evaluated from five to six images of three to five larvae per genotype), Fig. 2 L (n = 13–23 human cells evaluated from five images), and Fig. S1 P (n = 31–35 human cells evaluated from three to five images). We used Kruskal–Wallis test for Fig. 1 H (dot size; n = 20 cells evaluated from five to six images of three to five larvae per genotype), Fig. 3 D (n = 10 cells evaluated from four to six images of three to four larvae per genotype), and Fig. S2 E (n = 20 cells evaluated from four to six images of three to five larvae per genotype). Paired t test was used for analyzing dot numbers in Fig. 1 (L and N) (n = 10 cells evaluated from six to eight images of three to five larvae per genotype) and Fig. 2 G (n = 9 cells evaluated from four to five images of three to five larvae per genotype). U test was used for analyzing dot sizes in Fig. 1 (J, L, and N) (n = 10 cells evaluated from six to eight images of three to five larvae per genotype), Fig. 2 G (n = 9 cells evaluated from five to six images of three to five larvae per genotype) and Fig. 5 G (n = 10 cells evaluated from six to eight images of three to five larvae per genotype), and for analyzing dot numbers in Fig. 1 J (n = 10 cells evaluated from six to eight images of three to five larvae per genotype), Fig. 2 E (n = 30–45 cells evaluated from four to five images of three to five larvae per genotype), Fig. 3 G (n = 12–13 cells evaluated from four to five images of three to four larvae per genotype), Fig. S1 K (n = 43–45 cells evaluated from six to eight images of three to five larvae per genotype), and Fig. S2 F (n = 6 cells evaluated from five images of three larvae per genotype). U test was also used to compare colocalization ratios in Fig. S3 A (n = 10 cells evaluated from four to six images of three to five larvae per genotype). Error bars denote ± SEM in bar charts. In the box plot (Fig. 3 D), bars show the data ranging between the upper and lower quartiles; median is indicated as a horizontal black line within the box. P values for the relevant comparisons are shown in the panels.
Electron microscopy and acid phosphatase cytochemistry
Ultrastructural analyses of fat cells and nephrocytes were performed as described (Takáts et al., 2013; Lőrincz et al., 2016). Dissected fat bodies or proventriculi with the loosely attached garland nephrocytes were fixed in 3.2% PFA, 0.5% (nephrocytes) or 1% (fat cells) glutaraldehyde, 1% sucrose, and 0.028% CaCl2 in 0.1 N sodium cacodylate, pH 7.4, overnight at 4°C. Samples were then postfixed in 0.5% osmium tetroxide for 1 h and in half-saturated aqueous uranyl acetate for 30 min at RT, dehydrated in a graded series of ethanol, and embedded in Durcupan (Fluka) according to the manufacturer’s recommendations. 70-nm sections were stained in Reynolds lead citrate and viewed on a JEM-1011 transmission electron microscope (Jeol) equipped with a Morada digital camera (Olympus) using iTEM software (Olympus).
Acid phosphatase cytochemistry was performed as described (Lőrincz et al., 2014). In brief, dissected nephrocytes were fixed in 2% formaldehyde, 2% glutaraldehyde, 3 mM CaCl2, and 1% sucrose in 0.1 M Na-cacodylate (pH 7.4) for 1 h at RT, and then extensively washed. Buffer was changed to 0.05 M Na-acetate (pH 5.0, 3× 5 min at RT). Next, samples were incubated in Gömöri medium (5 mM Na-β-glycerophosphate and 4 mM lead nitrate dissolved in 0.05 M acetate buffer) or in substrate-free buffer (control experiment) for 2 h at RT. Samples were then washed for 3× 5 min in acetate buffer and processed for EM. Ultrathin sections were analyzed unstained.
Microscopy of compound eyes and whole larvae
Eyes and larvae were photographed on a Lumar V12 stereomicroscope (ZEISS) equipped with AxioCam ERc5s camera (ZEISS). For embedding and sectioning, adult heads were cut in half, fixed (3.2% PFA, 0.5% glutaraldehyde, 1% sucrose, and 0.028% CaCl2 in 0.1 N sodium cacodylate, pH 7.4) overnight at 4°C, postfixed in half-saturated aqueous uranyl acetate for 30 min, dehydrated in a graded series of ethanol, and embedded in Durcupan) according to the manufacturer’s recommendations. Osmium postfixation was omitted to preserve the color of pigment granules. 700-nm-thick sections were collected on glass slides and allowed to dry. Sections were then covered with a drop of staining solution: 0.2% Light Green SF (Pharmaceutical Raw Material Stocking) and 0.2% acetic acid in water, heated on a hot plate for 1–2 min, followed by rinsing with distilled water. Sections were covered with Eukitt quick-hardening mounting medium (Sigma-Aldrich) and photographed at RT using an AxioImager.Z1 microscope equipped with AxioCam ICc camera and EC Plan-Neofluar 100×/1.3-NA Oil objective using AxioVision 4.82 software (all ZEISS).
Online supplemental material
Fig. S1 shows additional Drosophila Rab2 and human RAB2A and RAB2B data. Fig. S2 shows endocytic uptake assays and quantification of acid phosphatase data. Fig. S3 shows quantification of Rab2 localization data and the effect of Rab2GTP expression on eye pigment formation.
Supplementary Material
Acknowledgments
We thank Sarolta Pálfia and Gina Puska for assistance.
We also thank our funders: Wellcome Trust (087518/Z/08/Z to G. Juhász), Hungarian Academy of Sciences (LP-2014/2 to G. Juhász), National Research, Development and Innovation Office of Hungary (GINOP-2.3.2-15-2016-00006 and K119842 to G. Juhász and PD112632 to K. Hegedűs), Italian Association for Cancer Research Investigator Grant (10168 and 18621 to G. Scita) and European Research Council (268836 to G. Scita), New National Excellence Program (UNKP-16-3-III to P. Lőrincz and UNKP-16-2-I to Z. Lakatos), and Italian Association for Cancer Research (S. Bisi).
The authors declare no competing financial interests.
Author contributions: P. Lőrincz and G. Juhász designed research. P. Lőrincz, S. Tóth, P. Benkő, Z. Lakatos, A. Boda, G. Glatz, K. Hegedűs, S. Takáts, S. Bisi, and M. Zobel performed experiments. P. Lőrincz, S. Tóth, P. Benkő, A. Boda, G. Glatz, S. Bisi, M. Zobel, G. Scita, and G. Juhász evaluated data. P. Lőrincz and G. Juhász wrote the paper with comments from all authors.
Footnotes
Abbreviations used:
- HOPS
- homotypic fusion and vacuole protein sorting
- LTR
- Lysotracker Red
- Y2H
- yeast two-hybrid
References
- Balderhaar H.J., and Ungermann C.. 2013. CORVET and HOPS tethering complexes: Coordinators of endosome and lysosome fusion. J. Cell Sci. 126:1307–1316. 10.1242/jcs.107805 [DOI] [PubMed] [Google Scholar]
- Caplan S., Hartnell L.M., Aguilar R.C., Naslavsky N., and Bonifacino J.S.. 2001. Human Vam6p promotes lysosome clustering and fusion in vivo. J. Cell Biol. 154:109–122. 10.1083/jcb.200102142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S., Jin E.J., Ozel M.N., Lu Z., Agi E., Wang D., Jung W.H., Epstein D., Meinertzhagen I.A., Chan C.C., and Hiesinger P.R.. 2013. Charcot-Marie-Tooth 2B mutations in rab7 cause dosage-dependent neurodegeneration due to partial loss of function. eLife. 2:e01064 10.7554/eLife.01064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Huang W., Lin T.H., Groulx J.F., Jean S., Nguyen J., Kuchitsu Y., Koyama-Honda I., Mizushima N., Fukuda M., and Kiger A.A.. 2017. Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. eLife. 6:e23367 10.7554/eLife.23367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham A.K., Sinka R., Torres I.L., Lilley K.S., and Munro S.. 2014. Toward a comprehensive map of the effectors of rab GTPases. Dev. Cell. 31:358–373. 10.1016/j.devcel.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz G., Gógl G., Alexa A., and Reményi A.. 2013. Structural mechanism for the specific assembly and activation of the extracellular signal regulated kinase 5 (ERK5) module. J. Biol. Chem. 288:8596–8609. 10.1074/jbc.M113.452235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P., Hu T., Zhang J., Jiang S., and Wang X.. 2010. Sequential action of Caenorhabditis elegans Rab GTPases regulates phagolysosome formation during apoptotic cell degradation. Proc. Natl. Acad. Sci. USA. 107:18016–18021. 10.1073/pnas.1008946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedűs K., Takáts S., Boda A., Jipa A., Nagy P., Varga K., Kovács A.L., and Juhász G.. 2016. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol. Biol. Cell. 27:3132–3142. 10.1091/mbc.E16-03-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Nishimura T., Sakamaki Y., Itakura E., Hatta T., Natsume T., and Mizushima N.. 2014. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell. 25:1327–1337. 10.1091/mbc.E13-08-0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiho H., Kajiho Y., Frittoli E., Confalonieri S., Bertalot G., Viale G., Di Fiore P.P., Oldani A., Garre M., Beznoussenko G.V., et al. 2016. RAB2A controls MT1-MMP endocytic and E-cadherin polarized Golgi trafficking to promote invasive breast cancer programs. EMBO Rep. 17:1061–1080. 10.15252/embr.201642032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter D., Raina V.B., Dwivedi D., Sindhwani A., Bahl S., and Sharma M.. 2015. The small GTPase Arl8b regulates assembly of the mammalian HOPS complex on lysosomes. J. Cell Sci. 128:1746–1761. 10.1242/jcs.162651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd V., Ramaswami M., and Krämer H.. 1998. Not just pretty eyes: Drosophila eye-colour mutations and lysosomal delivery. Trends Cell Biol. 8:257–259. 10.1016/S0962-8924(98)01270-7 [DOI] [PubMed] [Google Scholar]
- Lőrincz P., Lakatos Z., Maruzs T., Szatmári Z., Kis V., and Sass M.. 2014. Atg6/UVRAG/Vps34-containing lipid kinase complex is required for receptor downregulation through endolysosomal degradation and epithelial polarity during Drosophila wing development. BioMed Res. Int. 2014:851349 10.1155/2014/851349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz P., Lakatos Z., Varga Á., Maruzs T., Simon-Vecsei Z., Darula Z., Benkő P., Csordás G., Lippai M., Andó I., et al. 2016. MiniCORVET is a Vps8-containing early endosomal tether in Drosophila. eLife. 5:e14226 10.7554/eLife.14226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruzs T., Lőrincz P., Szatmári Z., Széplaki S., Sándor Z., Lakatos Z., Puska G., Juhász G., and Sass M.. 2015. Retromer ensures the degradation of autophagic cargo by maintaining lysosome function in Drosophila. Traffic. 16:1088–1107. 10.1111/tra.12309 [DOI] [PubMed] [Google Scholar]
- Mauvezin C., Ayala C., Braden C.R., Kim J., and Neufeld T.P.. 2014. Assays to monitor autophagy in Drosophila. Methods. 68:134–139. 10.1016/j.ymeth.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvezin C., Nagy P., Juhász G., and Neufeld T.P.. 2015. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 6:7007 10.1038/ncomms8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan D.G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F.P., Miranda de Stegmann D., Bhogaraju S., Maddi K., et al. 2015. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell. 57:39–54. 10.1016/j.molcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., and Klionsky D.J.. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075. 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountjoy J.R., Xu W., McLeod D., Hyndman D., and Oko R.. 2008. RAB2A: A major subacrosomal protein of bovine spermatozoa implicated in acrosomal biogenesis. Biol. Reprod. 79:223–232. 10.1095/biolreprod.107.065060 [DOI] [PubMed] [Google Scholar]
- Nagy P., Varga Á., Kovács A.L., Takáts S., and Juhász G.. 2015. How and why to study autophagy in DROSOPHILA: It’s more than just a garbage chute. Methods. 75:151–161. 10.1016/j.ymeth.2014.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis I.P., Simonsen A., Sagona A.P., Finley K., Gaumer S., Contamine D., Rusten T.E., Stenmark H., and Brech A.. 2008. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 180:1065–1071. 10.1083/jcb.200711108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Alemu E.A., Brech A., Bruun J.A., Lamark T., Overvatn A., Bjørkøy G., and Johansen T.. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188:253–269. 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircs K., Nagy P., Varga A., Venkei Z., Erdi B., Hegedus K., and Juhasz G.. 2012. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS One. 7:e44214 10.1371/journal.pone.0044214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols M.S., ten Brink C., Gosavi P., Oorschot V., and Klumperman J.. 2013. The HOPS proteins hVps41 and hVps39 are required for homotypic and heterotypic late endosome fusion. Traffic. 14:219–232. 10.1111/tra.12027 [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S., Akbar M.A., Ray S., Sevrioukov E.A., Haberman A.S., Rohrer J., and Krämer H.. 2005. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J. Cell Sci. 118:3663–3673. 10.1242/jcs.02502 [DOI] [PubMed] [Google Scholar]
- Riedel F., Gillingham A.K., Rosa-Ferreira C., Galindo A., and Munro S.. 2016. An antibody toolkit for the study of membrane traffic in Drosophila melanogaster. Biol. Open. 5:987–992. 10.1242/bio.018937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P., and Klumperman J.. 2009. Lysosome biogenesis and lysosomal membrane proteins: Trafficking meets function. Nat. Rev. Mol. Cell Biol. 10:623–635. 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- Saraste J. 2016. Spatial and functional aspects of ER-Golgi Rabs and tethers. Front. Cell Dev. Biol. 4:28 10.3389/fcell.2016.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukov E.A., He J.P., Moghrabi N., Sunio A., and Krämer H.. 1999. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol. Cell. 4:479–486. 10.1016/S1097-2765(00)80199-9 [DOI] [PubMed] [Google Scholar]
- Solinger J.A., and Spang A.. 2013. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 280:2743–2757. 10.1111/febs.12151 [DOI] [PubMed] [Google Scholar]
- Takáts S., Nagy P., Varga Á., Pircs K., Kárpáti M., Varga K., Kovács A.L., Hegedűs K., and Juhász G.. 2013. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201:531–539. 10.1083/jcb.201211160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Pircs K., Nagy P., Varga Á., Kárpáti M., Hegedűs K., Kramer H., Kovács A.L., Sass M., and Juhász G.. 2014. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol. Biol. Cell. 25:1338–1354. 10.1091/mbc.E13-08-0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kant R., Fish A., Janssen L., Janssen H., Krom S., Ho N., Brummelkamp T., Carette J., Rocha N., and Neefjes J.. 2013. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J. Cell Sci. 126:3462–3474. 10.1242/jcs.129270 [DOI] [PubMed] [Google Scholar]
- Wang Z., Miao G., Xue X., Guo X., Yuan C., Wang Z., Zhang G., Chen Y., Feng D., Hu J., and Zhang H.. 2016. The Vici syndrome protein EPG5 is a Rab7 effector that determines the fusion specificity of autophagosomes with late endosomes/lysosomes. Mol. Cell. 63:781–795. 10.1016/j.molcel.2016.08.021 [DOI] [PubMed] [Google Scholar]
- Wijdeven R.H., Janssen H., Nahidiazar L., Janssen L., Jalink K., Berlin I., and Neefjes J.. 2016. Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat. Commun. 7:11808 10.1038/ncomms11808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y., and Stenmark H.. 2015. Cellular functions of Rab GTPases at a glance. J. Cell Sci. 128:3171–3176. 10.1242/jcs.166074 [DOI] [PubMed] [Google Scholar]
- Zick M., and Wickner W.. 2016. Improved reconstitution of yeast vacuole fusion with physiological SNARE concentrations reveals an asymmetric Rab(GTP) requirement. Mol. Biol. Cell. 27:2590–2597. 10.1091/mbc.E16-04-0230 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.