Accurate spindle positioning depends on the action of the dynein motor protein at microtubules. Estrem et al. now show that increased dynein-mediated activity destabilizes microtubules by displacing dynactin from the plus end and suggests that the interplay between these opposing effects on microtubule stability at the cortex is required to move the spindle to the correct position.
Abstract
How dynein motors accurately move cargoes is an important question. In budding yeast, dynein moves the mitotic spindle to the predetermined site of cytokinesis by pulling on astral microtubules. In this study, using high-resolution imaging in living cells, we discover that spindle movement is regulated by changes in microtubule plus-end dynamics that occur when dynein generates force. Mutants that increase plus-end stability increase the frequency and duration of spindle movements, causing positioning errors. We find that dynein plays a primary role in regulating microtubule dynamics by destabilizing microtubules. In contrast, the dynactin complex counteracts dynein and stabilizes microtubules through a mechanism involving the shoulder subcomplex and the cytoskeletal-associated protein glycine-rich domain of Nip100/p150glued. Our results support a model in which dynein destabilizes its microtubule substrate by using its motility to deplete dynactin from the plus end. We propose that interplay among dynein, dynactin, and the stability of the microtubule substrate creates a mechanism that regulates accurate spindle positioning.
Introduction
During eukaryotic cell division, coordination between the mitotic spindle, which partitions duplicated chromosomes, and the cytokinetic contractile network, which partitions the cytoplasm, ensures that each daughter cell inherits a genome (Rappaport and Ebstein, 1965). In animal cells, the position of the spindle determines the site of cytokinesis, which is critical during asymmetric cell divisions that underlie development and tissue homeostasis for guiding the inheritance of cytoplasmic fate determinants and the positions of daughter cells. In unicellular organisms such as budding yeast, the spindle must be brought to a predetermined site of cytokinesis between the mother and bud. In all of these scenarios, the spindle is positioned through mechanisms that integrate polarity signals into force generation by the microtubule cytoskeleton (McNally, 2013).
Spindle movement in many species and cellular contexts is driven by cytoplasmic dynein, a large multisubunit protein complex that uses ATP hydrolysis to power motility toward the minus ends of microtubules. Dynein moves the spindle by generating pulling forces on astral microtubules, which are nucleated at microtubule-organizing centers and project into the cytoplasm, toward the cell cortex (Carminati and Stearns, 1997; Skop and White, 1998; Adames and Cooper, 2000; Grill et al., 2001; Kiyomitsu and Cheeseman, 2013). Observations in different organisms suggest that dynein can generate pulling forces either through end-on interactions with microtubules at the cortex or by sliding microtubules laterally along the cortex. In budding yeast, dynein is delivered on the plus ends of astral microtubules to the cortex, where it attaches to its cortical receptor Num1 (Lee et al., 2005). Then, dynein is activated, and its minus end–directed motility slides the astral microtubule along the cortex and past the anchored motor, drawing the spindle toward the cortex (Adames and Cooper, 2000; Lammers and Markus, 2015). In the Caenorhabditis elegans zygote, dynein accumulates at the anterior cell cortex and captures astral microtubules in an end-on manner (Labbé et al., 2003; Redemann et al., 2010). These microtubule ends quickly undergo catastrophe but remain at the cortex, and their depolymerization is thought to generate force to pull the spindle toward the anterior (Kozlowski et al., 2007). Whereas end-on interactions predominate during anaphase in C. elegans zygotes, lateral sliding interactions are observed during telophase and are highly abundant in embryos that generate free microtubule fragments by ectopic expression of the microtubule-severing protein katanin (Kozlowski et al., 2007; Gusnowski and Srayko, 2011). This suggests dynein can alternate between different modes of force generation. Consistent with this, experiments by Laan et al. (2012) demonstrate that dynein purified from budding yeast can generate force by capturing plus ends at a fabricated barrier, promoting catastrophe and maintaining attachment as the microtubules depolymerize. In this scenario, dynein is thought to promote catastrophe by holding the microtubule end close to the barrier. Microtubule ends can be induced to catastrophe by colliding with barriers, which is thought to impede the arrival of new tubulin subunits (Janson et al., 2003). Thus, dynein can generate force using either of two mechanisms—moving along the sides of microtubules and/or regulating plus-end dynamics—and the predominant mechanism may be determined by physiological context.
Spindle positioning requires the regulation of dynein by accessory subunits of the dynein complex (known as light, intermediate, and light intermediate chains) and interactions with extrinsic regulators (Yoder and Han, 2001; Lee et al., 2005; Pecreaux et al., 2006; Couwenbergs et al., 2007; Stuchell-Brereton et al., 2011). Dynactin is a multisubunit protein complex that links dynein to cargoes and promotes association with its microtubule track (Schroer, 2004; Culver-Hanlon et al., 2006; Ayloo et al., 2014; McKenney et al., 2014; Schlager et al., 2014). Dynactin is required for spindle positioning in diverse contexts and is thought to play important roles in linking dynein to the cell cortex and contributing to its motility (Skop and White, 1998; Moore et al., 2008, 2009; Siller and Doe, 2008). Although we have a detailed inventory of the proteins required for dynein-dependent spindle positioning, we do not understand the interplay between dynein and its dynamic microtubule track or how these activities might be differentially regulated to move the spindle in different contexts.
In this study, we investigate how dynein regulates the stability of its microtubule track and the role of microtubule stability in properly positioning the spindle in budding yeast. In yeast, microtubule dynamics occur exclusively at the plus ends; the minus ends are inert and anchored at the spindle pole bodies (SPBs; the centrosome equivalent in yeast; Byers and Goetsch, 1975; Maddox et al., 2000). We find that astral microtubules undergo a transition to depolymerization during dynein-dependent sliding along the cortex, and this plays a key role in properly positioning the spindle. Inhibiting depolymerization exacerbates dynein-dependent spindle movement, leading to aberrantly positioned spindles. We also find that dynein heavy chain destabilizes astral microtubules and demonstrate that this requires an active motor. In contrast, dynactin stabilizes astral microtubules, and our results indicate that dynein may induce the microtubule to catastrophe by depleting dynactin from the plus end. We propose that dynein’s regulation of plus-end stability is coordinated with its motility, creating a negative feedback loop that terminates spindle movement by destroying dynein’s microtubule track.
Results
Microtubule stability regulates dynein-dependent spindle movement
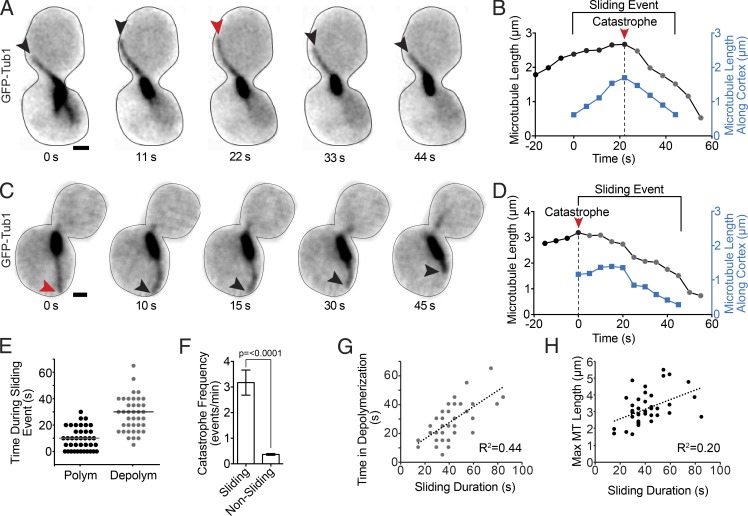
How does microtubule stability contribute to dynein’s role in positioning the mitotic spindle and nucleus? To address this question, we analyzed the dynamics of GFP-labeled microtubules (GFP-Tub1) during dynein-dependent microtubule sliding along the cell cortex. We defined sliding events as instances where astral microtubules contact the cell cortex and bend as they move along its concave surface, accompanied by movement of the spindle in the same direction as the microtubule (Fig. 1 A). To prevent anaphase spindle elongation from limiting movement, we arrested cells in S phase by treatment with hydroxyurea (HU). Sliding in arrested cells is similar to that seen in asynchronous cells, but the arrest allows us to measurement more events (Fig. S1, A–C). We examined microtubules during 39 sliding events in 30 WT cells. Fig. 1 B shows a representative example of microtubule length changes and changes in the association of the microtubule with the cell cortex during a sliding event. Our analysis reveals three prominent characteristics of microtubule dynamics during sliding events: (1) Sliding events are initiated by polymerizing microtubules (in 39 of 39 events, microtubules are polymerizing immediately before the onset of sliding). (2) During a sliding event, the microtubule undergoes catastrophe to switch from a polymerizing to a depolymerizing state. In 10 of 39 events catastrophe coincides with initiation (for example, see Fig. 1, C and D), whereas in 29 of 39 events catastrophe occurs after initiation (Fig. 1, A and B). (3) The microtubule spends most of the sliding event in a depolymerizing state (Fig. 1 E), and all sliding events analyzed end with depolymerizing microtubules (39 of 39 events analyzed). The rate of depolymerization during sliding is slower than that measured for microtubules in the cytoplasm (Fig. S1 C). The prominence of the depolymerizing state led us to ask whether microtubules are more likely to undergo catastrophe when plus ends are sliding along the cell cortex than when they are away from the cortex. Indeed, we find that the frequency of catastrophe is increased 8.6-fold during sliding events (3.17 ± 0.49 events per minute of sliding, compared with 0.37 ± 0.03 events per minute of nonsliding; P < 0.0001; Fig. 1 F). In contrast, rescues are rarely observed during sliding events (4 rescues in 39 events analyzed, with a mean of 28 s of depolymerization per event).
Figure 1.
Microtubule dynamics during spindle movement. (A) Time series images of GFP-Tub1 in a WT cell during a dynein-dependent sliding event. Each image is a maximum intensity projection from a confocal Z series. Black arrowheads point to the plus end of the sliding microtubule. The red arrowhead points to the plus end at the time of a catastrophe event. Bar, 1 µm. (B) Life plot of the microtubule depicted in A. Sliding Event denotes the time during which the microtubule is moving laterally along the cell cortex and the spindle is moving. Black and gray dots are the total length of the microtubule measured from the SPB to the microtubule plus end at 14 successive time points. Blue dots are the length on the microtubule laterally touching the cell cortex during nine successive time points. (C) Time series images of GFP-Tub1 in a WT cell during a dynein-dependent sliding event with the microtubule catastrophe at the start of the sliding event. Black arrowheads point to the plus end of the sliding microtubule. The red arrowhead points to the plus end at the time of a catastrophe event. Bar, 1 µm. (D) Life plot of the microtubule depicted in C. (E) Dot plot of time during a sliding event (seconds) the microtubule spent in polymerization (Polym) or depolymerization (Depolym). n = 39 for each. Horizontal bars show the mean of values. (F) Mean catastrophe frequency during sliding events, compared with all other times in pre-anaphase WT cells. Error bars are SEM, and p-value is determined by t test. Nonsliding microtubules, n = 24; sliding microtubules, n = 41. (G) Scatter plot depicting the coefficient of determination for microtubule time spent in depolymerization (seconds) and sliding duration. n = 33 sliding events. The dotted line is the best fit to a linear correlation. (H) Scatter plot depicting the coefficient of determination for maximum microtubule (MT) length during a sliding event and sliding duration. n = 35 sliding events.
Changes in microtubule stability could play a determining role in the duration of a sliding event and, consequently, the displacement of the spindle and nucleus. Indeed, our analysis reveals a strong positive correlation between the time the sliding microtubule spends in depolymerization and the duration of a sliding event (R2 = 0.44; Fig. 1 G). We also found a positive, albeit weaker, correlation with the maximum length of the sliding microtubule, i.e., the maximum length reached from the beginning to the end of the sliding event, and the duration of a sliding event (R2 = 0.20; Fig. 1 H). In separate experiments, we tested whether longer sliding events exhibit increased dynein at the microtubule plus end; however, our analysis does not reveal a correlation (R2 = 0.01). These results suggest that longer microtubules may sustain longer-lived sliding events.
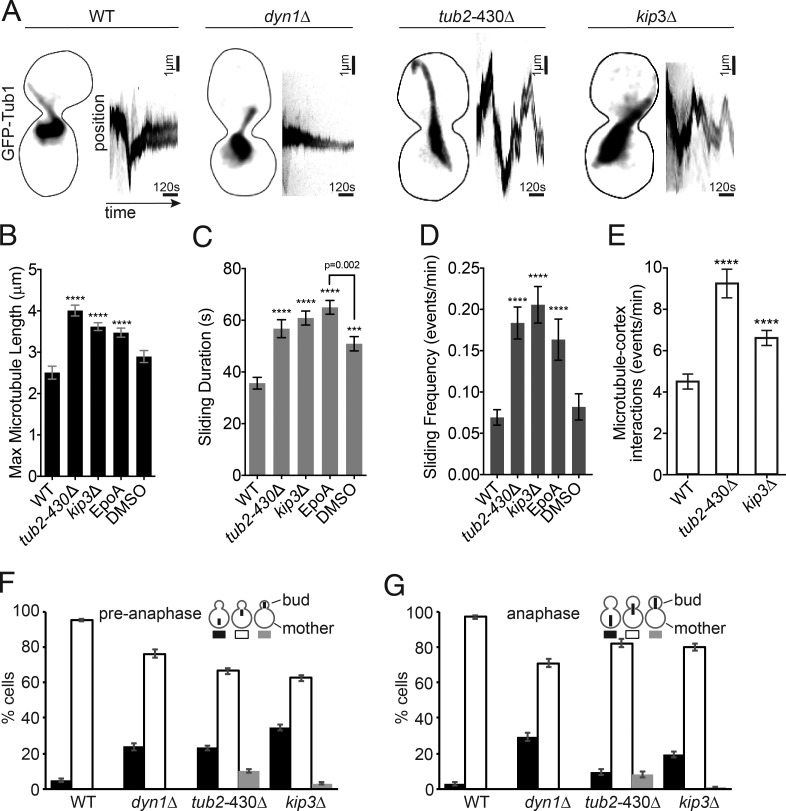
We tested the hypothesis that microtubule stability determines sliding behavior by using mutants and drugs to stabilize microtubules. We examined sliding events in two mutants: tub2-430Δ, which lacks the C-terminal 27 amino acids of β-tubulin, and kip3Δ, a deletion of the yeast kinesin 8. Both of these mutants exhibit longer and more stable microtubules and more pronounced spindle movements (Fig. 2 A; Gupta et al., 2006; Aiken et al., 2014). In contrast, dynein heavy chain–null mutants exhibit diminished spindle movement and no sliding events (Fig. 2 A). We find that the maximum length of sliding microtubules is increased in both tub2-430Δ and kip3Δ mutants, compared with WT controls (Fig. 2 B and Fig. S2 A). Sliding events are longer lived in both tub2-430Δ and kip3Δ mutants than in WT controls, but the velocity of spindle movement is unchanged (Fig. 2 C; Fig. S2, B and C; Videos 1–4). We also confirmed that mutants with longer astral microtubules do not increase the amount of dynein at the plus end (Fig. S2 D). These results are consistent with microtubule stability determining the duration of sliding events.
Figure 2.
Microtubule stability determines the duration of spindle movement. (A) Representative images of pre-anaphase cells with GFP-Tub1–labeled microtubules of the indicated genotypes. Each image is a maximum intensity projection from a confocal Z series. Bars, 1 µm. The corresponding kymographs track the fluorescence intensity of the spindle over time (10 min) showing spindle movement. (B) Mean of the maximum astral microtubule length reached during a sliding event in pre-anaphase cells. EpoA are WT cells treated with epothilone A. DMSO are WT cells treated with DMSO control. WT, n = 51 microtubules; tub2-430Δ, n = 97; kip3Δ, n = 208; EpoA, n = 106; DMSO, n = 42. (C) Mean sliding duration in seconds. WT, n = 52 microtubules; tub2-430Δ, n = 97; kip3Δ, n = 204; EpoA, n = 157; DMSO, n = 88. (D) Mean sliding frequency displayed in events per minute. WT, n = 91 microtubules; tub2-430Δ, n = 73; kip3Δ, n = 53; EpoA, n = 157; DMSO, n = 88. (E) Mean frequency of microtubule-cortex interactions per minute. WT, n = 39 cells; tub2-430Δ, n = 38; kip3Δ, n = 58. (B–E) Error bars are SEM. Asterisks denote a significant difference from WT. ***, P < 0.0006; ****, P < 0.0001; determined by t test. (F) Position of pre-anaphase spindles (<1.9 µm) in asynchronous cells. WT, n = 837 cells; dyn1Δ, n = 433; tub2-430Δ, n = 770; kip3Δ, n = 959. (G) Position of anaphase spindles (≥1.9 µm) in asynchronous cells. WT, n = 333 cells; dyn1Δ, n = 405; tub2-430Δ, n = 300; kip3Δ, n = 427. (F and G) Error bars are standard error of proportion.
In addition to changes in sliding duration, we find that sliding events occur more often in mutants that increase the stability of astral microtubules (Fig. 2 D and Fig. S2 E). tub2-430Δ and kip3Δ mutants also exhibit an increased frequency of astral microtubule plus ends colliding with the cell cortex (Fig. 2 E and Fig. S2 F). This suggests that plus-end interactions with the cell cortex may be a limiting step in the spindle movement mechanism. Interestingly, tub2-430Δ mutants convert these interactions into sliding events at a greater rate than observed in WT controls, whereas kip3Δ mutants convert microtubule–cortex interactions into sliding events at a rate that is similar to WT (Fig. S2 G). Why tub2-430Δ mutants specifically promote the conversion of microtubule–cortex interactions into sliding events is not immediately clear; however, we observed that, whenever plus ends collide with the cortex in these mutants, they tend to dwell rather than quickly depolymerizing (Fig. S2 H).
As an additional test of our hypothesis, we stabilized microtubules using the drug epothilone A (Bode et al., 2002; Prota et al., 2013). Treatment with 10 µM epothilone A caused WT cells to exhibit longer microtubules and longer-lived and more frequent sliding events (Figs. 2, B–D). Collectively, these results support our hypothesis that microtubule stability plays a determining role in the dynein-dependent sliding events that move the spindle and nucleus.
Then, we asked how increased microtubule sliding impacts the position of the mitotic spindle during cell division. First, we measured the position of bipolar, pre-anaphase spindles, using single–time point images of large populations of asynchronous cells. Whereas 95% of WT cells exhibit the spindle at the bud neck, both tub2-430Δ and kip3Δ mutants frequently exhibit spindles away from the bud neck, either in the distal end of the mother compartment or across the neck in the bud (Fig. 2 F). Spindle position is also disrupted in cells that have entered anaphase (Fig. 2 G). In contrast, ablating microtubule sliding by knocking out dynein heavy chain (dyn1Δ) increases the frequency of pre-anaphase and anaphase spindles that lag in the mother compartment, without the appearance of spindles in the bud (Fig. 2, F and G). We conclude that microtubule stability regulates the accuracy of spindle positioning, and increasing microtubule stability causes excessive microtubule sliding, which displaces the spindle from the nascent site of cytokinesis.
Dynein destabilizes astral microtubules
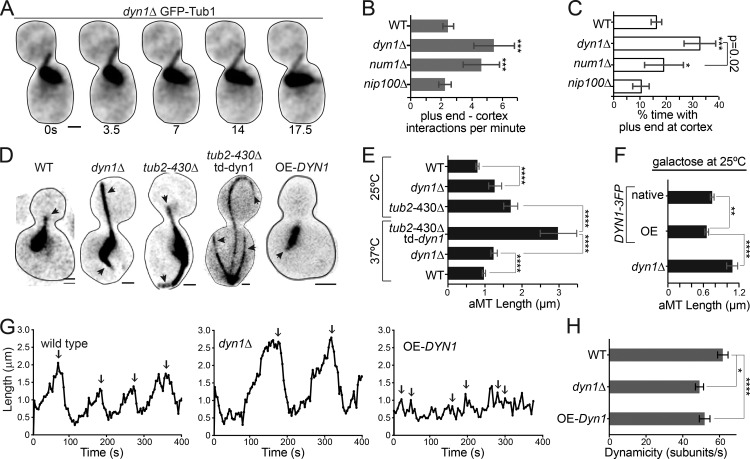
We hypothesized that dynein might also destabilize microtubule plus ends at the yeast cell cortex as part of the spindle-positioning mechanism. We tested this by comparing plus-end interactions with the cell cortex in WT controls and in mutants that disrupt dynein function. Whereas WT cells exhibit both lateral sliding interactions and plus-end interactions where only the end of the microtubule contacts the cortex, mutants that disrupt dynein only exhibit the latter (Fig. 3 A). Therefore, we compared the frequency and duration of plus-end interactions. In dyn1Δ-null mutants, plus-end interactions are more frequent and dwell longer than in WT controls (Fig. 3, B and C). To determine whether dynein must be attached to the cell cortex to regulate microtubule behavior, we examined num1Δ mutants, which lack dynein’s cortical receptor. In num1Δ mutants, dynein accumulates at the plus end but fails to attach to the cell cortex (Lee et al., 2003). We find that plus ends interact more frequently with the cortex in num1Δ mutants than in WT controls; however, the duration of these interactions is significantly shorter than we observed in dyn1Δ-null mutants (Fig. 3, B and C). We also examined nip100Δ mutants that lack the Nip100/p150glued subunit of the dynactin complex. In contrast to dyn1Δ and num1Δ mutants, nip100Δ mutants exhibit fewer and shorter-lived plus-end interactions with the cortex (Fig. 3, B and C). These results indicate that dynein, but not dynactin, destabilizes microtubules.
Figure 3.
Dynein regulates microtubule dynamics. (A) Time series images of GFP-Tub1 in a dynein-null cell in pre-anaphase to indicate microtubule dwell time at the cell cortex. A time scale (seconds) is shown below each image. Bar, 1 µm. (B) Median frequency of microtubule plus-end cortical interactions per minute. WT, n = 34 cells; dyn1Δ, n = 59; num1Δ, n = 56; nip100Δ, n = 30. Error bars are 95% c.i. (C) Median percent time with the microtubule plus end at the cell cortex. WT, n = 28 cells; dyn1Δ, n = 38; num1Δ, n = 31; nip100Δ, n = 20. Error bars are 95% c.i. (D) Representative images of microtubules labeled with GFP-Tub1 in pre-anaphase cells of the indicated genotypes. Arrowheads point to astral microtubules. Each image is a maximum intensity projection from a confocal Z series. Bars, 1 µm. (E) Median astral microtubule length in pre-anaphase cells at 25 and 37°C. 25°C: WT, n = 341 microtubules; dyn1Δ, n = 278; tub2-430Δ, n = 152. 37°C: tub2-430Δ and td-dyn1Δ, n = 46; dyn1Δ, n = 60; WT, n = 62. Error bars are 95% c.i. aMT, astral microtubule. (F) Median astral microtubule length in galactose. OE-DYN1, n = 343; native, n = 399; dyn1Δ, n = 221. Error bars are 95% c.i. (G) Representative life plots displaying the lengths of single microtubules over time. Arrows point to catastrophe events. (H) Mean dynamicity displayed in tubulin subunits per second. WT, n = 68 growth or shrinkage events; dyn1Δ, n = 47; OE-DYN1, n = 49. Error bars are SEM. Asterisks denote a significant difference from WT. *, P < 0.05; **, P < 0.005; ***, P < 0.0006; ****, P < 0.0001; determined by t test.
If dynein destabilizes microtubules, we predicted that altering dynein levels in the cell would lead to changes in microtubule length and dynamics. Indeed, dyn1Δ mutants show significantly longer astral microtubules, compared with WT controls (dyn1Δ = 1.3 ± 0.18 µm and WT = 0.8 ± 0.05 µm; median ± 95% confidence interval [c.i.]; P < 0.0001; Fig. 3, D and E; and Fig. S3 A). This phenotype was not specific to pre-anaphase cells; dyn1Δ mutants also exhibit longer astral microtubules during G1 (Fig. S3 C). To further confirm that loss of dynein increases astral microtubule length, we tested for an additive effect by combining the dyn1Δ mutation with the tub2-430Δ mutation. This combination is known to be lethal (Aiken et al., 2014); therefore, we examined strains that express tub2-430Δ combined with a conditional ts-degron allele of dynein heavy chain (td-dyn1). As expected, after dynein depletion, these cells exhibit astral microtubules that are significantly longer than either the dyn1Δ or tub2-430Δ mutants alone (tub2-430Δ td-dyn1 = 3 ± 0.48 µm; tub2-430Δ = 1.7 ± 0.19 µm; median ± 95% c.i.; P < 0.0001; Fig. 3, D and E; and Fig. S3 A). Therefore, loss of dynein heavy chain leads to longer astral microtubules.
To test whether elevating levels of dynein heavy chain would have the opposite effect and lead to shorter astral microtubules, we integrated a galactose-inducible promoter at the 5′ end of the DYN1 coding sequence, replacing the endogenous promoter. After 4-h induction with galactose, we observed significantly shorter astral microtubules in both pre-anaphase and G1 cells, compared with WT (Fig. 3, D and F; and Fig. S3, A and B).
To identify the changes in microtubule dynamics that lead to length differences in dynein mutants, we acquired time-lapse z-series images of pre-anaphase cells and measured astral microtubule lengths over time. Example life plots of individual astral microtubules are shown in Fig. 3 G. From these data, we determined mean rates of polymerization and depolymerization and the frequencies of catastrophe and rescue events (Table 1). We find that frequency of catastrophe events is decreased in mutants lacking dynein heavy chain, compared with WT controls (dyn1Δ = 0.9 ± 0.1 events/growth min and WT = 1.3 ± 0.16 events/growth min; P < 0.05; Fig. S3 E and Table 1). In addition, dyn1Δ mutants exhibited slower rates of both polymerization and depolymerization than WT controls (Table 1 and Fig. S3). To determine how changes in multiple parameters of microtubule dynamics might give rise to changes in microtubule length, we used our measurements to estimate the turnover of tubulin subunits at the dynamic plus ends, which can be defined by the dynamicity term (Toso et al., 1993). Astral microtubules in dyn1Δ mutants exhibit significantly lower dynamicity than WT controls, indicating that dynein heavy chain is necessary for the normal turnover of tubulin heterodimers at plus ends (Fig. 3 H and Fig. S3 B; P < 0.005). Cells overexpressing dynein heavy chain show very brief phases of polymerization, more frequent catastrophes, and faster rates of depolymerization (Fig. 3, G and H; Fig. S3; and Table 1). Together, these data suggest that dynein destabilizes astral microtubules in living cells.
Table 1. Microtubule dynamics measurements.
| Strain | Dynamicity | Polym rate | Polym duration | Depolym rate | Depolym duration | Rescue frequency | Catastrophe frequency |
|---|---|---|---|---|---|---|---|
| subunits/s | µm/min | min | µm/min | min | events/min | events/min | |
| WT (n = 68) | 62 ± 3 | 2.8 ± 0.2 | 34 ± 2 | 3.2 ± 0.2 | 26 ± 1 | 1.4 ± 0.2 | 1.3 ± 0.2 |
| Dyn1-3GFP (n = 22) | 68 ± 4 | 3.0 ± 0.3 | 26 ± 2 | 3.2 ± 0.2 | 20 ± 1 | 1.5 ± 0.4 | 1.1 ± 0.2 |
| dyn1Δ (n = 47) | 49 ± 2 | 1.8 ± 0.1 | 51 ± 3 | 2.5 ± 0.1 | 34 ± 1 | 1.1 ± 0.2 | 0.9 ± 0.1 |
| OE-DYN1 (n = 48) | 52 ± 3 | 2.2 ± 0.2 | 24 ± 3 | 1.9 ± 0.1 | 30 ± 4 | 1.8 ± 0.6 | 2.4 ± 0.5 |
| dyn1ΔMTBD (n = 31) | 56 ± 6 | 1.9 ± 0.1 | 44 ± 2 | 2.5 ± 0.1 | 32 ± 2 | 1.0 ± 0.1 | 0.7 ± 0.1 |
| dyn1-WA1 (n = 52) | 44 ± 2 | 2.0 ± 0.2 | 42 ± 4 | 2.3 ± 0.2 | 39 ± 3 | 1.3 ± 0.2 | 1.4 ± 0.2 |
| dyn1-WA3 (n = 87) | 40 ± 2 | 1.7 ± 0.1 | 51 ± 2 | 2.2 ± 0.1 | 41 ± 3 | 1.3 ± 0.2 | 1.0 ± 0.2 |
| pac1Δ (n = 60) | 48 ± 2 | 1.5 ± 0.1 | 52 ± 2 | 2.4 ± 0.1 | 32 ± 1 | 1.1 ± 0.2 | 0.8 ± 0.1 |
| num1Δ (n = 76) | 60 ± 4 | 1.8 ± 0.1 | 41 ± 2 | 2.3 ± 0.1 | 31 ± 2 | 1.2 ± 0.2 | 1.1 ± 0.2 |
| nip100Δ (n = 23) | 54 ± 4 | 1.7 ± 0.1 | 50 ± 3 | 2.6 ± 0.2 | 41 ± 3 | 0.8 ± 0.1 | 0.6 ± 0.1 |
| tub2-430Δ (n = 55) | 47 ± 3 | 1.8 ± 0.2 | 61 ± 5 | 2.5 ± 0.2 | 44 ± 4 | 0.9 ± 0.2 | 0.7 ± 0.1 |
Sample size (n) indicates the number of GFP-Tub1–labeled astral microtubules analyzed for each genotype. Values are mean ± SEM. Boldface indicates statistical significance (P < 0.05) compared with WT, determined by t test. Bold italics indicates statistical significance (P < 0.01) compared with WT. Depolym, depolymerization; Polym, polymerization.
Dynein motor activity destabilizes microtubules
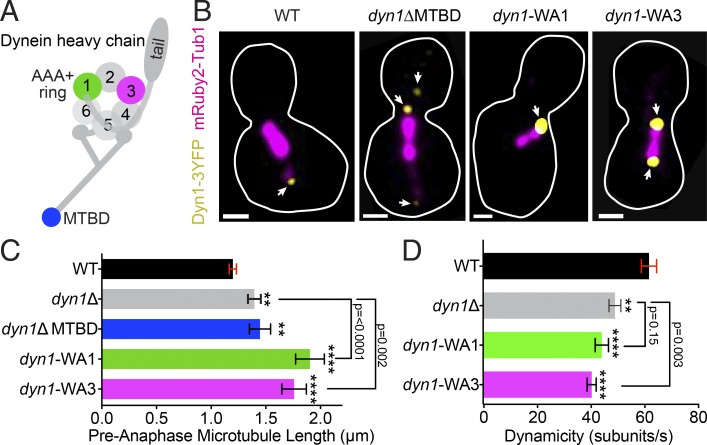
To investigate the mechanism through which dynein destabilizes microtubules, we used a series of mutants that ablate different domains of the heavy chain (Fig. 4 A). First, we asked whether microtubule-binding activity is required by examining a truncation allele of DYN1 that lacks the microtubule-binding domain (MTBD; residues 3,102–3,225; Lammers and Markus, 2015). The truncated protein still localizes to plus ends, similar to WT dynein heavy chain (Fig. 4 B; Lammers and Markus, 2015). Cells expressing the MTBD truncation exhibit longer astral microtubules and changes in dynamics that are equivalent to those seen in dyn1Δ-null mutants, indicating that dynein requires the MTBD to destabilize microtubules (dyn1Δ MTBD = 1.1 ± 0.2 µm; median ± 95% c.i.; P = 0.6 compared with dyn1Δ; Fig. 4 C, Fig. S3 A, and Table 1).
Figure 4.
Dynein requires a functional motor to alter microtubule dynamics. (A) Diagram of dynein heavy chain, including the N-terminal tail domain, ATPase ring, and MTBD. Colored regions are tested in our analysis. (B) Representative images of cells expressing Dyn1-3GFP (yellow) and mRuby2-Tub1 (magenta). Each image is a maximum intensity projection from a confocal Z series. Arrowheads point to the different locations of dynein foci. Bars, 1 µm. (C) Median astral microtubule length in pre-anaphase cells. WT, n = 594 microtubules; dyn1Δ, n = 123; dyn1Δ-MTBD, n = 158 microtubules; dyn1-WA1, n = 175; dyn1-WA3, n = 184. Error bars are 95% c.i. (D) Mean dynamicity displayed in tubulin subunits per second. Data for WT and dyn1Δ from Fig. 2 E are included for reference. dyn1-WA1, n = 52 growth or shrinkage events; dyn1-WA3, n = 87. Error bars are SEM. Asterisks denote a significant difference from WT. **, P < 0.005; ****, P < 0.0001; determined by t test.
To determine how microtubule destabilization is related to dynein’s motor activity, we examined the requirement for ATP binding at AAA1 (ATPase associated with various cellular activities 1) and AAA3, which control microtubule binding and are required for motility (Kon et al., 2004; DeWitt et al., 2015). We tested Walker A (WA) mutants in AAA1 or AAA3 at the endogenous DYN1 locus, referred to as dyn1-WA1 and dyn1-WA3, respectively. The dyn1-WA1 mutation increases dynein accumulation at plus ends, along the lengths of astral microtubules, and near the SPBs (Fig. 4 B; Markus et al., 2009). In contrast, the dyn1-WA3 mutation did not strongly alter dynein localization (Fig. 4 B). Astral microtubules in both dyn1-WA1 and dyn1-WA3 mutants are much longer than in WT controls (dyn1-WA1 = 1.3 ± 0.3 µm and dyn1-WA3 = 1.3 ± 0.2 µm; median ± 95% c.i.; Fig. 4 C and Fig. S3 A; P < 0.0001 compared with WT) and exhibit decreased dynamicity in our time-lapse analysis (dyn1-WA1 = 44.2 ± 2.5 subunits/s and dyn1-WA3 = 40.5 ± 2.5 subunits/s; Fig. 4 D; P < 0.0001 compared with WT). We conclude that the ATP-binding activities of AAA1 and AAA3 that are required for dynein motility are also required for destabilizing microtubules. Interestingly, we find that astral microtubules in dyn1-WA1 mutants are significantly longer than those observed in dyn1Δ-null mutants (Fig. 4 C), and both the dyn1-WA1 and dyn1-WA3 mutants have significantly altered dynamics compared with dyn1Δ (Fig. 4 D and Table 1). This raises the possibility that blocking ATP binding at AAA1 and AAA3 may cause a gain of function that stabilizes microtubules.
Dynactin stabilizes astral microtubules
Our results in Fig. 3 suggest that dynactin might have a role in stabilizing astral microtubules. In support of this notion, we find that astral microtubules are significantly shorter in nip100Δ mutants, which lack the Nip100/p150glued subunit of dynactin (Fig. 5, A and B; and Fig. S3 A). nip100Δ mutants also exhibit a unique phenotype in our dynamics assay: a significantly decreased frequency of rescue events (Table 1 and Fig. S3 D). This difference in the phenotypes of nip100Δ mutants and dyn1Δ mutants is specific to pre-anaphase cells. During anaphase, nip100Δ mutants and dyn1Δ mutants exhibit similar phenotypes: spindles that are mispositioned in the mother compartment and extending long astral microtubules into the bud (Moore et al., 2009). Our result suggests that Nip100/p150glued and dynein heavy chain have opposite effects on microtubule stability during pre-anaphase.
Figure 5.
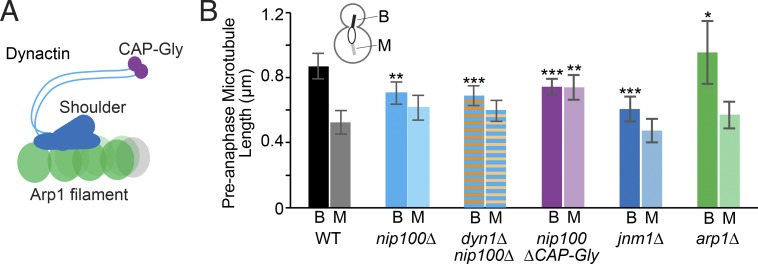
Components of the dynactin complex stabilize microtubules. (A) Diagram of the dynactin complex including the Arp1 filament and shoulder and arm domains to include Jnm1 and Nip100. Colored regions are tested in our analysis. (B) Median microtubule length measured in pre-anaphase cells (μm). Diagram shows cellular location for bud (B)-directed microtubules and mother (M)-directed microtubules. WT, n = 166 cells; nip100Δ, n = 192 cells; dyn1Δ nip100Δ, n = 152 cells; nip100ΔCAP-Gly, n = 313 cells; jnm1Δ, n = 174 cells; arp1Δ, n = 128 cells. Error bars are 95% c.i. Asterisks denote a significant difference from WT. *, P < 0.05; **, P < 0.005; ***, P < 0.0001; determined by t test.
Because dynactin is known to regulate dynein activity, we reasoned that nip100Δ mutants might exhibit shorter microtubules because of changes in dynein regulation. Alternatively, dyn1Δ mutants might exhibit longer microtubules because of changes in dynactin regulation. To distinguish between these possibilities, we examined dyn1Δ nip100Δ double mutants and found that astral microtubules are short, similar to the nip100Δ single mutants (Fig. 5 B and Fig. S3 A). Therefore, the long microtubule phenotype seen in dyn1Δ single mutants depends on dynactin, but the short microtubule phenotype in nip100Δ does not depend on dynein.
Then, we used a series of dynactin mutants to define the components of the dynactin complex that stabilize microtubules. Our results show that the shoulder subcomplex of dynactin, and specifically the cytoskeletal-associated protein glycine-rich (CAP-Gly) domain of Nip100/p150glued, is required to stabilize microtubules (Fig. 5, A and B). In contrast, the Arp1 filament is not required. In fact, we find that arp1Δ-null mutants have significantly longer astral microtubules, reminiscent of dynein-null mutants (Fig. 5, A and B).
Changes in dynein and dynactin localization during microtubule sliding
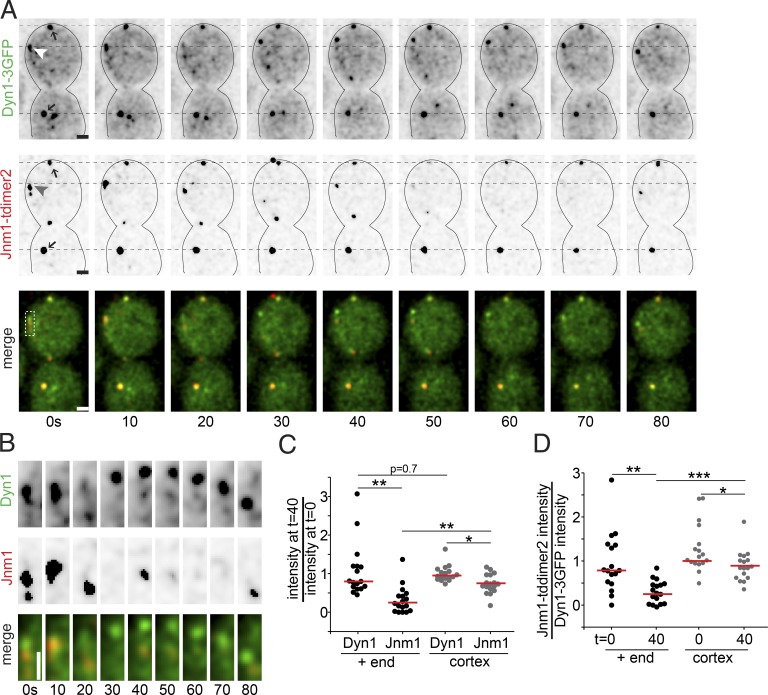
To reconcile the different effects of dynein and dynactin on microtubule stability, we sought to identify differences in localization of either complex at microtubule plus ends. For these experiments, we used a strain that simultaneously expresses native dynein heavy chain tagged with three copies of GFP and native Jnm1/p50 tagged with tdimer2. This allowed us to measure changes in the amounts of either protein at the same plus ends during 17 microtubule sliding events (Fig. 6, A and B, arrowheads). In the same cells, we also measured stationary cortical foci that contain both dynein and dynactin. These foci are stable over time and are therefore useful for determining the degree of signal loss caused by photobleaching (Fig. 6 A, arrows). We find that the majority of dynactin is lost from the plus end during the first 40 s of sliding (77 ± 17% loss; median ± 95% c.i.; Fig. 6 C). In contrast, the majority of dynein remains at the plus end over the same time period (20 ± 32% loss; median ± 95% c.i.; Fig. 6 C). Comparing changes in the ratio of GFP and tdimer2 signal intensities also suggests that the balance of dynein and dynactin at the plus end shifts over the course of a sliding event (Fig. 6 D).
Figure 6.
Dynein and dynactin localization during sliding events. (A) Time lapse images from a WT cell expressing Dyn1-3GFP and Jnm1-tdimer2 during a sliding event. Each image is a maximum intensity projection from a confocal Z series. Merged images show Dyn1-3GFP in green and Jnm1-tdimer2 in red. Arrowheads point to the microtubule plus end, and arrows point to stationary cortical foci. Dashed black lines are fiducial marks for assessing movement. Bars, 1 µm. (B) Localization of Dyn1-3GFP and Jnm1-tdimer2 at the plus end during the sliding event, from the boxed region in A. Bar, 1 µm. (C) Change in Dyn1-3GFP and Jnm1-tdimer2 signal during sliding events. Each dot represents the ratio of fluorescence intensity at t = 40 s of the sliding event divided by the intensity at t = 0 s. Black dots are measurements from the plus ends, and gray dots are measurements at stationary cortical foci. Red bars are the median. (D) Change in the ratio of Jnm1-tdimer2 to Dyn1-3GFP signal during sliding events. Each dot represents the fluorescence intensity of Jnm1-tdimer2 divided by the fluorescence intensity of Dyn1-3GFP at the same region. Black dots are measurements from the plus ends, and gray dots are measurements at stationary cortical foci. Red bars are the median. Cells were HU arrested, and 17 sliding events were analyzed. Asterisks denote a significant difference. *, P < 0.05; **, P < 0.005; ***, P < 0.0001; determined by t test.
In separate experiments, we measured dynein and Nip100/p150glued tagged with the same 3GFP fluor, but in different cells, to estimate the changes in stoichiometry at the plus end (Fig. S4). Our results show that, at the beginning of a sliding event, the fluorescence intensity of native Nip100/p150glued tagged with 3GFP is, on average, approximately half of that measured for dynein heavy chain (Fig. S4 G). This is similar to previous stoichiometry estimates of a 1:3 ratio of dynactin to dynein (Markus et al., 2011). By the end of the sliding event, the amount of plus-end Nip100 decreases by ∼58 ± 9% (median ± 95% c.i.; Fig. S4 H). In contrast, dynein heavy chain exhibits a smaller loss during sliding (15 ± 33%; median ± 95% c.i.) and was similar to that measured for Pac1/LIS1 (Fig. S4 H). Together, these results indicate that both dynactin and dynein are accumulated at the plus end when they reach the cortex, but during sliding, dynactin is depleted, and the balance of dynactin and dynein shifts.
Discussion
Dynein is necessary for spindle movement in many cellular contexts, but the mechanisms that regulate dynein to control the accuracy of spindle positioning are poorly defined. Our study provides three new insights into how dynein is regulated in budding yeast to accurately position the spindle at the nascent site of cytokinesis. First, we show that microtubule stability is a limiting factor of the initiation and duration of dynein-dependent spindle movement. Second, we provide the first evidence that dynein combines both lateral sliding and plus-end destabilization activities to position the spindle, and both require the motor activity of dynein heavy chain. Third, we show that dynactin plays a key role in stabilizing microtubules. We propose that the dynactin complex regulates the balance of dynein’s activities by stabilizing microtubules, thereby promoting microtubule–cortex interactions and transitioning those interactions into force production for spindle movement.
Dynein motor activity destabilizes microtubules
A previous study showed that yeast dynein heavy chain attached to a fabricated barrier can capture and destabilize microtubule plus ends in vitro (Laan et al., 2012). Comparing this study to our results in yeast points to interesting similarities as well as differences. In both scenarios, microtubules that interact with dynein at the cortex exhibit dramatic increases in the frequency of catastrophes (Fig. 1 F, and Fig. 3, A–C; Laan et al., 2012). Furthermore, in both scenarios, dynein’s catalytic activity is necessary to destabilize microtubules (Fig. 4; Laan et al., 2012). How dynein promotes catastrophe is an outstanding question. Laan et al. (2012) attributed the increased catastrophe frequency to dynein holding the plus end against the cortical barrier and preventing access to free tubulin subunits. Although a similar occlusion mechanism may contribute to catastrophe in yeast, this model is not sufficient to explain our results. First, our analysis comparing plus-end interactions at the cortex in num1Δ and dyn1Δ mutants indicates that dynein does not need to attach to the cortex to destabilize microtubule ends. Both mutants exhibit more frequent plus-end interactions at the cortex; however, these interactions are shorter lived in the num1Δ mutant than in the dyn1Δ mutant (Fig. 3, B and C). This suggests that dynein at the plus end in the num1Δ mutants may still destabilize the microtubules, albeit to a lesser degree than normal. Second, sliding microtubules exhibit more frequent catastrophes, even though the plus ends are presumably parallel to the cell cortex as they move along it (Fig. 1). Therefore, free tubulin subunits would be expected to have better access to the plus end of a sliding microtubule, and a catastrophe by a subunit occlusion model seems unlikely. We therefore favor an alternative model, where dynein motor activity promotes catastrophe by directly destabilizing the microtubule lattice.
Although dynein destabilizes microtubules in yeast, it is unlikely that this contributes force for spindle movement. We find that dynein-dependent spindle movement is initiated by polymerizing astral microtubules. In the majority of cases, these microtubules continue to polymerize as they slide along the cortex and move the spindle and remain in a polymerizing state for a mean of 12 s before undergoing catastrophe and switching to depolymerization (Fig. 1 E and unpublished data). This indicates that polymerizing microtubules support spindle movement. In addition, we find that increasing microtubule stability with mutants or drugs that inhibit depolymerization increases both the frequency and duration of spindle movement. These results are reminiscent of previous findings in C. elegans, where loss of the cortical protein EFA-6 increases the stability of astral microtubules and leads to increased cortical pulling forces by dynein, and studies of nuclear movements in Aspergillus mutants that alter microtubule dynamics (Grava and Philippsen, 2010; O’Rourke et al., 2010; Gibeaux et al., 2017). A simple interpretation of these results is that microtubule depolymerization is not required to move the spindle. This argues that microtubule depolymerization does not provide the energy needed for spindle movement, as has been proposed for kinetochore movement within the spindle (Asbury et al., 2006; Welburn et al., 2009). Instead, our results argue that stable microtubules promote spindle movement by enhancing lateral microtubule sliding by dynein. We speculate that stable microtubules could enhance sliding either by providing more binding sites for dynein to move along the lateral sides of the microtubule and/or by enabling the microtubule to withstand stresses at the cell cortex, e.g., bending and force from dynein motility.
Dynactin stabilizes microtubules
Dynactin is required for virtually all known cellular functions of cytoplasmic dynein and is regarded as an obligate positive regulator. Typically, null mutations in dynactin subunits elicit phenotypes that are equivalent to null mutations in dynein (Schroer, 2004). However, we find that dynein and dynactin have opposite effects on microtubule stability. In contrast to heavy chain mutants, null mutations in the Nip100/p150glued subunit of dynactin diminish the frequency of microtubule–cortex interactions and lead to abnormally short astral microtubules (Fig. 3, B and C; and Fig. 5 B). Importantly, double mutants combining null alleles of dynein heavy chain and NIP100 also exhibit shorter astral microtubules (Fig. 5 B). Therefore, NIP100 is epistatic to dynein heavy chain with regard to astral microtubule stability, and functional Nip100 must be required to generate the longer astral microtubules found in the single dynein heavy chain null (Fig. 3). These results support a model in which dynactin has a proximal role in stabilizing astral microtubules.
Our results indicate that astral microtubules are stabilized by the shoulder subcomplex of dynactin. The shoulder was first identified in electron microscopy analysis of purified vertebrate dynactin, where it lays across a short filament of the actin-related protein Arp1 (Schafer et al., 1994; Chowdhury et al., 2015; Urnavicius et al., 2015). The shoulder contains the subunits Nip100/p150glued, Jnm1/p50, and Ldb18/p24 (Schafer et al., 1994). The Nip100/p150glued subunit contains an N-terminal CAP-Gly domain, which binds to microtubules and suppresses catastrophes in vitro (Fig. 5 A; Waterman-Storer et al., 1995; Honnappa et al., 2006; Lazarus et al., 2013). We find that mutants lacking the CAP-Gly domain exhibit shorter microtubules, similar to the nip100Δ null (Fig. 5 B). Furthermore, mutants lacking the Jnm1/p50 subunit exhibit the same phenotype (Fig. 5 B). Interestingly, mutants lacking the Arp1 filament exhibit longer microtubules, reminiscent of dynein-null mutants (Fig. 5 B). This indicates that the shoulder stabilizes microtubules through a mechanism that involves the CAP-Gly domain of Nip100/p150glued but does not require the Arp1 filament. Although the shoulder is known to form a stable complex on its own (Moore et al., 2008), to our knowledge this is this first evidence of an independent cellular function for the shoulder.
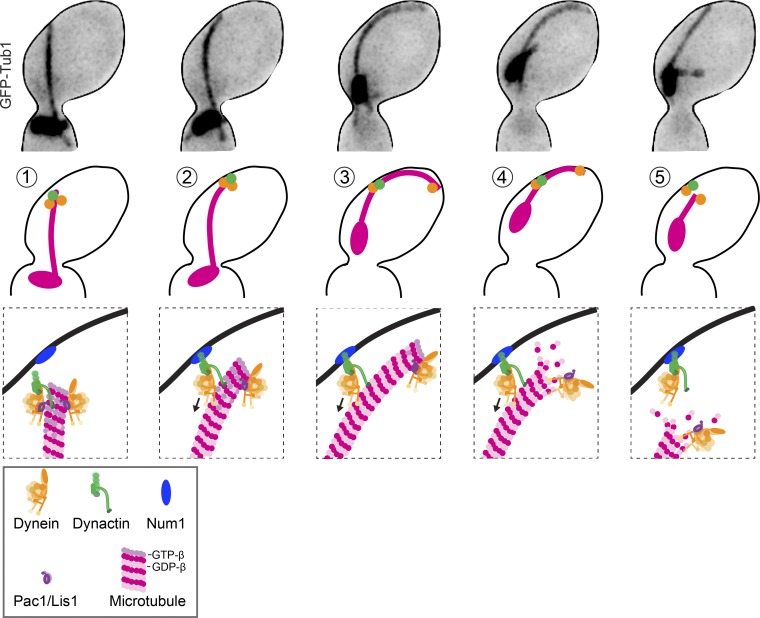
We propose that a balance of dynactin and dynein activities controls the initiation and magnitude of spindle movement. As depicted by our model in Fig. 7, dynein and dynactin are first recruited to the plus ends of astral microtubules, with an approximate stoichiometry of two dynein motors for every dynactin complex (Fig. 7, step 1; Markus et al., 2011). Dynactin stabilizes the plus end, promoting microtubule polymerization that eventually delivers dynein–dynactin to the cortex. At the cortex, 1:1 complexes of dynein/dynactin offload by binding to Num1 and begin to move along the microtubule toward the minus end (Fig. 7, steps 2 and 3). In this way, dynein motility depletes dynactin’s stabilizing activity from the plus end. Because the plus end begins with a two-times excess of dynein to dynactin, offloading equal amounts of dynein and dynactin causes an increase in the relative amount of dynein to dynactin that remains at the plus end. This shifts the balance toward the destabilizing activity of dynein and leads to catastrophe (Fig. 7, step 4). Then, the microtubule depolymerizes at a rate of 2.3 µm/min, which is 1.4× faster than dynein motility (unpublished data). This faster rate may allow depolymerization to catch up with dynein–dynactin along the microtubule lattice and terminate spindle movement by destroying its microtubule substrate (Fig. 7, step 5). In summary, our model proposes that the activation of dynein motility creates a negative feedback that leads to destruction of its microtubule track by titrating dynactin away from the plus end.
Figure 7.
Model for regulating dynein-dependent spindle movement. Images are from a cell expressing GFP-tubulin. Cartoons depict the predicted localization of dynein and its regulators during (1) the arrival of the plus end and the cortex, (2 and 3) the initiation of lateral microtubule sliding and spindle movement, (4) the switch to depolymerization during sliding, and (5) termination of sliding by loss of microtubule binding.
Materials and methods
Chemicals and reagents were from Thermo Fisher Scientific and Sigma-Aldrich, unless stated otherwise.
Yeast strains and manipulation
General yeast manipulation, media, and transformation were performed by standard methods (Amberg et al., 2005). A detailed list of strains is provided in Table S1. 3GFP fusions to Dyn1, Nip100, and Pac1 were generated using plasmids that integrate at the native locus (Lee et al., 2003; Moore et al., 2008). Jnm1-tdimer2 was generated by PCR-mediated tagging at the native locus (Sheff and Thorn, 2004). Strains expressing dynein WA mutants at endogenous levels were provided by S. Markus (Colorado State University, Fort Collins, CO) and were described previously (Markus et al., 2009, 2011). Deletion mutants were generated by conventional PCR-mediated methods (Petracek and Longtine, 2002). GFP-Tub1 and mRuby-Tub1 fusions were integrated and expressed ectopically so that the fusion proteins were expressed in addition to the native TUB1 (Song and Lee, 2001; Markus et al., 2015). For overexpression of dynein heavy chain, the GAL1/10 promoter was integrated 5′ of the DYN1 coding sequence, removing 160 bases of the native promoter, and a 3GFP tag was fused to the 3′ end of the DYN1 coding sequence using an integrating plasmid (Lee et al., 2003). Overexpression was achieved by first growing cells to early log-phase in media containing 2% sucrose and then adding galactose to 2% and returning to a 30°C shaking incubator for 4 h. Overexpression was confirmed by assessing the localization of the Dyn1-3GFP.
Microscopy and image analysis
Images were collected on a microscope (Ti-E; Nikon) equipped with a 1.45 NA 100× CFI Plan Apo objective, piezo electric stage (Physik Instrumente), spinning-disk confocal scanner unit (CSU10; Yokogawa Electric Corporation), 488- and 561-nm lasers (Agilent Technologies), and an electron-multiplying charge-coupled device camera (iXon Ultra 897; Andor Technology) using NIS Elements software (Nikon). Cells were grown asynchronously to early log-phase in nonfluorescent media and adhered to slide chambers coated with concanavalin A. Slide chambers were sealed with VALAP (vaseline, lanolin, and paraffin at 1:1:1). During acquisition, the temperature of the stage was 25°C except when imaging td-dyn1, during which the stage was heated to 37°C. Z series consisted of 13 images separated by 500 nm.
Analysis of microtubule–cortex interactions and microtubule sliding
Cells expressing an integrated fusion of GFP to the N terminus of α-tubulin (GFP-Tub1) were arrested with HU for 2 h and imaged for 10 min at a 4-s time interval between frames. Microtubule–cortex interactions were identified in 2D projections of Z-series images by the microtubule reaching the cell cortex, which was visualized by the signal from free GFP-tubulin subunits in the cytoplasm. Only interactions where the microtubule reached the edge of the 2D-projected cell cortex were scored. Therefore, our measurements likely underrepresent the total interactions occurring in 3D space. The dwell time of each interaction was defined as time elapsed between the first and last frame of the interaction. Microtubule sliding events were defined as interactions of microtubule plus ends with the cell cortex that transitioned into lateral microtubule movements along the cortex and caused the spindle to move. Termination of sliding was defined by the end of directed spindle movement and the movement of the microtubule away from the cell cortex.
Microtubule dynamics analysis
Microtubule dynamics were analyzed by measuring the lengths of astral microtubules labeled with GFP-Tub1 at 4- or 5-s intervals for 10 min. This analysis was conducted in pre-anaphase cells, which typically exhibit one or two astral microtubules emanating from each SPB. Assembly and disassembly events were defined as at least three contiguous data points that produced a length change ≥0.5 µm with a coefficient of determination ≥0.8. Data points that did not meet the criteria for assembly or disassembly were regarded as pauses. Catastrophes were defined as transitions to disassembly. Catastrophe frequencies were determined for individual astral microtubules by dividing the number of catastrophe events by the total lifetime, minus time spent in disassembly. Rescues were defined as transitions to assembly. Rescue frequencies were determined for individual astral microtubules by dividing the number of rescue events by the total lifetime, minus time spent in assembly. Microtubule dynamicity was calculated by the total change in length (growing and shrinking) divided by the change in time and expressed in tubulin subunits changed per second (Toso et al., 1993). At least 23 astral microtubules were analyzed for each genotype. Dynamics measurements for individual microtubules were pooled for the genotype and then compared with pooled data for different genotypes using a Student’s t test to assess whether the mean values for different datasets were significantly different.
Localization of dynein, dynactin, and Pac1
Two-color images of cells expressing Dyn1-3GFP and Jnm1-tdimer2 were collected in time-lapse, full-cell Z series separated by 300 nm. Then, images were analyzed using ImageJ (National Institutes of Health). Z series were collapsed into maximum intensity projections. We identified the astral microtubule plus end by observing changes in Dyn1-3GFP localization over time, as Dyn1 is known to localize to the plus ends of polymerizing and depolymerizing microtubules. Cortical foci were stationary in time-lapse imaging. Intensities of 3GFP or tdimer2 signals were measured within a 64-pixel region (472 × 472 nm). These values were adjusted for background signal by taking a fluorescence intensity measurement of the same size directly adjacent to the plus end of the microtubule and then subtracting that value from the value measured at the plus end. To determine the change in localization over the course of a microtubule–cortex interaction, we calculated the ratio of fluorescence intensity at the fifth time point of the interaction over the intensity at the first time point of interaction. Two-color images of cells expressing Dyn1-3GFP, Pac1-3GFP, or Nip100-3GFP with mRuby2-Tub1 were collected and analyzed similarly, with the exception that the astral microtubule plus end was identified using the mRuby2-Tub1 signal.
Spindle location analysis
Asynchronous cells expressing GFP-Tub1 were imaged with 13 Z slices separated by a 400-nm depth. Then, cells were sorted by pre-anaphase spindle or anaphase at the bud neck, in the mother cell or in the bud. A pre-anaphase spindle is defined as a bipolar spindle of <1.9 µm in length. An anaphase spindle is defined as a bipolar spindle ≥1.9 µm in length. Spindles were characterized as at the bud neck when there was at least one SPB in the neck region (between the mother and daughter cell) or spanning the bud neck region. Spindles were characterized as in the mother when both SPBs were in the half of the mother cytoplasm that is distal to the bud neck. Spindles were characterized as in the bud when both SPBs were across the bud neck and in the bud.
Drug treatment
Cells expressing GFP-labeled microtubules were grown to early log-phase in synthetic media at 30°C and then arrested in S phase by the addition of 200 mM HU. After 90 min in HU, 90% of cells contained short bipolar spindles. For treatment with epothilone, cells contained a pdr1-DBD-CYC8 mutation and GFP-labeled microtubules. The pdr1-DBD-CYC8 allele represses transcription of pleiotropic drug resistance genes and was generated at the native chromosomal locus using the pAGS1 integrating plasmid, provided by J. Nitiss (University of Illinois at Chicago College of Pharmacy, Rockford, IL; Stepanov et al., 2008). Cells were first arrested with HU for 90 min and then were treated with 10 µM epothilone A for 1 h before imaging.
Online supplemental material
Fig. S1 depicts measurements of microtubule dynamics during dynein-dependent sliding along the cell cortex. Fig. S2 shows that stabilizing microtubules with tub2-430Δ and kip3Δ mutants increases cortical contacts, sliding frequency, and sliding duration. Fig. S3 shows measurements of microtubule dynamics for all strains listed in Table 1. Fig. S4 shows that Nip100 labeled with 3GFP is depleted from the ends of sliding microtubules, whereas dynein labeled with 3GFP remains at the plus end. Video 1 shows microtubule sliding and spindle movement of a WT cell expressing α-tubulin labeled with GFP. Video 2 shows a dyn1Δ-mutant cell expressing α-tubulin labeled with GFP. Video 3 shows a tub2-430Δ-mutant cell expressing α-tubulin labeled with GFP. Video 4 shows a kip3Δ-mutant cell expressing α-tubulin labeled with GFP. Table S1 is available as an Excel file and shows the strains used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Steven Markus for providing multiple dynein mutant strains and sharing valuable advice, Dr. John Nitiss for providing the integrating plasmid for generating the pdr1-DBD-CYC8 allele, and Gabriella Li for sharing preliminary results.
This work was supported by the National Institutes of Health grant R01 GM112893 to J.K. Moore. C. Estrem was supported by the University of Colorado Anschutz Medical Campus (Pre-doctoral Training Program in Molecular Biology grant NIH-T32-GM008730 and a Butcher Innovation Award provided by Chancellor Elliman).
The authors declare no competing financial interests.
Author contributions: J.K. Moore and C. Estrem conceived and designed experiments. J.K. Moore and C. Estrem constructed strains and performed image acquisition and data analysis. C.P. Fees wrote custom Matlab code for analyzing microtubules dynamics and provided data analysis advice. J.K. Moore and C. Estrem prepared the figures and wrote the paper.
Footnotes
Abbreviations used:
- AAA
- ATPase associated with various cellular activities
- CAP-Gly
- cytoskeletal-associated protein glycine-rich
- c.i.
- confidence interval
- HU
- hydroxyurea
- MTBD
- microtubule-binding domain
- SPB
- spindle pole body
References
- Adames N.R., and Cooper J.A.. 2000. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149:863–874. 10.1083/jcb.149.4.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken J., Sept D., Costanzo M., Boone C., Cooper J.A., and Moore J.K.. 2014. Genome-wide analysis reveals novel and discrete functions for tubulin carboxy-terminal tails. Curr. Biol. 24:1295–1303. 10.1016/j.cub.2014.03.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D.C., Burke D.J., and Strathern J.N.. 2005. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 230 pp. [Google Scholar]
- Asbury C.L., Gestaut D.R., Powers A.F., Franck A.D., and Davis T.N.. 2006. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA. 103:9873–9878. 10.1073/pnas.0602249103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayloo S., Lazarus J.E., Dodda A., Tokito M., Ostap E.M., and Holzbaur E.L.. 2014. Dynactin functions as both a dynamic tether and brake during dynein-driven motility. Nat. Commun. 5:4807 10.1038/ncomms5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode C.J., Gupta M.L.J. Jr., Reiff E.A., Suprenant K.A., Georg G.I., and Himes R.H.. 2002. Epothilone and paclitaxel: unexpected differences in promoting the assembly and stabilization of yeast microtubules. Biochemistry. 41:3870–3874. 10.1021/bi0121611 [DOI] [PubMed] [Google Scholar]
- Byers B., and Goetsch L.. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati J.L., and Stearns T.. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629–641. 10.1083/jcb.138.3.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Ketcham S.A., Schroer T.A., and Lander G.C.. 2015. Structural organization of the dynein-dynactin complex bound to microtubules. Nat. Struct. Mol. Biol. 22:345–347. 10.1038/nsmb.2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couwenbergs C., Labbé J.C., Goulding M., Marty T., Bowerman B., and Gotta M.. 2007. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J. Cell Biol. 179:15–22. 10.1083/jcb.200707085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver-Hanlon T.L., Lex S.A., Stephens A.D., Quintyne N.J., and King S.J.. 2006. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat. Cell Biol. 8:264–270. 10.1038/ncb1370 [DOI] [PubMed] [Google Scholar]
- DeWitt M.A., Cypranowska C.A., Cleary F.B., Belyy V., and Yildiz A.. 2015. The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release. Nat. Struct. Mol. Biol. 22:73–80. 10.1038/nsmb.2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaux R., Politi A.Z., Philippsen P., and Nédélec F.. 2017. Mechanism of nuclear movements in a multinucleated cell. Mol. Biol. Cell. 28:645–660. 10.1091/mbc.E16-11-0806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grava S., and Philippsen P.. 2010. Dynamics of multiple nuclei in Ashbya gossypii hyphae depend on the control of cytoplasmic microtubules length by Bik1, Kip2, Kip3, and not on a capture/shrinkage mechanism. Mol. Biol. Cell. 21:3680–3692. 10.1091/mbc.E10-06-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S.W., Gönczy P., Stelzer E.H., and Hyman A.A.. 2001. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 409:630–633. 10.1038/35054572 [DOI] [PubMed] [Google Scholar]
- Gupta M.L.J. Jr., Carvalho P., Roof D.M., and Pellman D.. 2006. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8:913–923. 10.1038/ncb1457 [DOI] [PubMed] [Google Scholar]
- Gusnowski E.M., and Srayko M.. 2011. Visualization of dynein-dependent microtubule gliding at the cell cortex: implications for spindle positioning. J. Cell Biol. 194:377–386. 10.1083/jcb.201103128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Okhrimenko O., Jaussi R., Jawhari H., Jelesarov I., Winkler F.K., and Steinmetz M.O.. 2006. Key interaction modes of dynamic +TIP networks. Mol. Cell. 23:663–671. 10.1016/j.molcel.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Janson M.E., de Dood M.E., and Dogterom M.. 2003. Dynamic instability of microtubules is regulated by force. J. Cell Biol. 161:1029–1034. 10.1083/jcb.200301147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., and Cheeseman I.M.. 2013. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 154:391–402. 10.1016/j.cell.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T., Nishiura M., Ohkura R., Toyoshima Y.Y., and Sutoh K.. 2004. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 43:11266–11274. 10.1021/bi048985a [DOI] [PubMed] [Google Scholar]
- Kozlowski C., Srayko M., and Nedelec F.. 2007. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 129:499–510. 10.1016/j.cell.2007.03.027 [DOI] [PubMed] [Google Scholar]
- Laan L., Pavin N., Husson J., Romet-Lemonne G., van Duijn M., López M.P., Vale R.D., Jülicher F., Reck-Peterson S.L., and Dogterom M.. 2012. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 148:502–514. 10.1016/j.cell.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé J.C., Maddox P.S., Salmon E.D., and Goldstein B.. 2003. PAR proteins regulate microtubule dynamics at the cell cortex in C. elegans. Curr. Biol. 13:707–714. 10.1016/S0960-9822(03)00251-3 [DOI] [PubMed] [Google Scholar]
- Lammers L.G., and Markus S.M.. 2015. The dynein cortical anchor Num1 activates dynein motility by relieving Pac1/LIS1-mediated inhibition. J. Cell Biol. 211:309–322. 10.1083/jcb.201506119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus J.E., Moughamian A.J., Tokito M.K., and Holzbaur E.L.. 2013. Dynactin subunit p150(Glued) is a neuron-specific anti-catastrophe factor. PLoS Biol. 11:e1001611 10.1371/journal.pbio.1001611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., Oberle J.R., and Cooper J.A.. 2003. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 160:355–364. 10.1083/jcb.200209022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., Kaiser M.A., and Cooper J.A.. 2005. The offloading model for dynein function. J. Cell Biol. 168:201–207. 10.1083/jcb.200407036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Bloom K.S., and Salmon E.D.. 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2:36–41. 10.1038/71357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S.M., Punch J.J., and Lee W.L.. 2009. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr. Biol. 19:196–205. 10.1016/j.cub.2008.12.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S.M., Plevock K.M., St Germain B.J., Punch J.J., Meaden C.W., and Lee W.L.. 2011. Quantitative analysis of Pac1/LIS1-mediated dynein targeting: Implications for regulation of dynein activity in budding yeast. Cytoskeleton (Hoboken). 68:157–174. 10.1002/cm.20502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S.M., Omer S., Baranowski K., and Lee W.L.. 2015. Improved plasmids for fluorescent protein tagging of microtubules in Saccharomyces cerevisiae. Traffic. 16:773–786. 10.1111/tra.12276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney R.J., Huynh W., Tanenbaum M.E., Bhabha G., and Vale R.D.. 2014. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 345:337–341. 10.1126/science.1254198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F.J. 2013. Mechanisms of spindle positioning. J. Cell Biol. 200:131–140. 10.1083/jcb.201210007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K., Li J., and Cooper J.A.. 2008. Dynactin function in mitotic spindle positioning. Traffic. 9:510–527. 10.1111/j.1600-0854.2008.00710.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K., Sept D., and Cooper J.A.. 2009. Neurodegeneration mutations in dynactin impair dynein-dependent nuclear migration. Proc. Natl. Acad. Sci. USA. 106:5147–5152. 10.1073/pnas.0810828106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S.M., Christensen S.N., and Bowerman B.. 2010. Caenorhabditis elegans EFA-6 limits microtubule growth at the cell cortex. Nat. Cell Biol. 12:1235–1241. 10.1038/ncb2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecreaux J., Röper J.C., Kruse K., Jülicher F., Hyman A.A., Grill S.W., and Howard J.. 2006. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr. Biol. 16:2111–2122. 10.1016/j.cub.2006.09.030 [DOI] [PubMed] [Google Scholar]
- Petracek M.E., and Longtine M.S.. 2002. PCR-based engineering of yeast genome. Methods Enzymol. 350:445–469. 10.1016/S0076-6879(02)50978-2 [DOI] [PubMed] [Google Scholar]
- Prota A.E., Bargsten K., Zurwerra D., Field J.J., Díaz J.F., Altmann K.H., and Steinmetz M.O.. 2013. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science. 339:587–590. 10.1126/science.1230582 [DOI] [PubMed] [Google Scholar]
- Rappaport R., and Ebstein R.P.. 1965. Duration of stimulus and latent periods preceding furrow formation in sand dollar eggs. J. Exp. Zool. 158:373–382. 10.1002/jez.1401580311 [DOI] [PubMed] [Google Scholar]
- Redemann S., Pecreaux J., Goehring N.W., Khairy K., Stelzer E.H., Hyman A.A., and Howard J.. 2010. Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos. PLoS One. 5:e12301 10.1371/journal.pone.0012301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.A., Gill S.R., Cooper J.A., Heuser J.E., and Schroer T.A.. 1994. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J. Cell Biol. 126:403–412. 10.1083/jcb.126.2.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager M.A., Hoang H.T., Urnavicius L., Bullock S.L., and Carter A.P.. 2014. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 33:1855–1868. 10.15252/embj.201488792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer T.A. 2004. Dynactin. Annu. Rev. Cell Dev. Biol. 20:759–779. 10.1146/annurev.cellbio.20.012103.094623 [DOI] [PubMed] [Google Scholar]
- Sheff M.A., and Thorn K.S.. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Siller K.H., and Doe C.Q.. 2008. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev. Biol. 319:1–9. 10.1016/j.ydbio.2008.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop A.R., and White J.G.. 1998. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr. Biol. 8:1110–1117. 10.1016/S0960-9822(98)70465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., and Lee K.S.. 2001. A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis. J. Cell Biol. 152:451–470. 10.1083/jcb.152.3.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanov A., Nitiss K.C., Neale G., and Nitiss J.L.. 2008. Enhancing drug accumulation in Saccharomyces cerevisiae by repression of pleiotropic drug resistance genes with chimeric transcription repressors. Mol. Pharmacol. 74:423–431. 10.1124/mol.107.044651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton M.D., Siglin A., Li J., Moore J.K., Ahmed S., Williams J.C., and Cooper J.A.. 2011. Functional interaction between dynein light chain and intermediate chain is required for mitotic spindle positioning. Mol. Biol. Cell. 22:2690–2701. 10.1091/mbc.E11-01-0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso R.J., Jordan M.A., Farrell K.W., Matsumoto B., and Wilson L.. 1993. Kinetic stabilization of microtubule dynamic instability in vitro by vinblastine. Biochemistry. 32:1285–1293. 10.1021/bi00056a013 [DOI] [PubMed] [Google Scholar]
- Urnavicius L., Zhang K., Diamant A.G., Motz C., Schlager M.A., Yu M., Patel N.A., Robinson C.V., and Carter A.P.. 2015. The structure of the dynactin complex and its interaction with dynein. Science. 347:1441–1446. 10.1126/science.aaa4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Karki S., and Holzbaur E.L.. 1995. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1). Proc. Natl. Acad. Sci. USA. 92:1634–1638. 10.1073/pnas.92.5.1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P., Grishchuk E.L., Backer C.B., Wilson-Kubalek E.M., Yates J.R. III, and Cheeseman I.M.. 2009. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev. Cell. 16:374–385. 10.1016/j.devcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder J.H., and Han M.. 2001. Cytoplasmic dynein light intermediate chain is required for discrete aspects of mitosis in Caenorhabditis elegans. Mol. Biol. Cell. 12:2921–2933. 10.1091/mbc.12.10.2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.