Monis et al. demonstrate that the BLOC-1 protein complex is required for trafficking polycystin-2, but not fibrocystin or smoothened, from endosomes to primary cilia. This is the first demonstration of a role for BLOC-1 in ciliary assembly and emphasizes the complexity of distinct ciliary membrane protein trafficking routes.
Abstract
Primary cilia perceive the extracellular environment through receptors localized in the ciliary membrane, but mechanisms directing specific proteins to this domain are poorly understood. To address this question, we knocked down proteins potentially important for ciliary membrane targeting and determined how this affects the ciliary trafficking of fibrocystin, polycystin-2, and smoothened. Our analysis showed that fibrocystin and polycystin-2 are dependent on IFT20, GMAP210, and the exocyst complex, while smoothened delivery is largely independent of these components. In addition, we found that polycystin-2, but not smoothened or fibrocystin, requires the biogenesis of lysosome-related organelles complex-1 (BLOC-1) for ciliary delivery. Consistent with the role of BLOC-1 in sorting from the endosome, we find that disrupting the recycling endosome reduces ciliary polycystin-2 and causes its accumulation in the recycling endosome. This is the first demonstration of a role for BLOC-1 in ciliary assembly and highlights the complexity of pathways taken to the cilium.
Introduction
Many vertebrate cells possess a single nonmotile primary cilium that extends from the cell surface. These cilia perceive the extracellular environment by localizing specific receptors to the ciliary membrane. To date, more than 25 different receptors have been found to be ciliary localized (Hilgendorf et al., 2016). These include the important cystoproteins polycystin-1, polycystin-2, and fibrocystin, which are defective in polycystic kidney disease (PKD) and the hedgehog receptors patched-1 and smoothened (Pazour et al., 2002b; Yoder et al., 2002; Corbit et al., 2005; Rohatgi et al., 2007; Harris and Torres, 2009; Follit et al., 2010). Heritable mutations in genes that encode proteins essential for the structure or function of primary cilia cause a broad class of human diseases called the ciliopathies (Sattar and Gleeson, 2011). The ciliopathies include a wide variety of developmental and degenerative diseases that reflect the important and diverse roles cilia play in organ development and tissue homeostasis.
Cilia have no protein synthesis capability, and thus all components are synthesized in the cytoplasm and trafficked into the organelle (Nachury et al., 2010). Nonmembrane proteins are thought to be transported from a pool at the base of the cilium into the cilium by intraflagellar transport (IFT). The IFT system consists of kinesin-2 and dynein-2 motors and a large adaptor complex made up of IFT complex A, IFT complex B, and the BBSome (Rosenbaum and Witman, 2002; Nachury et al., 2007). The involvement of IFT in the trafficking of membrane proteins has not been fully addressed.
The IFT complex B subunit IFT20 is localized at both the primary cilium and the Golgi apparatus, where it is in a complex with the golgin protein GMAP210 (Follit et al., 2006, 2008). Finding IFT20 at the Golgi complex suggested that IFT20 might be involved in trafficking of membrane proteins from the Golgi apparatus to the primary cilium. Complete loss of IFT20 blocked ciliary assembly precluding analysis of membrane protein trafficking to the organelle. However, cells with a partial loss of IFT20 (which could still ciliate) had reduced ciliary polycystin-2, consistent with a role for IFT20 in transport of this membrane protein (Follit et al., 2006). The golgin GMAP210, which anchors IFT20 to the Golgi membrane, is not required for ciliary assembly, but cells lacking it have reduced ciliary polycystin-2, suggesting that the Golgi pool of IFT20 is important for sorting ciliary membrane proteins (Follit et al., 2008). Photoreceptor outer segments, which are cilia, have very high demands for membrane protein transport to maintain the structure. In mouse, it is estimated that ∼4,300 opsin molecules need to be transported per minute into the cilium to maintain the organelle, while as many as 50,000 are needed per minute in fish and frogs (Young, 1967; Besharse and Horst, 1990; Williams, 2002). Loss of IFT20 or other IFT proteins leads to opsin mislocalization and photoreceptor degeneration (Keady et al., 2011; Crouse et al., 2014). Interestingly, acute deletion of IFT20 causes opsin accumulation at the Golgi complex, whereas acute deletion of IFT140 causes opsin accumulation in the inner segment plasma membrane (Keady et al., 2011; Crouse et al., 2014). These data are consistent with a model in which IFT20 is important for sorting or trafficking of membrane proteins from the Golgi apparatus to the base of the cilium, where they engage the rest of the IFT system (Follit et al., 2006).
It is not clear that all membrane proteins are trafficked to the cilium by the same route. Early work on opsin transport in frogs and mastigoneme transport in Ochromonas danica suggested that these proteins traffic in vesicles directly from the Golgi apparatus to the base of the cilium, where the vesicles dock to the plasma membrane just outside of the cilium before the proteins are transported into the organelle (Bouck, 1971; Papermaster et al., 1985; Deretic et al., 1995). More recent work on smoothened transport suggests that this protein is trafficked to the plasma membrane and laterally moves into the cilium (Milenkovic et al., 2009). Agglutinin transport in Chlamydomonas reinhardtii uses a similar mechanism (Hunnicutt et al., 1990; Cao et al., 2015). A third pathway whereby proteins are first transported to the plasma membrane followed by endocytosis and delivery to the base of the cilium by the recycling pathway has been proposed, but no proteins are known to take this route (Weisz and Rodriguez-Boulan, 2009; Nachury et al., 2010).
The finding of opsin transport defects when IFT20 is perturbed suggests a role for IFT20-GMAP210 in the direct trafficking pathway from the Golgi complex to the base of the cilium. However, the role of IFT20 in trafficking additional membrane proteins from the Golgi complex and/or through other organelles has not been tested. To assess their importance in trafficking of ciliary membrane proteins, we use a fluorescence-based pulse-chase assay to measure the dynamics of smoothened, polycystin-2, and fibrocystin delivery to the cilium after perturbing the IFT20-GMAP210 complex. We find that this complex is important for fibrocystin and polycystin-2 trafficking but has a minimal role in smoothened delivery.
To understand how IFT20 functions to traffic membrane proteins to cilia, we examined interactome data (Rual et al., 2005; Wang et al., 2011) and found connections between IFT20 and the exocyst and the biogenesis of lysosome-related organelles complex-1 (BLOC-1). Using our fluorescence-based pulse-chase assay, we find minimal roles for these complexes in smoothened transport, while the exocyst is important for both fibrocystin and polycystin-2 delivery. Interestingly, BLOC-1 is required only for the delivery of polycystin-2. Consistent with this, we find that loss of function BLOC-1 mouse models have a mild cystic kidney phenotype. Given the role of BLOC-1 in recycling endosome trafficking, we used a dominant-negative MyoVb construct to perturb trafficking through the recycling endosome (Lapierre et al., 2001; Volpicelli et al., 2002). The construct blocks polycystin-2 trafficking to cilia but does not affect fibrocystin or smoothened delivery to cilia. Polycystin-2 trafficking is also perturbed by expression of a Rab11a dominant-negative mutation that disrupts the recycling endosome. Our studies uncover a new endosomal BLOC-1-dependent pathway for delivery of membrane proteins to cilia and demonstrate that membrane proteins use different routes for delivery to the primary cilium.
Results
Fibrocystin and polycystin-2 are dependent on IFT20 and GMAP210 for ciliary trafficking, whereas smoothened delivery to cilia is largely independent of this complex
We previously showed that IFT20 and GMAP210 are important for localization of polycystin-2 to cilia but their involvement in trafficking of other ciliary membrane proteins is not known. To extend our understanding of IFT20-GMAP210 in ciliary trafficking, we used a pulse-chase assay to compare polycystin-2 with two other ciliary membrane proteins when IFT20 or GMAP210 was perturbed. The proteins fibrocystin and smoothened were chosen because of their diverse structures and because of their importance to human disease. Fibrocystin is a single-pass transmembrane protein mutated in human autosomal recessive PKD, smoothened is a seven-transmembrane receptor involved in hedgehog signaling, and polycystin-2 is multispan membrane protein mutated in human autosomal dominant PKD (Alcedo et al., 1996; Mochizuki et al., 1996; van den Heuvel and Ingham, 1996; Ward et al., 2002, 2003).
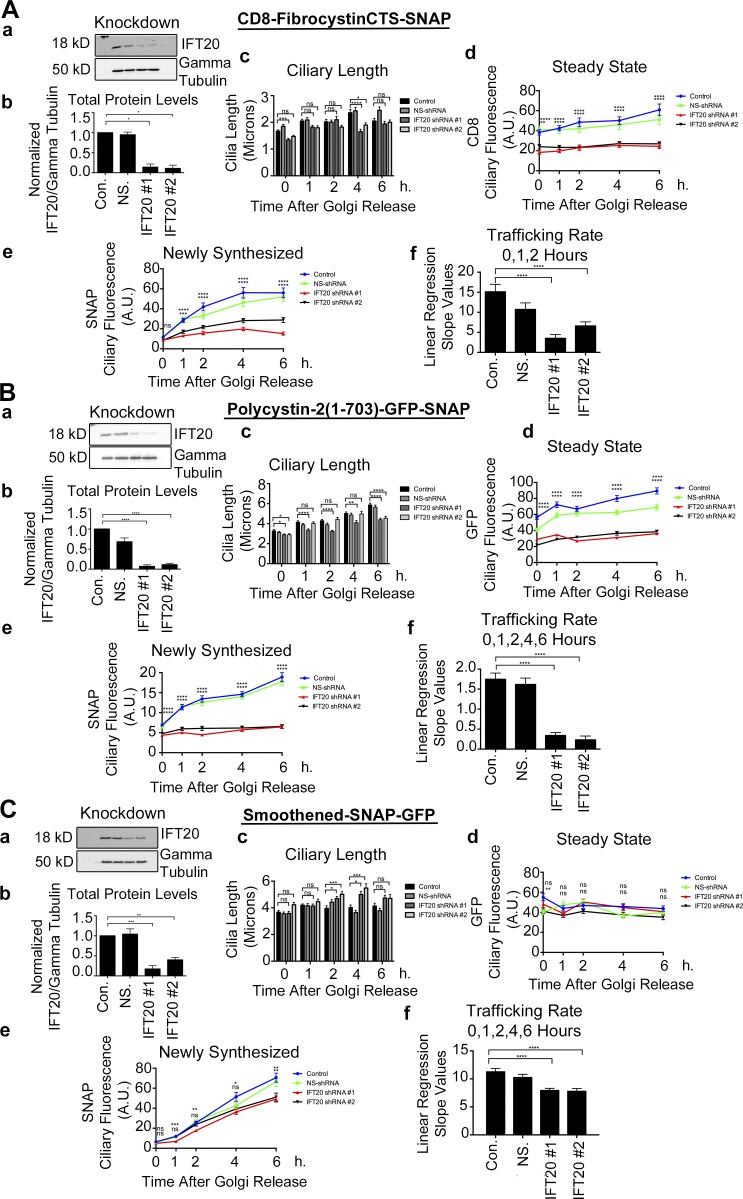
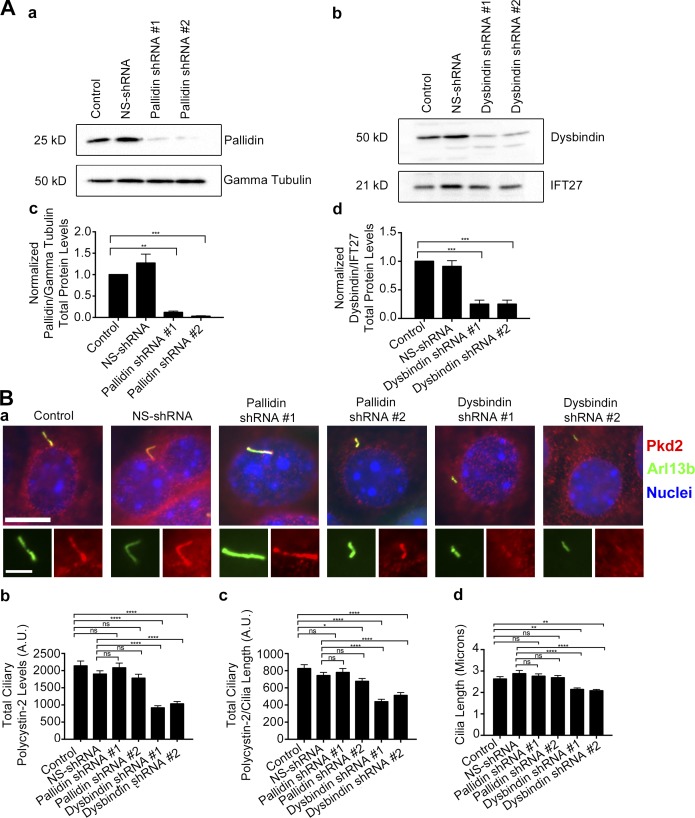
Our pulse-chase assay is based on the SNAP tag, which can be derivatized with a variety of modified benzylguanines (Follit and Pazour, 2013; Follit et al., 2014). This allows one to first block all existing protein with a nonfluorescent derivative and then follow newly synthesized protein with a fluorescent derivative (Sun et al., 2011). The fibrocystin construct consisted of the extracellular domain of CD8 fused to fibrocystin just N-terminal to the transmembrane domain with a SNAP tag placed just before the stop codon. The polycystin-2 construct consists of the first 703 amino acids fused to SNAP and GFP at the C-terminal end. Similarly, full-length smoothened was tagged with SNAP and GFP at the C-terminal end (Fig. S1, Aa–Ca). The SNAP tag is used to quantify newly synthesized protein, while the CD8 epitope tag (included in the extracellular domain of CD8) and GFP are used to monitor total protein levels at steady state. Each of these chimeric proteins is robustly localized to cilia (Fig. S1, Ab–Cb). Inner medullary collecting duct (IMCD3) Flp-In cell lines that uniformly expressed each of the proteins were then infected with lentiviral shRNA constructs to knock down the levels of proteins of interest. Reduction of protein was verified by immunoblot, and the amount remaining was typically about ∼10%–20% of the controls (Figs. 1, S2, S3, and S4).
Figure 1.
IFT20 knockdown affects fibrocystin and polycystin-2 ciliary trafficking but only modestly affects smoothened trafficking to the primary cilium. Quantification of (A) CD8-fibrocystinCTS-SNAP, (B) polycystin-2(1-703)-GFP-SNAP, and (C) smoothened-SNAP-GFP trafficking from the Golgi apparatus to the primary cilium during lenti-shRNA knockdown of IFT20. (A–C, subpanel a) Selected immunoblot images of IFT20 knockdown and γ tubulin loading control. (A–C, subpanel b) Quantification of knockdown. Mean protein levels plotted from three independent experiments for each condition (control [Con.], NS-shRNA [NS.], shRNA#1, and shRNA#2) and normalized to their corresponding γ tubulin loading control. shRNA-mediated knockdown of IFT20 results in >90% reduction of total protein abundance. (A–C, subpanel c) Mean cilia length, (A–C, subpanel d) mean CD8 or GFP ciliary fluorescence, and (A–C, subpanel e) mean SNAP ciliary fluorescence plotted from three independent experiments in which 30 cilia were quantified for each condition (control, NS-shRNA, shRNA#1, and shRNA#2) at each time point (n = 90 total cilia per time point). CD8 or GFP and SNAP pixel intensity measurements were taken at individual time points starting at the time of temperature shift from 19°C to 37°C (0 h). Data were analyzed using one-way ANOVA and the Bonferroni multiple-comparisons test. The control condition was compared with the shRNA#1 and shRNA#2 conditions to determine statistical significance. (A–C, subpanel f) Linear regression analysis of selected time points of newly synthesized membrane protein delivery to the cilium was completed. Slope values between control and experimental shRNA groups were compared with one another to determine statistical significance. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars represent SEM.
The assay starts with blocking all the SNAP-tag binding sites with a cell-permeable nonfluorescent benzylguanine. This substrate is then washed out of the media, and the cells are allowed to synthesize new protein for 1.5 h. The cells are then treated with cycloheximide to inhibit further protein synthesis and shifted to 19°C to prevent the Golgi apparatus from releasing proteins. This causes an accumulation of newly synthesized protein at the Golgi apparatus (Ang et al., 2004). While still keeping the cells in cycloheximide, the cells are shifted back to 37°C, allowing newly synthesized protein to be released from the Golgi complex and trafficked to the cilium. The cells were then fixed at indicated time points after temperature shift back to 37°C, and the amount of newly synthesized and total tagged proteins in the cilium was measured.
For each experiment, we quantified the effectiveness of the shRNA knockdown by charge-coupled device detection of chemiluminescence and displayed results normalized to the untransfected control sample using γ tubulin or IFT27 as a loading control. For each trafficking experiment, we measured cilia length, ciliary SNAP fluorescence, and either ciliary GFP fluorescence or CD8 immunofluorescence at 0, 1, 2, 4, and 6 h after shift from 19°C to 37°C. The data were then plotted with respect to time, and the trafficking rate was determined from the slope of the curve derived from the SNAP fluorescence. All experiments are plotted and displayed similarly, with the control cell displayed in blue, the nonsilencing shRNA in green, and the two shRNAs targeting the gene of interest in red and black.
Under control conditions, the proteins showed different kinetics of delivery to the cilium (Fig. 1, Ae, Af, Be, Bf, Ce, and Cf; and Fig. S1 D). In all cases, newly synthesized protein could be detected in cilia within 1 h of temperature shift. Rates of delivery of fibrocystin and smoothened were similar to each other but substantially higher than polycystin-2 (Fig. S1 D). Fibrocystin typically reached a plateau within 2–4 h after temperature shift, while smoothened and polycystin-2 were still increasing at the 6-h time point. We were unable to follow cells for a longer time, as the cells became unhealthy because of the cycloheximide treatment.
Initially we examined the functions of IFT20 and GMAP210 on the transport of our three model ciliary membrane proteins. As we observed previously, only a small amount of IFT20 is needed to support ciliary assembly, as knockdown to ∼20% of normal had a minimal effect on cilia length (Fig. 1, Ac–Cc). Knockdown of GMAP210 had no effect on cilia length (Fig. S2, Ac–Cc). In all experiments, we noted that ciliary length increased after shift to 37°C. This is likely due to recovery from ciliary shortening that occurs when cells are placed at 19°C (Fig. S1 E). The knockdown of IFT20 and GMAP210 reduced the steady-state levels of ciliary fibrocystin and polycystin-2 and also reduced the rate of delivery of these proteins to cilia (Fig. 1, Ad, Bd, Ae, Be, Af, and Bf; Fig. 2, D and E; and Fig. S2, Ad, Bd, Ae, and Be). In contrast, the steady-state levels of ciliary smoothened and its rate of delivery to cilia were not greatly affected by the reduction in either IFT20 or GMAP210 (Fig. 1, Cd–Cf; Fig. 2 F; and Fig. S2, Cd and Ce). The differences in rates of delivery of the three model proteins and the fact that IFT20 and GMAP210 are not needed for the delivery of smoothened support a model whereby proteins can take different pathways to the cilium.
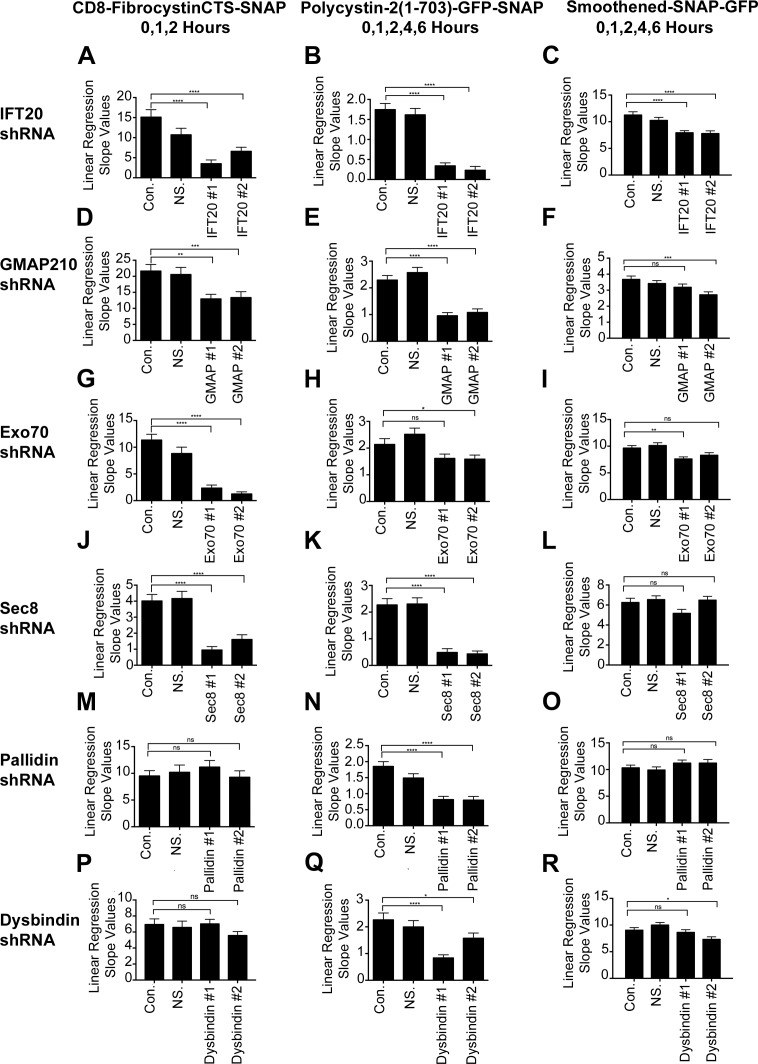
Figure 2.
Fibrocystin, polycystin-2, and smoothened trafficking rates to the primary cilium. (A–C) Linear regression slope value analysis of newly synthesized CD8-FibrocystinCTS-SNAP, Polycystin-2-GFP-SNAP, or Smoothened-SNAP-GFP trafficking from the Golgi apparatus to the primary cilium when IFT20, GMAP210 (D–F), Exo70 (G–I), Sec8 (J–L), pallidin (M–O), or dysbindin (P–R) is knocked down using lenti-shRNAs. Linear regression analysis of newly synthesized membrane protein delivery to the cilium was performed on selected time points: 0, 1, and 2 h for CD8-fibrocystinCTS-SNAP; 0, 1, 2, 4, and 6 h for polycystin-2-GFP-SNAP; and 0, 1, 2, 4, and 6 h for smoothened-SNAP-GFP. The time points were chosen by determining which data points remained linear before (or if) reaching a plateau in the control groups (Fig. 1; and Figs. S2, S3, and S4). The slope values obtained from this analysis represent the trafficking rates of newly synthesized membrane protein delivery to the cilium. Slope values between control and experimental shRNA groups were compared with one another to determine statistical significance. shRNAs of interest are listed vertically on the left side, and their corresponding membrane proteins are listed horizontally. Error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. The IFT20 shRNA linear regression analysis (A–C) from Fig. 1 is shown again for completion.
IFT20 interacts with the BLOC-1 subunit pallidin and the exocyst subunits Exo70 and Sec8
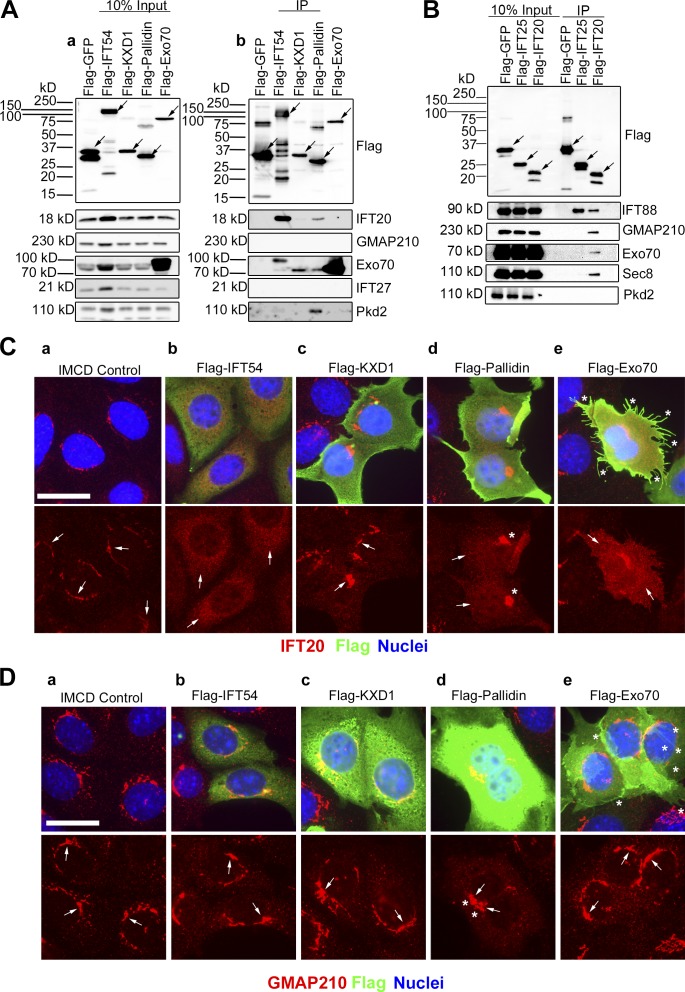
To gain insight into the mechanism of how IFT20 and GMAP210 direct proteins to the ciliary membrane, we examined large-scale yeast-two hybrid screens for new IFT20-interacting proteins (Rual et al., 2005; Wang et al., 2011). From this, we identified the exocyst subunit Exo70, the BLOC-1 subunit pallidin, and a KxDL motif-containing protein named KXD1 as candidate IFT20-binding proteins (Rual et al., 2005; Wang et al., 2011). To determine if these bind to IFT20, the coding sequences were cloned with N-terminal Flag tags, expressed in IMCD3 cells, and immunoprecipitated with anti-Flag resin (Fig. 3 A). Flag-GFP and Flag-IFT54 were used as negative and positive controls (Follit et al., 2009). As expected, Flag-GFP did not pull down IFT20, but Flag-IFT54 showed a strong interaction with IFT20 (Fig. 3 Ab). Flag-Pallidin brought down endogenous IFT20, while no binding of IFT20 was observed with Flag-KXD1 and Flag-Exo70 (Fig. 3 Ab). Interestingly, Flag-Pallidin also precipitated endogenous polycystin-2 (Fig. 3 Ab). Endogenous Exo70 was brought down by Flag-Pallidin (Fig. 3 Ab). This is consistent with work showing BLOC-1 interacting with the exocyst components Sec6 and Sec8 (Gokhale et al., 2012). Flag-KXD1 pulled down Exo70 (Fig. 3 Ab), which may be explained by KXD1 interacting with BLOC-1 (Hayes et al., 2011; Yang et al., 2012).
Figure 3.
IFT20 interacts with the BLOC-1 subunit pallidin and the exocyst subunits Exo70 and Sec8. (A) Flag-GFP (negative IFT20 binding control), Flag-IFT54 (positive IFT20 binding control), Flag-KXD1, Flag-Pallidin, and Flag-Exo70 were expressed in mouse IMCD3 cells. The cells were lysed, immunoprecipitated, with Flag antibody and analyzed by immunoblotting. (Aa) The left group is the starting material before immunoprecipitation (10% input), and (Ab) the right group is the precipitated material (IP). Flag-tagged proteins are marked with arrows on the Flag Western blot. Antibodies used for the Western blots are listed on the right side. (B) Flag-GFP (negative binding control), Flag-IFT25 (positive binding control for IFT complex B protein IFT88), and Flag-IFT20 were expressed in mouse IMCD3 cells. The cells were lysed, immunoprecipitated, with Flag antibody and analyzed by immunoblotting. The left group is the starting material before immunoprecipitation (10% input), and the right group is the precipitated material. Proteins are marked with arrows. Antibodies used for the Western blots are listed on the right side. (C) Selected images of IMCD3 cells expressing the Flag-fusion proteins that are listed horizontally at the top of the figure. Flag-fusion proteins were detected with Flag antibody staining (green), endogenous IFT20 antibody staining (red), and nuclei detected with DAPI (blue). (Ca) IFT20 is localized at the Golgi membranes in IMCD3 control cells (arrows). (Cb) Increased pixel intensity of IFT20 in the cytoplasm (arrows) and complete loss of IFT20 from the perinuclear Golgi pool in Flag-IFT54-expressing cells. (Cc) Flag-KXD1-expressing cells show IFT20 localization at the Golgi membranes (arrows) and no IFT20 displacement into the cytoplasm. (Cd) Compacted Golgi membranes (asterisks) and increased pixel intensity of IFT20 in the cytoplasm (arrows) and partial displacement from the Golgi membranes in Flag-Pallidin-expressing cells. (Ce) Up-regulated filopodia (asterisks) formation and increased IFT20 pixel intensity (arrows), with partial IFT20 displacement from Golgi membranes in cells expressing Flag-Exo70. Bar, 10 µm. (D) Selected images of IMCD3 cells expressing the Flag-fusion proteins that are listed horizontally at the top of the figure. Flag-fusion proteins were detected with Flag antibody staining (green), endogenous GMAP210 antibody staining (red), and nuclei detected with DAPI (blue). (Da) GMAP210 is localized at the Golgi membranes in IMCD3 control cells, (Db) Flag-IFT54-expressing cells, and (Dc) Flag-KXD1-expressing cells (arrows). (Dd) Compacted Golgi membranes (asterisks) and GMAP210 are localized at the Golgi membranes (arrows) in Flag-Pallidin-expressing cells. (De) Up-regulated filopodia (asterisks) formation and GMAP210 are localized at the Golgi membranes (arrows) in cells expressing Flag-Exo70. Bar, 10 µm.
Because no interaction was observed between Flag-Exo70 and endogenous IFT20, we tested to see if Flag-IFT20 could precipitate endogenous Exo70 or Sec8 exocyst subunits (Fig. 3 B). Flag-GFP and Flag-IFT25 were negative controls, and neither interacted with either of the exocyst components. As positive controls, Flag-IFT20 and Flag-IFT25 interacted as expected with IFT88 and Flag-IFT20 with GMAP210 (Fig. 3 B; Follit et al., 2008, 2009; Keady et al., 2011). Importantly, Flag-IFT20 also brought down endogenous Exo70 and Sec8 (Fig. 3 B). The reason for the discrepancy between the two immunoprecipitations is unknown but not uncommon in these types of studies. However, our finding of an interaction between IFT20 and both Exo70 and Sec8 is consistent with prior work showing that the Sec10 subunit of the exocyst could coprecipitate IFT20 and IFT88 (Fogelgren et al., 2011).
Previously we showed that overexpressing the IFT20-binding proteins IFT54 or GMAP210 displaces IFT20 from the Golgi membranes. Importantly, overexpressing other IFT proteins that do not directly bind IFT20 did not have any effect on the distribution of IFT20 in cells (Follit et al., 2009). As a further test for evidence of interaction with IFT20, we overexpressed Flag-KXD1, Flag-Pallidin, Flag-Exo70, and, as a positive control, Flag-IFT54 (Fig. 3 C). Flag-IFT54 overexpression completely displaced IFT20 from the Golgi stacks (Fig. 3 Cb). High expression of Flag-KXD1 did not displace IFT20 from the Golgi (Fig. 3 Cc), consistent with a failure to interact in the immunoprecipitation assay. Flag-Pallidin overexpression resulted in partial displacement of IFT20 into the cytoplasm and for unknown reasons caused the Golgi apparatus to compact (Fig. 3 Cd). Flag-Exo70 overexpression also caused displacement of IFT20 from the Golgi and induced filopodia formation, similar to what has been described (Fig. 3, Ce and De; Zuo et al., 2006). To ensure that this result was not due to disruption of the Golgi complex by overexpression of our test proteins, we stained with the Golgi marker GMAP210. GMAP210 was not displaced by overexpression of any of the proteins, and Golgi structure was normal except for the compaction observed when Flag-Pallidin was expressed (Fig. 3 D). Together the immunoprecipitation and displacement results provide strong evidence that IFT20 interacts with the BLOC-1 subunit pallidin and the exocyst subunit Exo70.
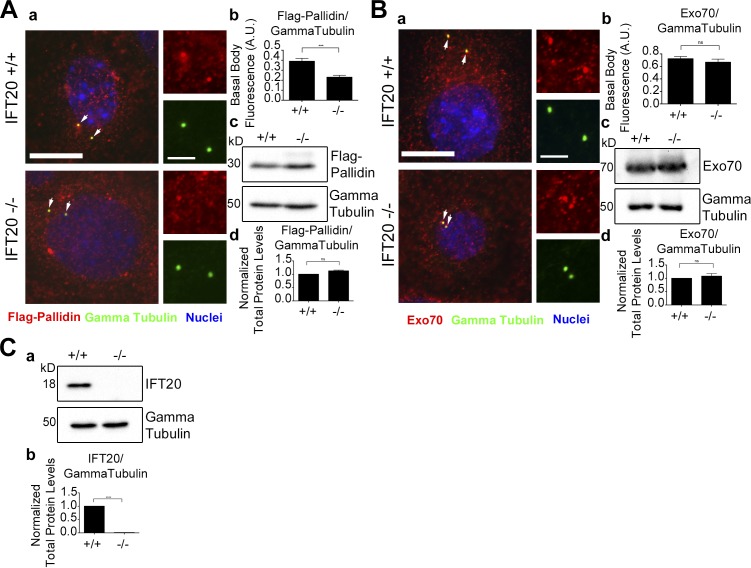
The localization of pallidin at the basal body is partially dependent on IFT20
Because IFT20 binds Exo70 and pallidin, we determined whether their cellular localization is dependent on IFT20. To do this, wild-type and Ift20−/− mouse embryonic kidney (MEK) cells (Jonassen et al., 2008) were transfected with Flag-Pallidin or stained for endogenous Exo70 (Fig. 4). In wild-type cells, Flag-Pallidin partially localizes at the basal body (Fig. 4 Aa). This is consistent with prior work showing other BLOC-1 subunits localizing to the centrosome (Wang et al., 2004). Interestingly, there is a significant decrease in the amount of basal body–localized Flag-Pallidin in the Ift20−/− MEKs, while there is no difference in the total Flag-Pallidin levels in the cell (Fig. 4 A). Endogenous Exo70 partially localizes at the basal body, but there was no difference between control and Ift20−/− cells (Fig. 4 B).
Figure 4.
Localization of pallidin at the basal body is dependent on IFT20. (A) Wild-type and Ift20−/− MEK cells expressing Flag-Pallidin. (Aa) Flag antibody staining (red), γ tubulin antibody staining (green), and nuclei detected with DAPI (blue). Flag-Pallidin localizes at the basal body (arrows). (Ab) Mean steady-state levels of Flag-Pallidin at the basal body are decreased in the Ift20-null cells. (Ac) Selected immunoblot images of total Flag-Pallidin and γ tubulin loading control protein levels. (Ad) No difference in the mean total protein levels of Flag-Pallidin in the Ift20−/− MEK cells compared with the control. (B) Endogenous Exo70 levels at the basal body in wild-type and Ift20−/− MEK cells. (Ba) MEK cells stained for endogenous Exo70 (red), γ tubulin (green), and nuclei detected with DAPI (blue). Endogenous Exo70 localizes at the basal body (arrows). (Bb) No difference in the mean steady-state levels of Exo70 at the basal body in the Ift20−/− cells compared with the control. (Bc) Selected immunoblot images of total endogenous Exo70 and γ tubulin loading control protein levels. (Bd) No difference in the mean total protein levels of Exo70 in the Ift20−/− cells compared with the control. (C) Total protein levels of endogenous IFT20 in wild-type and Ift20−/− MEK cells. (Ca) Selected immunoblot images of total endogenous IFT20 and γ tubulin loading control. (Cb) Mean immunoblot pixel intensity quantification showing no IFT20 protein present in the Ift20−/− MEK cells. n = 50 basal bodies per experimental group. Error bars are standard error of the mean. Data were analyzed using the unpaired Student’s t test. ***, P < 0.001; ****, P < 0.0001. Bars, 10 µm. Insets are 190% enlargements of the centrosome regions.
There does not appear to be any effect on IFT20 localization when either pallidin or Exo70 is knocked down (Fig. S6), indicating that IFT20 localization is not dependent on either protein.
The exocyst complex is involved in the ciliary trafficking of fibrocystin and polycystin-2
Previous work had demonstrated a role for exocyst subunit Sec10 in ciliary assembly (Zuo et al., 2009). Because we had found that IFT20 was able to precipitate the exocyst subunits Exo70 and Sec8, we tested the exocyst’s role in trafficking of our model ciliary proteins by knocking down these subunits (Fig. 2, G–L; and Fig. S3). Knockdown of Exo70 to about ∼20% of normal had modest effects on ciliary length (Fig. S3, Ac–Cc), while knockdown of Sec8 to ∼20% of controls decreased ciliary length to ∼40% of normal (Fig. S3, Dc–Fc). Knockdown of these exocyst subunits reduced the steady-state levels of ciliary fibrocystin and polycystin-2 (Fig. S3, Ad, Bd, Dd, and Ed) and reduced the rate of delivery of these proteins to cilia (Fig. 2, G, H, J, and K; and Fig. S3, Ae, Be, De, and Ee). The reduction of Exo70 or Sec8 did not affect the rate of delivery of newly synthesized smoothened to the cilium (Fig. 2, I and L; and Fig. S3, Ce and Fe), and the steady-state level of ciliary smoothened was only modestly or not affected (Fig. S3, Cd and Fd). This further strengthens the idea that distinct membrane proteins are using different trafficking pathways and specific molecular machineries for ciliary delivery.
BLOC-1 is important for the trafficking of polycystin-2 to primary cilia
BLOC-1 has not been previously implicated in ciliary trafficking, but our finding that pallidin bound IFT20 and polycystin-2 suggests involvement. To test this idea, we assayed ciliary transport after knocking down pallidin and another BLOC-1 subunit dysbindin. Knockdown of pallidin to 10%–15% of normal had no effect on ciliary length (Fig. S4, Ac–Cc). Knockdown of dysbindin to ∼10% of normal affects ciliary assembly (Fig. S4, Dc–Fc). Knockdown of either dysbindin or pallidin had no effect on steady-state levels or rates of delivery of fibrocystin and smoothened to cilia (Fig. 2, M, O, P, and R; and Fig. S4, Ad, Cd, Dd, Fd, Ae, Ce, De, and Fe). In contrast, knockdown of either dysbindin or pallidin reduced the rates of delivery of polycystin-2 to cilia, and dysbindin knockdown decreased steady-state levels of ciliary polycystin-2 (Fig. 2, N and Q; and Fig. S4, Be, Ed, and Ee). The amount of reduction in steady-state ciliary polycystin-2 varied between the two dysbindin shRNAs but correlated with the more effective knockdown, causing a larger decrease in ciliary polycystin-2 levels (Fig. 2 Q and Fig. S4, Ea, Eb, Ed, and Ee). Our protein interaction data and pulse-chase trafficking data give strong evidence that BLOC-1 is specifically involved in polycystin-2 delivery to cilia. This emphasizes that polycystin-2 is taking a specific pathway to the cilium that is dependent on BLOC-1 machinery.
Perturbation of the exocyst or BLOC-1 complexes decreases endogenous polycystin-2 levels at the primary cilium
To further verify our finding that the exocyst complex and BLOC-1 are involved in trafficking polycystin-2 to the primary cilium, we performed knockdowns in MEK cells and measured endogenous polycystin-2 ciliary levels. MEK cells were chosen because they have more polycystin-2 in their cilia than IMCD3 cells, and because they are mouse cells, the shRNAs developed previously are effective in them.
Similar to what we observed in IMCD3 cells, Exo70 knockdown had little to no effect on ciliary assembly in MEK cells, but Sec8 knockdown decreased ciliary length (Fig. S5, Bd and Cd). Importantly, knockdown of either Exo70 or Sec8 resulted in decreased steady-state ciliary levels of polycystin-2 (Fig. S5, Ba–Bc), confirming our results with an exogenous chimeric protein. Additionally, knockdown of Sec8 but not Exo70 reduced the ciliary levels of Arl13b at the cilium (Fig. S5, Ca–Cc). Arl13b is a peripheral membrane protein anchored to the cilium by palmitoylated cysteines (Sun et al., 2004; Cevik et al., 2010; Li et al., 2010). It is not clear why the two subunits have different effects on ciliary Arl13b levels, but Sec8 is a core exocyst subunit, while Exo70 is a peripheral membrane-docking subunit (TerBush and Novick, 1995; TerBush et al., 1996; He et al., 2007; Wu et al., 2010). It is possible that core subunits are more important for delivery of Arl13b or that higher levels of Sec8 are needed than Exo70 and the Exo70 levels were not reduced below the critical level.
To confirm the results obtained with the exogenous chimeric protein, we examined the involvement of BLOC-1 in trafficking of endogenous polycystin-2 to primary cilia. Again, MEK cells were used because of their high levels of ciliary polycystin-2. In these cells, knockdown reduced pallidin and dysbindin to 10%–15% of normal (Fig. 5, Ac and Ad). The dysbindin knockdown caused a decrease in cilia length and reduced the steady-state level of ciliary polycystin-2 (Fig. 5 B). Similar to what we saw in IMCD3 cells, pallidin knockdown did not affect cilia length or the levels of ciliary polycystin-2 (Fig. 5 B). This continues to strengthen the evidence that the exocyst complex and BLOC-1 are involved in the trafficking of polycystin-2 to cilia.
Figure 5.
Dysbindin knockdown decreases endogenous ciliary polycystin-2 levels. (A) Immunoblot and quantification of total protein levels of MEK cells expressing either pallidin or dysbindin lenti-shRNAs. (Aa) Selected immunoblot images showing pallidin knockdown and (Ab) dysbindin knockdown with either γ tubulin or IFT27 loading control. (Ac) Mean quantification of pallidin and (Ad) dysbindin knockdown under each condition (control, NS-shRNA, shRNA#1, and shRNA#2). Intensity was normalized to their corresponding γ tubulin or IFT27 loading control. shRNA-mediated knockdown of either pallidin or dysbindin results in >90% reduction of total protein abundance. (B) Immunostaining and quantification of ciliary polycystin-2 and cilia length in MEK cells expressing either pallidin or dysbindin lenti-shRNAs. (Ba) Selected images of polycystin-2 (Pkd2) antibody staining (red), Arl13b antibody staining (green), and nuclei are detected with DAPI (blue). Bars, 10 µm. Insets are 170% enlargements of the cilia. (Bb) Mean steady-state ciliary polycystin-2 is decreased and (Bc) mean ciliary polycystin-2 per micrometer is reduced in cells expressing dysbindin lenti-shRNAs. (Bd) Mean cilia length is reduced in cells expressing dysbindin shRNAs. n = 107 cilia per experimental group. Error bars are SEM. Data were analyzed using one-way ANOVA and the Bonferroni multiple-comparisons test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Dtnbp1sdy/sdy and Pldnpa/pa mice possess a cystic kidney phenotype
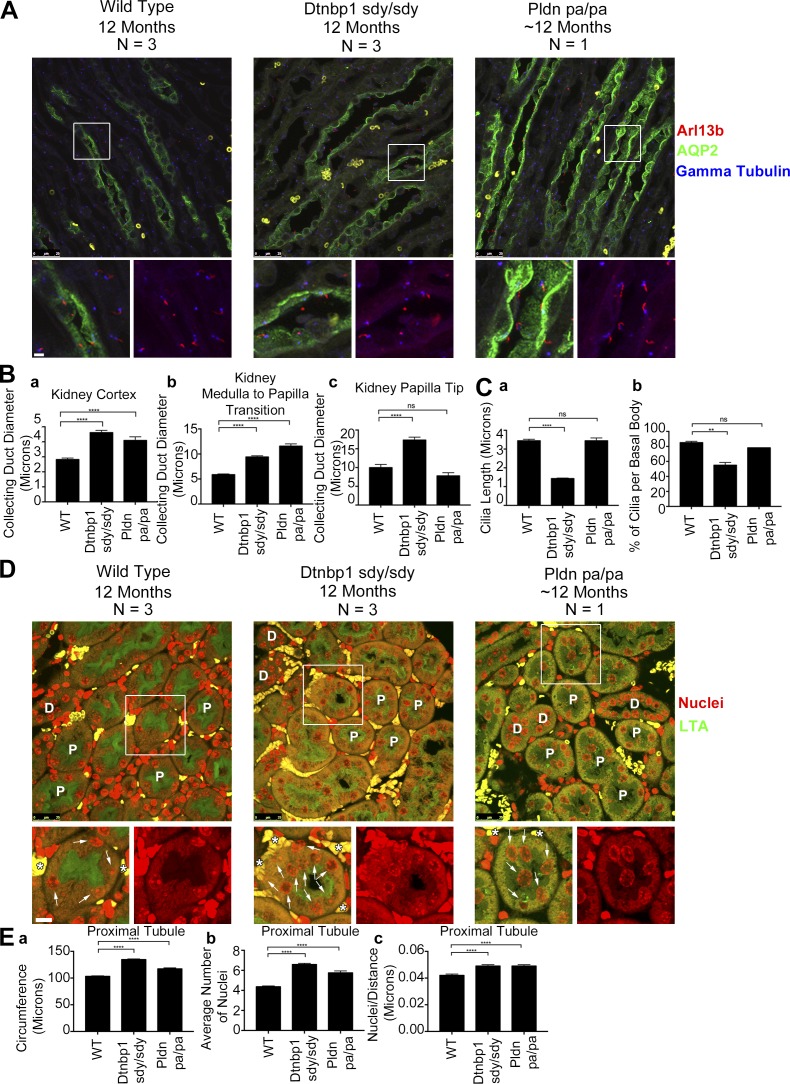
The sandy (Dtnbp1sdy/sdy) and pallid mice (Pldnpa/pa) are loss-of-function models of dysbindin and pallidin (Huang et al., 1999; Li et al., 2003). Given that our data suggest that BLOC-1 is involved in the trafficking of polycystin-2 to primary cilia, we hypothesized that the Dtnbp1sdy/sdy and Pldnpa/pa mice would have cystic kidneys. Hematoxylin and eosin staining of 5-wk-old Dtnbp1sdy/sdy and 8-wk-old Pldnpa/pa kidneys showed no prominent cyst formation, but the collecting ducts in the papilla were slightly distended in both mouse models (Fig. S7, A, Ba, and Bb). Proximal tubule circumference was also slightly increased in the Dtnbp1sdy/sdy mice, and both the Dtnbp1sdy/sdy and Pldnpa/pa mice had a slight increase in the mean number of proximal tubule nuclei per cross section (Fig. S7 C). Additionally, the Dtnbp1sdy/sdy and Pldnpa/pa mice had a moderate decrease in ciliary length, and Dtnbp1sdy/sdy mice showed a modest decrease in the percentage of cilia per basal body (Fig. S7, Bc and Bd).
Given that the first cohort of Dtnbp1sdy/sdy and Pldnpa/pa mice were relatively young, we aged a second cohort for 12 mo to see whether they would present a more severe cystic kidney phenotype. These animals suffer from lung fibrosis, and very few animals survive to 1 yr of age (McGarry et al., 2002). However, we were able to collect three Dtnbp1sdy/sdy and one Pldnpa/pa animal at this age. None of the animals showed prominent cysts, but the collecting ducts were obviously distended (Fig. 6 A). Quantification showed a significant increase in the Dtnbp1sdy/sdy collecting duct diameter measured in the kidney cortex, at the junction between the outer and inner medulla, and in the inner medulla at the tip of the papilla (Fig. 6 B). The Pldnpa/pa also showed increased diameter of collecting ducts, but only one animal was examined. Proximal tubules were also dilated in the two models. In both models, the circumferences were slightly larger, and the number of nuclei per cross section was increased (Fig. 6, D and E). In addition, we noted abnormal position of the proximal tubule nuclei in the two models. Proximal tubule cells typically have well-separated nuclei located near the basal surface. In the mutants, nuclei distribute throughout the cell, including at the apical surface, and often are found next to each other (Fig. 6 D).
Figure 6.
Dtnbp1sdy/sdy and Pldnpa/pa mice have mildly cystic kidneys. (A) Selected confocal images of 12-mo-old wild-type, Dtnbp1sdy/sdy, and Pldnpa/pa mouse kidney sections stained with cilia marker Arl13b antibody (red), collecting duct marker AQP2 antibody (green), and basal body marker γ tubulin antibody (blue). Bars, 25 µm. Insets are 285% enlargements. (B) Quantification of mean collecting duct diameters in three regions of the kidney; (Ba) cortex (wild-type and Dtnbp1sdy/sdy n = 183 collecting ducts per group, Pldnpa/pa n = 61 collecting ducts), (Bb) medulla to papilla transition (wild-type and Dtnbp1sdy/sdy n = 183 collecting ducts per group, Pldnpa/pa n = 61 collecting ducts), and (Bc) tip of the papilla (wild-type and Dtnbp1sdy/sdy n = 33 collecting ducts per group, Pldnpa/pa n = 11 collecting ducts). There is an increase in collecting duct diameter in the Dtnbp1sdy/sdy and Pldnpa/pa cortex and medulla-to-papilla transition. Additionally, the Dtnbp1sdy/sdy papilla tip collecting duct diameter is wider. (C) Quantification of mean kidney cilia length and the percentage of cilia per basal body. (Ca) Wild-type and Dtnbp1sdy/sdy n = 300 cilia per group, Pldnpa/pa n = 100 cilia. Cilia in the Dtnbp1sdy/sdy kidneys are shorter compared with control. (Cb) Wild-type and Dtnbp1sdy/sd n = 315 basal bodies per group, Pldnpa/pa n = 105 basal bodies. Percentage cilia per basal body is reduced in the Dtnbp1sdy/sdy kidneys. (D) Selected confocal images of 12-mo-old wild-type, Dtnbp1sdy/sdy, and Pldnpa/pa kidney sections stained with proximal tubule marker Lotus tetragonolobus agglutinin (LTA; green) and nuclei with Dapi (red). Most tubules are proximal, and some are marked with “P.” The few distal convoluted tubules are marked with “D.” Arrows label proximal tubule nuclei, and asterisks indicate red blood cells. Bars, 25 µm. Insets are 172% enlargements. (E) Quantification of mean kidney proximal tubule nuclei circumference, mean number of proximal tubule nuclei, and nuclei number per proximal tubule circumference. Wild-type and Dtnbp1sdy/sdy n = 225 proximal tubules per group, Pldnpa/pa n = 75 proximal tubules. (Ea) Dtnbp1sdy/sdy and Pldnpa/pa proximal tubules have an increased circumference. (Eb) Dtnbp1sdy/sdy and Pldnpa/pa proximal tubules have an increase in the mean number of nuclei. (Ec). Increase in nuclei number per proximal tubule circumference. n = 3 mice per wild-type and Dtnbp1sdy/sdy groups and n = 1 mouse per Pldnpa/pa group. Tissue from the wild-type and Dtnbp1sdy/sdy mice was harvested at precisely 12 mo of age. Two of the three Pldnpa/pa mice died of a known lung fibrosis phenotype during the aging process. Tissue from the remaining Pldnpa/pa mouse was harvested at 11 mo and 19 d of age. Error bars are SEM. Data were analyzed using one-way ANOVA and the Bonferroni multiple-comparisons test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
There is no difference in cilia assembly and the percentage of cilia per basal body in the Pldnpa/pa mice (Fig. 6 C). However, the Dtnbp1sdy/sdy mice have a decrease in both cilia length and the percentage of cilia per basal body (Fig. 6 C). The Dtnbp1sdy/sdy and Pldnpa/pa mouse phenotype strongly correlates with the biochemical and cell biology data and supports the conclusion that BLOC-1 is involved in trafficking polycystin-2 to primary cilia.
Polycystin-2 but not fibrocystin or smoothened is retained in the recycling endosome when the C-terminal tail of MyoVb is overexpressed
The BLOC-1 complex is thought to function at the recycling endosome tubules that extend from sorting endosomes to direct cargos into the recycling endosome and lysosome (Di Pietro et al., 2006; Salazar et al., 2006; Setty et al., 2007; Ryder et al., 2013; Delevoye et al., 2016; Dennis et al., 2016). Because the recycling endosome is localized at the base of the cilium (Westlake et al., 2011), we questioned whether the function of BLOC-1 in trafficking polycystin-2 to cilia might involve its trafficking to or through the recycling endosome. To disrupt trafficking through the recycling endosome, we expressed a fragment of MyoVb that acts as a dominant negative on trafficking through this compartment (Lapierre et al., 2001; Volpicelli et al., 2002). The MyoVb dominant-negative fragment contains the C-terminal Rab11-binding domain but lacks the motor domain, allowing the fragment to bind Rab11-containing vesicles and interfere with their transport. GFP-MyoVb C-terminal tail colocalizes with the recycling endosome marker Rab11 (Fig. S8 A) but not with the Golgi markers IFT20 (Fig. S8 B) and GMAP210 (Fig. S8 C). Expression of the MyoVb fragment induces compaction of the Golgi complex (Fig. S8, B and C) but had only a small effect on cilia length (Fig. 7, Ad and Bd).
Figure 7.
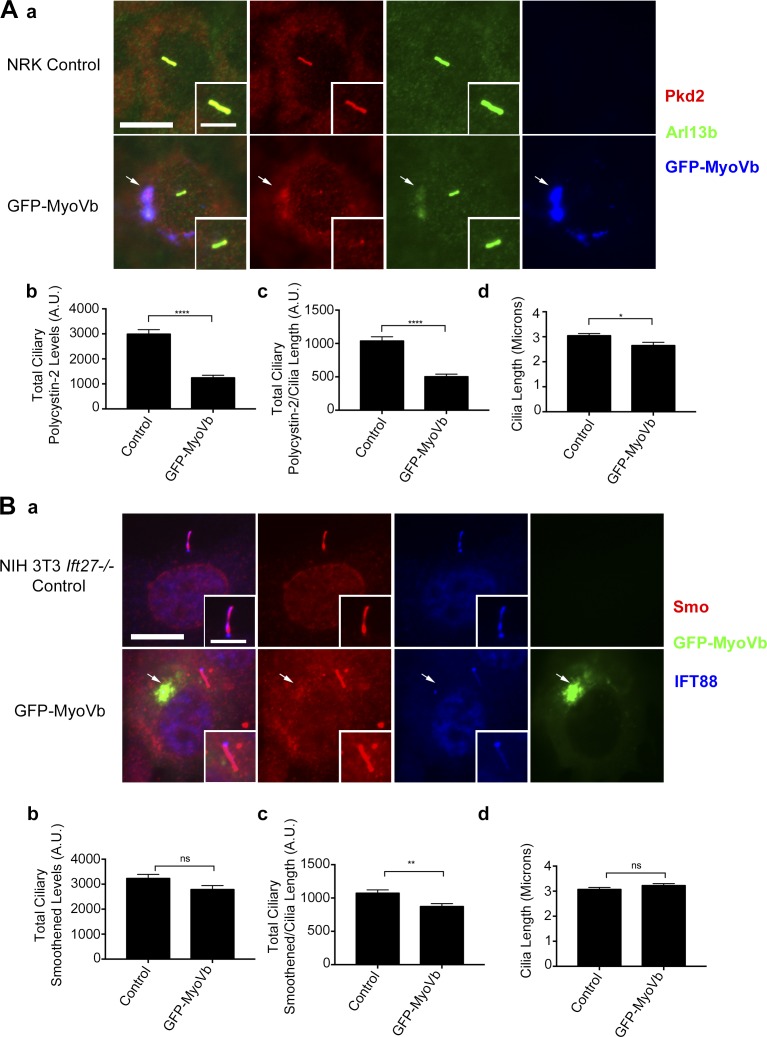
Ciliary levels of polycystin-2 but not smoothened are reduced in GFP-MyoVb-overexpressing cells. (A) NRK cells expressing GFP-MyoVb C-terminal tail. (Aa) Polycystin-2 (Pkd2) antibody staining (red), Arl13b antibody staining (green), and GFP-MyoVb (blue). Polycystin-2 and Arl13b colocalize with GFP-MyoVb at the recycling endosome (arrows). Bars, 10 µm. Insets are 150% enlargements of the cilia. (Ab) Mean steady-state ciliary polycystin-2 is decreased, (Ac) ciliary polycystin-2/cilia is reduced, and (Ad) there is no difference in mean cilia length in NRK cells expressing GFP-MyoVb. (B) NIH 3T3 Ift27−/− cells expressing GFP-MyoVb C-terminal tail. (Ba) Smoothened (Smo) antibody staining (red), GFP-MyoVb (green), and IFT88 antibody staining (blue). Modest levels of smoothened colocalizes with GFP-MyoVb at the recycling endosome (arrows). Bar, 10 µm. Insets are 150% enlargements of the cilia. (Bb) No difference in mean steady-state ciliary smoothened, (Bc) total ciliary smoothened/length is decreased, and (Bd) there is no difference in mean ciliary length in GFP-MyoVb-expressing cells. n = 102 cilia per experimental group. Error bars are SEM. Data were analyzed using the unpaired Student’s t test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
We were unable to package the MyoVb expression vector into lentivirus, as it appears to interfere with some step of the process, and had to rely on electroporation to deliver the construct. MEK cells are difficult to transfect, so we used normal rat kidney (NRK) cells, which also have high levels of ciliary polycystin-2 and are easy to transfect. NRK cells transfected with GFP-MyoVb accumulated polycystin-2 at the recycling endosome (Fig. 7 Aa and Fig. S9, A and B) and had a significant decrease in the steady-state levels of ciliary polycystin-2 (Fig. 7, Ab and Ac). Additionally, we observed a moderate accumulation of endogenous Arl13b at the recycling endosome upon GFP-MyoVb overexpression (Fig. 7 Aa).
To examine the effect of the MyoVb construct on trafficking of smoothened to the cilium, we used Ift27−/− NIH 3T3 cells, as the lack of IFT27 causes smoothened to accumulate to high levels in cilia (Eguether et al., 2014). Expression of GFP-MyoVb C-terminal tail in these cells resulted in a moderate accumulation of smoothened in the recycling endosome (Fig. 7 Ba and Fig. S9, C and D), but there was no difference in ciliary smoothened levels (Fig. 7, Bb and Bc).
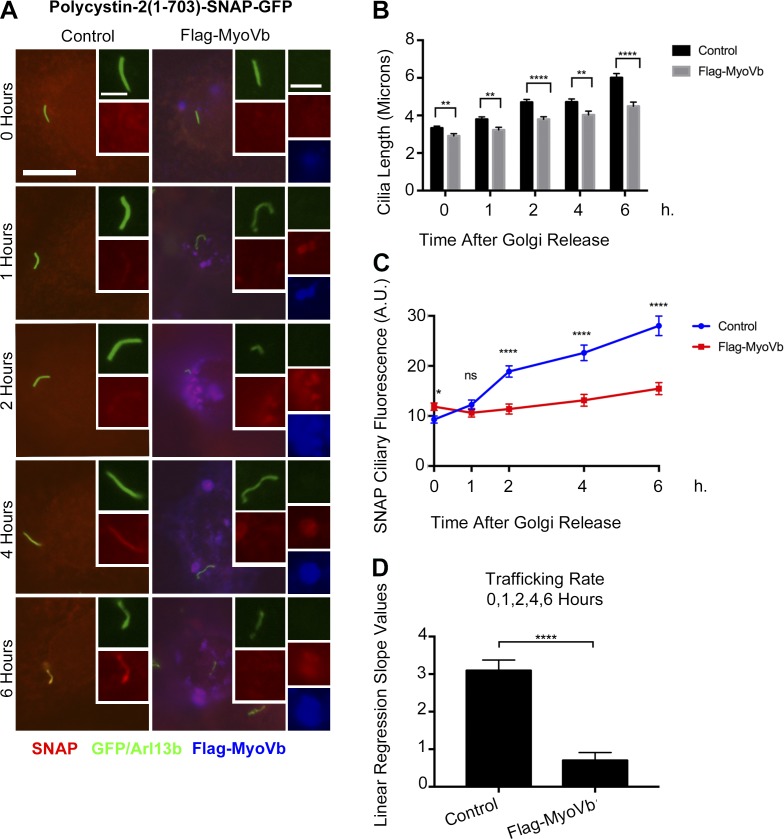
We went on to use a Flag-MyoVb construct in our pulse-chase trafficking assays for fibrocystin, polycystin-2, and smoothened. We observed a bolus of polycystin-2 colocalizing with the Flag-MyoVb in the recycling endosome after Golgi release (Fig. 8 A). There is a decrease in both the ciliary length (Fig. 8 B) and ciliary trafficking of polycystin-2 (Fig. 8, C and D) in the Flag-MyoVb-overexpressing cells. However, trafficking of fibrocystin and smoothened were not affected by the expression of the MyoVb dominant negative construct (Fig. S11).
Figure 8.
Overexpression of MyoVb perturbs polycytin-2 trafficking to the primary cilium. (A) Selected trafficking images of polycystin-2-GFP-SNAP IMCD Flp-In cells overexpressing Flag-MyoVb C-terminal tail. GFP and Arl13b antibody staining (green), SNAP TMR STAR (red), and Flag-MyoVb (blue). Insets depict newly synthesized polycystin-2-GFP-SNAP trafficking to the cilium in control or Flag-MyoVb-overexpressing cells. Insets also show accumulation of newly synthesized polycystin-2-GFP-SNAP in the recycling endosome in the Flag-MyoVb-overexpressing cells. Insets are 200% enlargements of the cilia and 170% enlargements of the recycling endosome. Bars, 10 µm. (B) Mean cilia length of polycystin-2-GFP-SNAP IMCD Flp-In control and Flag-MyoVb-overexpressing cells after the Golgi release. Cilia are shorter in Flag-MyoVb-overexpressing cells. (C) Mean SNAP ciliary fluorescence of Polycystin-2-GFP-SNAP delivery to the cilium after Golgi release. Polycystin-2 ciliary trafficking decreased in Flag-MyoVb-overexpressing cells. (D) Linear regression slope values of newly synthesized polycystin-2-GFP-SNAP in IMCD Flp-In control and Flag-MyoVb-overexpressing cells after the Golgi release using data points taken at 0, 1, 2, 4, and 6 h. Mean SNAP ciliary fluorescence and mean ciliary length were plotted from three independent experiments in which 30 cilia were quantified for each condition (control and Flag-MyoVb) at each time point (n = 90 total cilia per time point). Error bars represent SEM. Data were analyzed using the unpaired Student’s t test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
To determine whether knockdown of the exocyst or BLOC-1 components results in accumulation of endogenous polycystin-2 in endosome cellular compartments, we stained our shRNA MEK cell lines with the recycling endosome marker Rab11 (Fig. S10). We did not detect accumulation of polycystin-2 in Rab11-positive endosomes in our exocyst or BLOC-1 knockdowns (Fig. S10 B).
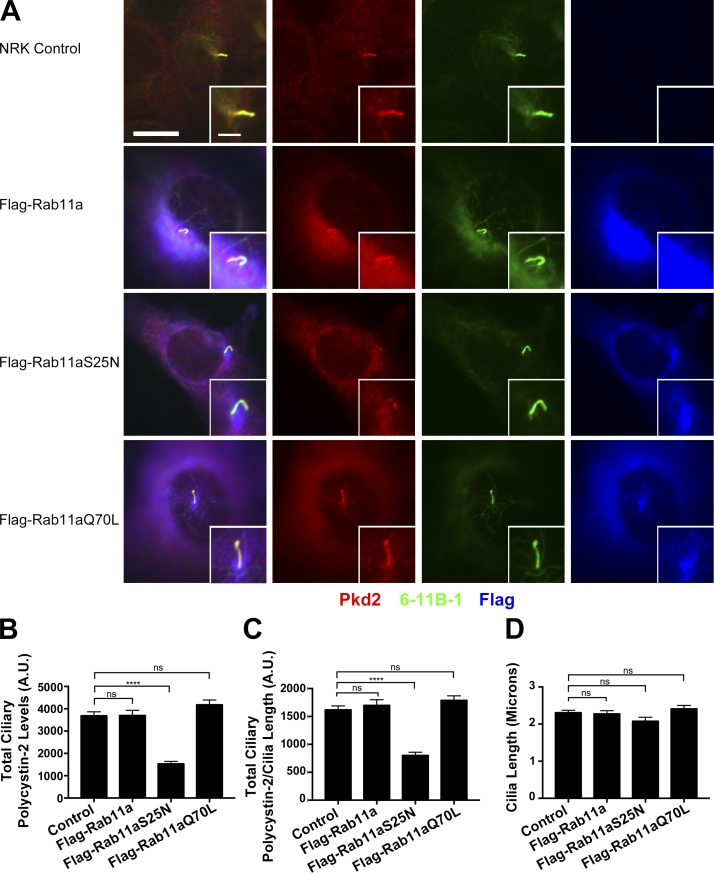
Ciliary polycystin-2 is reduced by overexpression of dominant-negative Rab11a
To obtain more evidence that polycystin-2 is using the recycling endosome for trafficking to the primary cilium, we measured ciliary polycystin-2 levels in cells expressing wild-type or mutant forms of Rab11a that are thought to disrupt the recycling endosome (Hehnly and Doxsey, 2014). Overexpressing wild-type Rab11a, or Rab11aQ70L, which is thought to block GTP hydrolysis and keep the protein in a constitutively active GTP-bound form, had no effect on ciliary levels of polycystin-2. However, overexpressing Rab11aS25N, which is thought to act as a dominant negative by binding guanine exchange factors, reduced ciliary polycystin-2 levels (Fig. 9, A–C). None of these constructs affected ciliary length (Fig. 9 D). Our finding that ciliary polycystin-2 levels are reduced in cells expressing Rab11aS25N is consistent with dominant-negative forms of Rab11 disrupting the recycling endosome (Westlake et al., 2011; Hehnly and Doxsey, 2014) and further supports our model that polycystin-2 is trafficked to cilia through the recycling endosome.
Figure 9.
Rab11aS25N expression perturbs polycystin-2 trafficking to the primary cilium. (A) Selected images of NRK cells expressing Flag-Rab11a, Flag-Rab11aS25N, or Flag-Rab11aQ70L stained for polycystin-2 (Pkd2) antibody (red), acetylated tubulin (6-11B-1) antibody (green), and Flag antibody (blue). Bars, 10 µm. Insets are 200% enlargements of the cilia. (B) Quantification of mean total ciliary polycystin-2 levels. Ciliary polycystin-2 is reduced in Flag-Rab11aS25N-expressing cells. (C) Quantification of mean total ciliary polycystin-2/cilia length. Ciliary polycystin-2 per cilium length is reduced in Flag-Rab11aS25N-expressing cells. (D) Quantification of mean cilia length. There is no difference in ciliary assembly when the Rab11a constructs are expressed. n = 85 cilia per experimental group. Error bars are SEM. Data were analyzed using one-way ANOVA and the Bonferroni multiple-comparisons test. ****, P < 0.0001.
Discussion
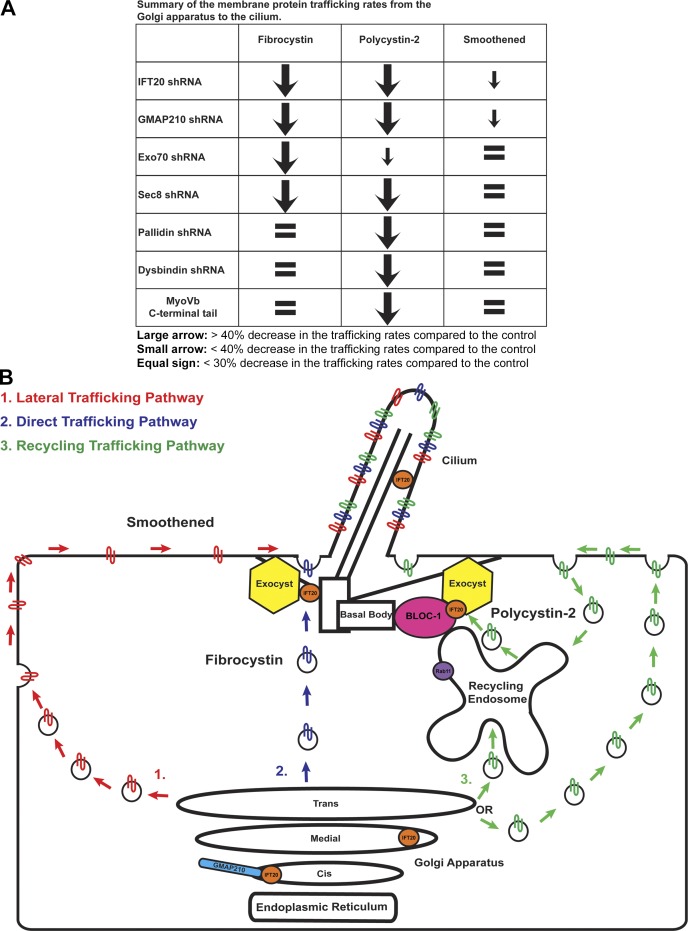
Cilia monitor the extracellular environment through specific receptors concentrated in their membranes, but we know little about how membrane proteins reach this organelle. Our prior work showed that IFT20 was required for delivery of polycystin-2 to primary cilia and rhodopsin to photoreceptor outer segments. IFT20 localizes to the Golgi complex by interaction with the golgin protein GMAP210 and is part of the IFT complex localized at cilia. This dual localization suggests that IFT20 could sort or traffic vesicles destined for the cilium, but details of how it accomplishes this are lacking. In our present work, we find that IFT20 interacts with the exocyst and the BLOC-1 complexes, and we find that polycystin-2, fibrocystin, and smoothened each has unique requirements for delivery to cilia. Fibrocystin requires IFT20-GMAP210 and the exocyst. Polycystin-2 requires IFT20-GMAP210, the exocyst, and BLOC-1. However, smoothened delivery is largely independent of all of these proteins.
The exocyst was required for the delivery of polycystin-2 and fibrocystin to cilia. This complex consists of eight proteins (Sec3/Exoc1, Sec5/Exoc2, Sec6/Exoc3, Sec8/Exoc4, Sec10/Exoc5, Sec15/Exoc6, Exo70/Exoc7, and Exo84/Exoc8) conserved from yeast to mammals (TerBush et al., 1996; Kee et al., 1997; Heider et al., 2016). The exocyst tethers vesicles at target sites before membrane fusion (Heider and Munson, 2012; Luo et al., 2014). In polarized cells, the exocyst localizes along sites of secretion on the lateral membrane and at the base of the cilium (Grindstaff et al., 1998; Rogers et al., 2004; Mazelova et al., 2009). Knockdown of Sec10 disrupts ciliogenesis in Madin-Darby canine kidney cells, and targeted deletion of Sec10 in the mouse kidney causes ciliary defects and cyst formation (Zuo et al., 2009; Fogelgren et al., 2015; Seixas et al., 2016). We found that IFT20 interacted with the Exo70 and Sec8 subunits. However, it is likely that IFT20 interacts with the entire exocyst complex, as the Sec10 subunit also interacts with IFT20, and a large-scale proteomic study found IFT20 binding to multiple components (Exoc3, Exoc7, Exoc5, and Exoc6B; Fogelgren et al., 2011; Huttlin et al., 2015). We found that the localization of the exocyst at the base of the cilium is independent of IFT20, indicating that the exocyst is not delivered by IFT. This suggests the exocyst may capture or anchor vesicles containing ciliary cargos at the base of the cilium before membrane fusion. Because the exocyst can interact with IFT proteins, it may facilitate connections between ciliary membrane proteins and the IFT complex before entry into the cilium. However, the exocyst also interacts with BLOC-1 proteins, so it may have roles in capturing ciliary cargos from the endosome (Fig. 10).
Figure 10.
Fibrocystin, polycystin-2, and smoothened take different trafficking routes to the primary cilium. (A) Summary of the trafficking rates of membrane protein delivery from the Golgi apparatus to the cilium. Membrane proteins are listed horizontally. shRNAs and MyoVb C-terminal tail expression are listed on the left side. (B) IFT20 localizes at the cis Golgi compartment where it interacts with the vesicle tethering golgin GMAP210. IFT20 also localizes to the medial Golgi compartment, the cilium, and the basal body. Subunits of the exocyst localize to the basal body. Exocyst subunits Exo70 and Sec8 interact with IFT20. BLOC-1 also interacts with the exocyst. The exocyst tethers vesicles containing ciliary membrane protein cargo to the base of the cilium before SNARE-mediated fusion. We propose that fibrocystin is using the direct trafficking pathway because it is affected only by knockdown of the IFT20-GMAP210 complex and the exocyst. Smoothened is likely using the lateral trafficking pathway because its delivery to cilia is not greatly affected by the knockdown of the IFT20-GMAP210 complex, and it is not affected by the knockdown of the exocyst or BLOC-1. The BLOC-1 subunit pallidin localizes to the basal body and its localization is partially dependent on IFT20. Pallidin interacts with IFT20 and polycystin-2. Knockdown of the BLOC-1 subunits pallidin and dysbindin affects the trafficking of polycystin-2 but not fibrocystin or smoothened. MyoVb C-terminal tail overexpression reduced ciliary polycystin-2 levels and caused its accumulation at the recycling endosome. Additionally, expression of dominant-negative Rab11aS25N perturbs polycystin-2 ciliary trafficking. This suggests that polycystin-2 is trafficked to the cilium through the recycling endosome. IFT20, BLOC-1, and the exocyst are acting together to deliver polycystin-2 cargo from the recycling endosome to the primary cilium.
The BLOC-1 complex, which was required for trafficking of polycystin-2 to cilia contains eight proteins (pallidin, dysbindin, muted, snapin, cappuccino, BLOS1, BLOS2, and BLOS3; Falcón-Pérez et al., 2002; Moriyama and Bonifacino, 2002; Ciciotte et al., 2003; Li et al., 2003; Starcevic and Dell’Angelica, 2004; Mullin et al., 2011). This complex is best known for trafficking of proteins from the endosome system to the lysosome and lysosome-related organelles such as melanosomes and platelet dense-granules (Di Pietro et al., 2006; Setty et al., 2007, 2008; Sitaram et al., 2012). BLOC-1 defects cause Hermansky-Pudlak syndrome, in which patients exhibit severe hypopigmentation and defects in platelet aggregation (Morgan et al., 2006). Recently BLOC-1 has been connected to schizophrenia, as a leading susceptibility gene encodes the dysbindin subunit, and brain tissue from schizophrenia patients has reduced dysbindin mRNA and protein levels (Talbot et al., 2004; Weickert et al., 2008; Mullin et al., 2011). The involvement of BLOC-1 in schizophrenia is not clear, but BLOC-1 is involved in dopamine and glutamate release (Numakawa et al., 2004; Kumamoto et al., 2006). Additionally, BLOC-1 regulates membrane protein targeting to synaptic vesicles and delivery of dopamine receptors to the cell surface (Iizuka et al., 2007; Ji et al., 2009; Newell-Litwa et al., 2009; Marley and von Zastrow, 2010; Larimore et al., 2011).
Before our work, the BLOC-1 complex had no ciliary connections. However, we found interactions of BLOC-1 components with IFT20 and polycystin-2 and a requirement for this complex in the transport of polycystin-2 to cilia. Our finding that BLOC-1 is required for ciliary trafficking of polycystin-2 to cilia suggests that defects in BLOC-1-encoding genes should cause cystic kidneys. Previous studies showed that BLOC-1 mouse models have lower rates of lysosomal enzyme secretion into urine, but no other analysis of kidney structure or function has been done (Novak and Swank, 1979). We found that young Dtnbp1sdy/sdy and Pldnpa/pa mice had dilations of the collecting ducts, suggesting that they may become cystic with age. A second cohort of mice was aged 12 months. The animals have lung fibrosis, and most did not survive the year (McGarry et al., 2002). The survivors did not have prominent cystic disease, unlike the phenotype of Pkd2 and Ift20 mutant animals (Wu et al., 1998; Jonassen et al., 2008). However, both Dtnbp1sdy/sdy and Pldnpa/pa animal models showed significant dilation of the collecting ducts and proximal tubules. Additionally, Dtnbp1sdy/sdy mice have reduced ciliation and shortened cilia, which is similar to what we observed in cell culture after knockdown. This suggests that that BLOC-1 defects trigger increased proliferation of tubule cells, but compensatory mechanisms keep the tubules from greatly expanding. We also noted increased numbers of abnormally positioned nuclei in the Dtnbp1sdy/sdy and Pldnpa/pa proximal tubules. The disrupted nuclei positioning in the Dtnbp1sdy/sdy and Pldnpa/pa animals may be indicative of a loss of cell polarization within the kidney tubule. Little is known about this phenotype but aberrant nuclei positioning is associated with pathology in muscular dystrophy and hearing loss (Sullivan et al., 1999; Zhang et al., 2007; Lei et al., 2009; Gundersen and Worman, 2013; Horn et al., 2013).
BLOC-1 localizes to recycling endosome tubules that extend from sorting endosomes, where it controls tubule formation and trafficking of membrane proteins to melanosomes (Di Pietro et al., 2006; Salazar et al., 2006; Setty et al., 2007; Ryder et al., 2013; Delevoye et al., 2016; Dennis et al., 2016). Given that BLOC-1 is involved in polycystin-2 trafficking, we hypothesized that polycystin-2 is traversing the recycling endosome compartment before delivery to the cilium. To perturb trafficking through the recycling endosome, we expressed the C-terminal tail of the myosin motor MyoVb (Lapierre et al., 2001; Volpicelli et al., 2002). This MyoVb fragment interacts with Rab11 but lacks motor activity (Lapierre et al., 2001). This construct caused polycystin-2 to accumulate in the recycling endosome and reduced the amount on cilia. In addition, expression of the Rab11aS25N dominant-negative form of Rab11a perturbs ciliary polycystin-2 trafficking. Although this is the first demonstration of a protein traveling through the recycling endosome to reach the cilium, the recycling endosome is involved in the early steps of cilia formation and the recycling endosome remains at the centrosome after ciliary assembly (Westlake et al., 2011; Hehnly et al., 2012; Hehnly and Doxsey, 2014). In addition to our data, other connections between the endosome system and trafficking of polycystin-2 to cilia are beginning to emerge. The lipid kinase PI3K-C2α localizes to the pericentriolar-recycling endosome at the base of the cilium and knockdown of this kinase reduced ciliary polycystin-2 (Franco et al., 2016). Similarly, knockdown of the endosome protein SDCCAG3 reduces ciliary polycystin-2 (Yu et al., 2016).
Summary
Our work uncovered previously unappreciated complexity in membrane protein trafficking pathways to cilia (Fig. 10). These pathways include an IFT20-independent pathway whereby proteins travel to the plasma membrane before lateral diffusion into the cilium, a second IFT20- and exocyst-dependent pathway from the Golgi complex to the ciliary base, and a third endosomal pathway whereby proteins traverse the recycling endosome on the way to the cilium. This complexity suggests that through evolution, the cell customized preexisting cellular trafficking pathways and machineries to generate distinct ciliary membrane protein trafficking routes.
Materials and methods
Cell culture
NRK, IMCD3 (Rauchman et al., 1993) and IMCD3 Flp-In (Mukhopadhyay et al., 2010) cells were grown in 47.5% DMEM (4.5 g/liter glucose) 47.5% F12, 5% FBS, 100 U/ml penicillin, and 100 g/ml streptomycin (all from Gibco) at 37°C in 5% CO2. MEK cells were grown in the same culture medium described above but with 10% FBS. NIH 3T3 cells were cultured in 90% DMEM (4.5 g/liter glucose), 10% FBS, 100 U/ml penicillin, and 100 g/ml streptomycin (Gibco). Cells were serum-starved for 48 h to induce ciliation in their described culture medium but with 0.25% FBS.
DNA constructs
The open reading frames of MmKXD1 (NM_029366.2), MmPallidin (NM_019788.3), and MmExo70 (AF014461.1) were amplified from mouse kidney cDNA using primers that placed them in frame with the Flag epitope pJAF113 (Follit et al., 2009) to generate constructs WJM1, WJM2, and FX70, respectively. Flag-GFP (JAF146), Flag-MmIFT54 (FX34), Flag-MmIFT25 (JAF143), and Flag-MmIFT20 (JAF134) were previously described (Follit et al., 2009). The OcMyoVb (AF176517.1) C-terminal tail was amplified from GFP-OcMyoVb C-terminus (Lapierre et al., 2001; Volpicelli et al., 2002) and cloned into pJAF113 to generate WJM30. Flag-MmRab11a (NM_017382.5) was amplified and Gibson-assembled into a lentiviral pHAGE vector to generate GP730. Gibson assembly mutagenesis was used to generate Flag-MmRab11aS25N (GP731) and Flag-MmRab11aQ70L (GP733). The coding sequences of HsCD8-MmFibrocystinCTS-SNAP (JAF271), HsPolycystin-2(1-703)-GFP-SNAP (pJag689) (a gift from J. Shah, Harvard Medical School, Boston, MA), and MmSmoothened-SNAP-GFP (JAF250) were amplified and TA-cloned into pCR8-GW-Topo vector (Invitrogen) to make entry clones. The entry clones were gateway-cloned into the destination vector pEF5B-FRT-DEST using Gateway LR Clonase II Enzyme Mix (Invitrogen) to generate the respective Flp-In expression clones WJM8, WJM15, and WJM6. All DNA constructs were verified by sequencing.
Flp-In system
The Flp-In system comprises a construct encoding the Flp recombinase pCAGGS-FlpE (Addgene) and the Flp-In expression clone in the pEF5B-FRT-DEST backbone, a gift from M. Nachury (Stanford University School of Medicine, Stanford, CA). pCAGGS-FlpE and the expression clone were electroporated (Bio-Rad Laboratories) into IMCD3 Flp-In cells (a gift from P. Jackson, Stanford University School of Medicine, Stanford, CA), in the following proportions: pCAGGS-FlpE, 9; expression clone: 1. Starting 48 h after electroporation, cells were drug-selected with 4 µg/ml blasticidin for 72 h. Colonies were recovered after 1–2 wk in culture and screened by immunofluorescence.
Lenti-shRNA production
All lenti-shRNAs are in the pGIPZ vector backbone (GE Healthcare). The lenti-shRNA system comprises two packaging vectors (psPAX2 and pMD2.G; Addgene). The virus was packaged by cotransfecting 1.2 × 106 human embryonic kidney 293T cells per well of a six-well cell culture plate with the lenti-shRNA pGIPZ vector and the two packaging vectors in the following proportions: pGIPZ, 2; psPAX2, 2; pMD2.G, 1. Effectene (QIAGEN) transfection reagent was used. After 48 h, the virus was harvested and filtered through a 0.45-µm filter. Virus was concentrated using Lenti-x concentrator (Takara Bio Inc.), and the pellet was resuspended in 80% DMEM (4.5 g/liter glucose), 20% FBS, 100 U/ml penicillin, 100 g/ml streptomycin, and 5 µg/ml polybrene (Sigma-Aldrich). Virus was finally added to cells plated at 1.25 × 105 cells per well of a six-well cell culture plate. Cells were infected for 72 h before being drug-selected with 1 µg/ml puromycin for 72 h.
Full hairpin sequence (mature antisense sequences are in boldface type): IFT20 shRNA #1: 5′-TGCTGTTGACAGTGAGCGCGGTCTAATTGAGCTTGTTGATTAGTGAAGCCACAGATGTAATCAACAAGCTCAATTAGACCATGCCTACTGCCTCGGA-3′; IFT20 shRNA #2: 5′-TGCTGTTGACAGTGAGCGAAAGGACTTTGTGGACAAAATTTAGTGAAGCCACAGATGTAAATTTTGTCCACAAAGTCCTTGTGCCTACTGCCTCGGA-3′; GMAP shRNA #1: 5′-TGCTGTTGACAGTGAGCGCCGGTTGACAGTGATAACAATTTAGTGAAGCCACAGATGTAAATTGTTATCACTGTCAACCGATGCCTACTGCCTCGGA-3′; GMAP shRNA #2: 5′-TGCTGTTGACAGTGAGCGACAGCTGTTTGCAGAAGATCAATAGTGAAGCCACAGATGTATTGATCTTCTGCAAACAGCTGCTGCCTACTGCCTCGGA-3′; Exo70 shRNA #1: 5′-TGCTGTTGACAGTGAGCGACTGGCTAAAGGTGACTGACTATAGTGAAGCCACAGATGTATAGTCAGTCACCTTTAGCCAGCTGCCTACTGCCTCGGA-3′; Exo70 shRNA #2: 5′-TGCTGTTGACAGTGAGCGACGCCATCTTCCTACACAACAATAGTGAAGCCACAGATGTATTGTTGTGTAGGAAGATGGCGCTGCCTACTGCCTCGGA-3′; Sec8 shRNA #1: 5′-TGCTGTTGACAGTGAGCGAATCGTGGAGAAGACAGTACAATAGTGAAGCCACAGATGTATTGTACTGTCTTCTCCACGATGTGCCTACTGCCTCGGA-3′; Sec8 shRNA #2: 5′-TGCTGTTGACAGTGAGCGAATCGTTCAGCACTACACAGAATAGTGAAGCCACAGATGTATTCTGTGTAGTGCTGAACGATCTGCCTACTGCCTCGGA-3′; Pallidin shRNA #1: 5′-TGCTGTTGACAGTGAGCGCACCAAGTTGTGTTACTAGATATAGTGAAGCCACAGATGTATATCTAGTAACACAACTTGGTTTGCCTACTGCCTCGGA-3′; Pallidin shRNA #2: 5′-TGCTGTTGACAGTGAGCGAGGCGATAGAGAGATCGATAAATAGTGAAGCCACAGATGTATTTATCGATCTCTCTATCGCCCTGCCTACTGCCTCGGA-3′; Dysbindin shRNA #1: 5′-TGCTGTTGACAGTGAGCGACAGGTGCTTAGAGGTTTTCAATAGTGAAGCCACAGATGTATTGAAAACCTCTAAGCACCTGGTGCCTACTGCCTCGGA-3′; Dysbindin shRNA #2: 5′-TGCTGTTGACAGTGAGCGCCCGAAGTACTCTGCTGGACTATAGTGAAGCCACAGATGTATAGTCCAGCAGAGTACTTCGGTTGCCTACTGCCTCGGA-3′.
pHAGE lentivirus production
Lentivirus production using the pHAGE system was previously described (Eguether et al., 2014). In brief, the pHAGE system uses four packaging vectors (Tat, Rev, Gag/Pol, and VSV-G). The virus was packaged by cotransfecting 1.2 × 106 HEK293T cells per well of a six-well cell culture plate with the backbone vector and the four packaging vectors in the following proportions: (backbone, 5; Tat, 0.5; Rev, 0.5; Gag/Pol, 0.5; VSV-G, 1) using Effectene (QIAGEN) transfection reagent. After 48 h, the virus was harvested and filtered through a 0.45-µm filter. Virus was concentrated using Lenti-x concentrator (Takara Bio Inc.) and resuspended in 47.5% DMEM (4.5 g/liter glucose) 47.5% F12, 10% FBS, 100 U/ml penicillin, 100 g/ml streptomycin, and 5 µg/ml Polybrene (Sigma-Aldrich). Virus was added to cells plated at 1.25 × 105 cells per well of a six-well cell culture plate. After 48 h, the medium was changed and the cells drug selected with 50 µg/ml Nourseothricin N-acetyl transferase (Sigma-Aldrich; Kochupurakkal and Iglehart, 2013) for 72 h.
Fluorescence pulse-chase trafficking assay
The fluorescence pulse-chase assay was previously described (Follit and Pazour, 2013; Follit et al., 2014). In brief, cells expressing DNA constructs fused to the SNAP tag were incubated with 0.04 µM cell-permeable nonfluorescent benzylguanine block (SNAP-Cell Block; New England Biolabs, Inc.) for 20 min to block all binding sites on the SNAP tag. The cells were then washed three times with complete growth media and were allowed to synthesize new protein for 1.5 h. Next, a final concentration of 20 mM Hepes, pH 7.4, and 150 µg/ml cycloheximide was added. The temperature was shifted to 19°C for 2 h to allow accumulation of newly synthesized protein at the Golgi apparatus. The cells were shifted back to 37°C, allowing newly synthesized protein to be released from the Golgi. Cells were fixed at the indicated time points and labeled with 0.3 µM fluorescent SNAP-TMR STAR (New England Biolabs, Inc.).
Immunofluorescence
Immunofluorescence microscopy was described previously (Follit et al., 2006). Cells were grown on coverslips and fixed for 15 min with 2% paraformaldehyde in PHEM (0.05 M Pipes, 0.025 M Hepes, 0.01 M EGTA, and 0.01 M MgCl2, pH 7.2) followed by membrane permeabilization with 0.1% Triton X-100 in PHEM with 2% paraformaldehyde for 2 min. In some instances, cells were treated with 0.05% SDS in PBS for 5 min to recover antigens. To visualize basal bodies, cells were preextracted with 0.1% Triton X-100 in PHEM for 1 min, followed by fixation in cold 100% methanol for 10 min. Cells were washed with 1× PBS and blocked in 1% BSA in 1× TBST (150 mM NaCl, 1% Tween 20, and 50 mM Tris, pH 7.5) for 1 h. Primary antibodies were diluted in blocking solution and incubated for 2 h at room temperature. Cells were washed four times with blocking solution over 20 min. Next, they were incubated with 1:2,000 dilutions of either Alexa Fluor 488–, 568–, 594–, 647–, and 680–conjugated IgG anti-mouse or IgG anti-rabbit (Thermo Fisher Scientific), SNAP TMR STAR (New England Biolabs, Inc.) for 1 h. Cells were then washed again with blocking solution, followed by three washes with 1× PBS. Coverslips were mounted onto slides with prolong medium (Molecular Probes).
Immunoprecipitations and immunoblotting
Immunoprecipitation assays were previously described (Follit et al., 2009). Cells were washed once with cold 1× PBS + 1 mM PMSF. They were lysed with CelLytic buffer (Sigma-Aldrich) supplemented with 0.1% NP-40 (Sigma-Aldrich), 0.1% CHAPSO (Bio-Rad Laboratories), and 1× Complete Protease Inhibitors (Roche), then rotated at 4°C for 10 min. After centrifugation, the supernatant was incubated with prewashed Anti-Flag M2 affinity gel beads (Sigma-Aldrich) for 90 min at 4°C. The beads were washed three times with TBST (300 mM NaCl, 1% Tween 20, and 50 mM Tris, pH 7.5), followed by three washes with TBS (150 mM NaCl and 50 mM Tris, pH 7.5). Flag-tagged proteins were eluted from the beads using 200 µg/ml 3× Flag peptide (Sigma-Aldrich).
Immunoblotting was performed as previously described (Pazour et al., 1998). Total-cell lysates were harvested by washing the cells once with cold 1× PBS, then scraping the cells into protein-denaturing buffer and passing it through a 22-gauge needle. Samples were run on SDS-PAGE and transferred to polyvinylidene difluoride membrane at 4°C overnight. Polyvinylidene difluoride was equilibrated with 1× TBST (150 mM NaCl, 1% Tween 20, and 50 mM Tris, pH 7.5) for 10 min, and membranes were blocked with 5% nonfat dry milk in 1× TBST supplemented with 1% fish gelatin (Sigma-Aldrich) for 30 min. Primary antibodies were diluted in the blocking solution as recommended by the manufacturer and incubated with the membrane for 2 h at room temperature. Goat anti–rabbit IgG (H+L) or goat anti–mouse IgG (H+L), HRP-conjugated secondary antibodies (Thermo Fisher Scientific), and Dura Western Substrate (Thermo Fisher Scientific) were used to detect the primary antibodies. Immunoblot images were acquired using either a LAS-3000 imaging system (Fujifilm) or a Molecular Imager Chemi Doc XRS+ imaging system (Bio-Rad Laboratories).
Primary antibodies used include anti-MmPallidin (a gift from E. Dell'Angelica, University of California, Los Angeles School of Medicine, Los Angeles, CA; Nazarian et al., 2006), anti-MmDTNBP1 (11132-1-AP; Proteintech), anti-MmExo70 (70X13F3; Sigma-Aldrich), anti-MmSec8 (14G1; Stressgen), anti–γ tubulin (GTU88; Sigma-Aldrich), anti–γ tubulin (a gift from S. Doxsey, University of Massachusetts Medical School, Worcester, MA; Zheng et al., 1995), anti-SpAcetylated tubulin (6-11B-1; Sigma-Aldrich), anti-Flag (F1804; Sigma-Aldrich), anti-MmArl13b (N295B/66; Davis/National Institutes of Health NeuroMab Facility), anti-MmSmoothened (E-5; Santa Cruz Biotechnology, Inc.), anti-MmAQP2 (SAB5200110; Sigma-Aldrich), anti-MmRab11 (47; BD), anti-MmRab11 (20229-1-AP; Proteintech), anti-MmIFT20 (Pazour et al., 2002a), anti-MmGMAP210 (Follit et al., 2008), anti-MmPkd2 (Pazour et al., 2002b), anti-MmIFT27 (Follit et al., 2009), and anti-MmIFT88 (Pazour et al., 2002a).
Mouse breeding
All mouse work was approved by the Institutional Animal Care and Use Committee at Emory University. All mice were in C57BL/6 background and are described in Larimore et al. (2014).
Histology
Harvesting and fixing tissues
Wild-type and Dtnbp1sdy/sdy and Pldnpa/pa mice were sacrificed with CO2 narcosis followed by cervical dislocation. Kidneys were harvested and fixed in 4% paraformaldehyde in 1× PBS overnight at 4°C, followed by paraffin embedding (Jonassen et al., 2008).
Hematoxylin and eosin staining
Paraffin tissue sections were dewaxed using SafeClear (Thermo Fisher Scientific) and rehydrated with graded aqueous solutions of ethanol. The sections were stained in CAT Hematoxylin solution (Biocare Medical) for 8 min followed by washing in running tap water for 5 min and differentiation in 2% hydrochloric acid in 70% ethanol for 30 s. The sections were washed in running tap water for 1 min before Bluing in Tacha’s Bluing Solution (Biocare Medical) for 1 min followed by washing in running tap water for 5 min. Next, the samples were dipped ten times in 95% ethanol, counterstained in Edgar Degas Eosin solution (Biocare Medical) for 1 min and washed three times in 100% ethanol for 1 min. Last, the sections were cleared in two changes of xylene for 5 min and mounted with Permount (Thermo Fisher Scientific; San Agustin et al., 2016).
Immunohistochemistry
Paraffin sections were dewaxed and rehydrated as described above. Antigen retrieval was performed in 10 mM sodium citrate, pH 6.0, in an autoclave for 40 min at 121°C. Samples were cooled to ambient temperature and equilibrated in 1× TBS (50 mM TBS, pH 7.4) for 5 min, followed by blocking in 4% nonimmune goat serum, 0.1% cold water fish skin gelatin (Sigma-Aldrich), and 0.1% Triton X-100, in 1× TBST (150 mM NaCl, 1% Tween 20, and 50 mM Tris, pH 7.5) for 30 min. Sections were washed in 1× TBST and incubated with primary antibodies or 488-conjugated Lotus tetragonolobus agglutinin (Vector Laboratories) diluted in 0.1% cold water fish skin gelatin in 1× TBST overnight at 4°C. Next, samples were washed with 1× TBST and incubated with 1:1,000 dilutions of secondary antibodies diluted in 0.1% cold water fish skin gelatin in 1× TBST for 1 h. Last, the sections were washed with 1× TBST, rinsed with 1× TBS, dipped in 1× TBS supplemented with 1 mg/ml DAPI for 5 s, and mounted with Prolong Gold (Thermo Fisher Scientific; San Agustin et al., 2016).
Microscopy
Wide-field images were captured using an Orca ER camera on a Zeiss Axiovert 200 M microscope equipped with either an EC plan-Neofluar 40×/1.3 NA oil or Plan-Apochromat 100×/1.4 NA oil objective (ZEISS) using Openlab software (PerkinElmer). Immunohistochemistry images were captured using a DFC365 FX camera (Leica Biosystems) on a TCS SPE DM 500 Q confocal microscope (Leica Biosystems) equipped with an ACS Apo 40X/1.15 NA oil objective (Leica Biosystems). Z-stacks were acquired using Leica Biosystems Application Suite Advanced Fluorescence software and converted to single planes by maximum projection with ImageJ software (National Institutes of Health). Bright-field images were acquired using an Axioskop 2 Plus (ZEISS) using an N-Achroplan 2.5×/0.07 NA air ZEISS objective equipped with an Axiocam HRC color digital camera and Axiovision acquisition software. All images were acquired at room temperature.
Data analysis
Quantification of fluorescent pixel intensity, immunoblot pixel intensity, and length were measured using the measurement tools of ImageJ. Linear regression and statistical analysis were determined using GraphPad Prism software. Linear regression analysis of newly synthesized membrane protein delivery to the cilium was performed on selected time points: 0, 1, and 2 h for CD8-fibrocystinCTS-SNAP; 0, 1, 2, 4, and 6 h for polycystin-2-GFP-SNAP; and 0, 1, 2, 4, and 6 h for smoothened-SNAP-GFP. The time points were chosen by determining which data points remained linear before (or if) reaching a plateau in the control groups. The slope values obtained from this analysis represent the trafficking rates of newly synthesized membrane protein delivery to the cilium. Slope values between control and experimental groups were compared with one another to determine statistical significance. Data were subjected to either unpaired Student’s t test or one-way ANOVA and Bonferroni or Tukey multiple comparisons test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Data distribution was assumed to be normal but was not formally tested.
Online supplemental material
Fig. S1 depicts schematics of the ciliary membrane protein CD8/GFP and SNAP tag constructs, ciliary membrane protein trafficking rates, and cilia length after temperature shift to 19°C. Fig. S2 depicts ciliary membrane protein trafficking assays during GMAP210 shRNA knockdown. Fig. S3 shows ciliary membrane protein trafficking assays during exocyst shRNA knockdown. Fig. S4 shows ciliary membrane protein trafficking assays during BLOC-1 shRNA knockdown. Fig. S5 depicts steady-state ciliary levels of polycystin-2 and Arl13b during exocyst shRNA knockdown. Fig. S6 depicts steady-state basal body levels of IFT20 during pallidin or Exo70 shRNA knockdown. Fig. S7 shows kidney collecting duct diameter and proximal tubule circumference in young Dtnbp1sdy/sdy and Pldnpa/pa mice. Fig. S8 shows that dominant-negative MyoVb colocalizes with Rab11 and causes the Golgi complex to compact. Fig. S9 shows that polycystin-2 accumulates in Rab11-positive endosomes when dominant-negative MyoVb is expressed. Fig. S10 illustrates that polycystin-2 is not detected in Rab11-positive endosomes when either the exocyst or BLOC-1 is knocked down. Fig. S11 shows that dominant-negative MyoVb expression does not affect fibrocystin or smoothened trafficking to the primary cilium.
Supplementary Material
Acknowledgments
We thank Dr. Jovenal San Agustin for assistance with histology and Drs. Maxence Nachury, Peter Jackson, Jagesh Shah, Esteban Dell'Angelica, and Stephen Doxsey for sharing reagents.
This work was supported by funding from the National Institutes of Health grants GM060992 and DK103632 to G.J. Pazour and GM077569 and NS088503 to V. Faundez.
The authors declare no competing financial interests.
Author contributions: W.J. Monis and G.J. Pazour conceived the project and designed experiments. W.J. Monis performed all experimental research and data analysis. V. Faundez contributed advice, mouse tissue, and other reagents. W.J. Monis, V. Faundez, and G.J. Pazour wrote the manuscript.
Footnotes
Abbreviations used:
- BLOC-1
- biogenesis of lysosome-related organelles complex-1
- IFT
- intraflagellar transport
- IMCD3
- inner medullary collecting duct
- MEK
- mouse embryonic kidney
- NRK
- normal rat kidney
- PKD
- polycystic kidney disease
References
- Alcedo J., Ayzenzon M., Von Ohlen T., Noll M., and Hooper J.E.. 1996. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell. 86:221–232. 10.1016/S0092-8674(00)80094-X [DOI] [PubMed] [Google Scholar]
- Ang A.L., Taguchi T., Francis S., Fölsch H., Murrells L.J., Pypaert M., Warren G., and Mellman I.. 2004. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 167:531–543. 10.1083/jcb.200408165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besharse J.C., and Horst C.J.. 1990. The photoreceptor connecting cilium: a model for the transition zone. In Ciliary and Flagellar Membranes. Bloodgood R.A., editor. Plenum, New York: 389–417. [Google Scholar]
- Bouck G.B. 1971. The structure, origin, isolation, and composition of the tubular mastigonemes of the Ochromonas flagellum. J. Cell Biol. 50:362–384. 10.1083/jcb.50.2.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M., Ning J., Hernandez-Lara C.I., Belzile O., Wang Q., Dutcher S.K., Liu Y., and Snell W.J.. 2015. Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. eLife. 4:4 10.7554/eLife.05242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik S., Hori Y., Kaplan O.I., Kida K., Toivenon T., Foley-Fisher C., Cottell D., Katada T., Kontani K., and Blacque O.E.. 2010. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J. Cell Biol. 188:953–969. 10.1083/jcb.200908133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciciotte S.L., Gwynn B., Moriyama K., Huizing M., Gahl W.A., Bonifacino J.S., and Peters L.L.. 2003. Cappuccino, a mouse model of Hermansky-Pudlak syndrome, encodes a novel protein that is part of the pallidin-muted complex (BLOC-1). Blood. 101:4402–4407. 10.1182/blood-2003-01-0020 [DOI] [PubMed] [Google Scholar]
- Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., and Reiter J.F.. 2005. Vertebrate smoothened functions at the primary cilium. Nature. 437:1018–1021. 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- Crouse J.A., Lopes V.S., Sanagustin J.T., Keady B.T., Williams D.S., and Pazour G.J.. 2014. Distinct functions for IFT140 and IFT20 in opsin transport. Cytoskeleton. 71:302–310. 10.1002/cm.21173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C., Heiligenstein X., Ripoll L., Gilles-Marsens F., Dennis M.K., Linares R.A., Derman L., Gokhale A., Morel E., Faundez V., et al. 2016. BLOC-1 brings together the actin and microtubule cytoskeletons to generate recycling endosomes. Curr. Biol. 26:1–13. 10.1016/j.cub.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M.K., Delevoye C., Acosta-Ruiz A., Hurbain I., Romao M., Hesketh G.G., Goff P.S., Sviderskaya E.V., Bennett D.C., Luzio J.P., et al. 2016. BLOC-1 and BLOC-3 regulate VAMP7 cycling to and from melanosomes via distinct tubular transport carriers. J. Cell Biol. 214:293–308. 10.1083/jcb.201605090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic D., Huber L.A., Ransom N., Mancini M., Simons K., and Papermaster D.S.. 1995. rab8 in retinal photoreceptors may participate in rhodopsin transport and in rod outer segment disk morphogenesis. J. Cell Sci. 108:215–224. [DOI] [PubMed] [Google Scholar]
- Di Pietro S.M., Falcón-Pérez J.M., Tenza D., Setty S.R., Marks M.S., Raposo G., and Dell’Angelica E.C.. 2006. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol. Biol. Cell. 17:4027–4038. 10.1091/mbc.E06-05-0379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T., San Agustin J.T., Keady B.T., Jonassen J.A., Liang Y., Francis R., Tobita K., Johnson C.A., Abdelhamed Z.A., Lo C.W., and Pazour G.J.. 2014. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev. Cell. 31:279–290. 10.1016/j.devcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón-Pérez J.M., Starcevic M., Gautam R., and Dell’Angelica E.C.. 2002. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J. Biol. Chem. 277:28191–28199. 10.1074/jbc.M204011200 [DOI] [PubMed] [Google Scholar]
- Fogelgren B., Lin S.Y., Zuo X., Jaffe K.M., Park K.M., Reichert R.J., Bell P.D., Burdine R.D., and Lipschutz J.H.. 2011. The exocyst protein Sec10 interacts with polycystin-2 and knockdown causes PKD-phenotypes. PLoS Genet. 7:e1001361 10.1371/journal.pgen.1001361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogelgren B., Polgar N., Lui V.H., Lee A.J., Tamashiro K.K., Napoli J.A., Walton C.B., Zuo X., and Lipschutz J.H.. 2015. Urothelial defects from targeted inactivation of exocyst Sec10 in mice cause ureteropelvic junction obstructions. PLoS One. 10:e0129346 10.1371/journal.pone.0129346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., and Pazour G.J.. 2013. Analysis of ciliary membrane protein dynamics using SNAP technology. Methods Enzymol. 524:195–204. 10.1016/B978-0-12-397945-2.00011-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., Tuft R.A., Fogarty K.E., and Pazour G.J.. 2006. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 17:3781–3792. 10.1091/mbc.E06-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., San Agustin J.T., Xu F., Jonassen J.A., Samtani R., Lo C.W., and Pazour G.J.. 2008. The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genet. 4:e1000315 10.1371/journal.pgen.1000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., Xu F., Keady B.T., and Pazour G.J.. 2009. Characterization of mouse IFT complex B. Cell Motil. Cytoskeleton. 66:457–468. 10.1002/cm.20346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., Li L., Vucica Y., and Pazour G.J.. 2010. The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J. Cell Biol. 188:21–28. 10.1083/jcb.200910096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., San Agustin J.T., Jonassen J.A., Huang T., Rivera-Perez J.A., Tremblay K.D., and Pazour G.J.. 2014. Arf4 is required for mammalian development but dispensable for ciliary assembly. PLoS Genet. 10:e1004170 10.1371/journal.pgen.1004170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco I., Margaria J.P., De Santis M.C., Ranghino A., Monteyne D., Chiaravalli M., Pema M., Campa C.C., Ratto E., Gulluni F., et al. 2016. Phosphoinositide 3-kinase-C2α regulates polycystin-2 ciliary entry and protects against kidney cyst formation. J. Am. Soc. Nephrol. 27:1135–1144. 10.1681/ASN.2014100967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A., Larimore J., Werner E., So L., Moreno-De-Luca A., Lese-Martin C., Lupashin V.V., Smith Y., and Faundez V.. 2012. Quantitative proteomic and genetic analyses of the schizophrenia susceptibility factor dysbindin identify novel roles of the biogenesis of lysosome-related organelles complex 1. J. Neurosci. 32:3697–3711. 10.1523/JNEUROSCI.5640-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindstaff K.K., Yeaman C., Anandasabapathy N., Hsu S.C., Rodriguez-Boulan E., Scheller R.H., and Nelson W.J.. 1998. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell. 93:731–740. 10.1016/S0092-8674(00)81435-X [DOI] [PubMed] [Google Scholar]
- Gundersen G.G., and Worman H.J.. 2013. Nuclear positioning. Cell. 152:1376–1389. 10.1016/j.cell.2013.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P.C., and Torres V.E.. 2009. Polycystic kidney disease. Annu. Rev. Med. 60:321–337. 10.1146/annurev.med.60.101707.125712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M.J., Bryon K., Satkurunathan J., and Levine T.P.. 2011. Yeast homologues of three BLOC-1 subunits highlight KxDL proteins as conserved interactors of BLOC-1. Traffic. 12:260–268. 10.1111/j.1600-0854.2010.01151.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Xi F., Zhang X., Zhang J., and Guo W.. 2007. Exo70 interacts with phospholipids and mediates the targeting of the exocyst to the plasma membrane. EMBO J. 26:4053–4065. 10.1038/sj.emboj.7601834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H., and Doxsey S.. 2014. Rab11 endosomes contribute to mitotic spindle organization and orientation. Dev. Cell. 28:497–507. 10.1016/j.devcel.2014.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H., Chen C.T., Powers C.M., Liu H.L., and Doxsey S.. 2012. The centrosome regulates the Rab11- dependent recycling endosome pathway at appendages of the mother centriole. Curr. Biol. 22:1944–1950. 10.1016/j.cub.2012.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider M.R., and Munson M.. 2012. Exorcising the exocyst complex. Traffic. 13:898–907. 10.1111/j.1600-0854.2012.01353.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider M.R., Gu M., Duffy C.M., Mirza A.M., Marcotte L.L., Walls A.C., Farrall N., Hakhverdyan Z., Field M.C., Rout M.P., et al. 2016. Subunit connectivity, assembly determinants and architecture of the yeast exocyst complex. Nat. Struct. Mol. Biol. 23:59–66. 10.1038/nsmb.3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgendorf K.I., Johnson C.T., and Jackson P.K.. 2016. The primary cilium as a cellular receiver: organizing ciliary GPCR signaling. Curr. Opin. Cell Biol. 39:84–92. 10.1016/j.ceb.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn H.F., Brownstein Z., Lenz D.R., Shivatzki S., Dror A.A., Dagan-Rosenfeld O., Friedman L.M., Roux K.J., Kozlov S., Jeang K.T., et al. 2013. The LINC complex is essential for hearing. J. Clin. Invest. 123:740–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Kuo Y.M., and Gitschier J.. 1999. The pallid gene encodes a novel, syntaxin 13-interacting protein involved in platelet storage pool deficiency. Nat. Genet. 23:329–332. 10.1038/15507 [DOI] [PubMed] [Google Scholar]
- Hunnicutt G.R., Kosfiszer M.G., and Snell W.J.. 1990. Cell body and flagellar agglutinins in Chlamydomonas reinhardtii: the cell body plasma membrane is a reservoir for agglutinins whose migration to the flagella is regulated by a functional barrier. J. Cell Biol. 111:1605–1616. 10.1083/jcb.111.4.1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., et al. 2015. The BioPlex Network: a systematic exploration of the human interactome. Cell. 162:425–440. 10.1016/j.cell.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka Y., Sei Y., Weinberger D.R., and Straub R.E.. 2007. Evidence that the BLOC-1 protein dysbindin modulates dopamine D2 receptor internalization and signaling but not D1 internalization. J. Neurosci. 27:12390–12395. 10.1523/JNEUROSCI.1689-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Yang F., Papaleo F., Wang H.X., Gao W.J., Weinberger D.R., and Lu B.. 2009. Role of dysbindin in dopamine receptor trafficking and cortical GABA function. Proc. Natl. Acad. Sci. U S A. 106:19593–19598. 10.1073/pnas.0904289106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen J.A., San Agustin J., Follit J.A., and Pazour G.J.. 2008. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J. Cell Biol. 183:377–384. 10.1083/jcb.200808137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady B.T., Le Y.Z., and Pazour G.J.. 2011. IFT20 is required for opsin trafficking and photoreceptor outer segment development. Mol. Biol. Cell. 22:921–930. 10.1091/mbc.E10-09-0792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y., Yoo J.S., Hazuka C.D., Peterson K.E., Hsu S.C., and Scheller R.H.. 1997. Subunit structure of the mammalian exocyst complex. Proc. Natl. Acad. Sci. U S A. 94:14438–14443. 10.1073/pnas.94.26.14438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochupurakkal B.S., and Iglehart J.D.. 2013. Nourseothricin N-acetyl transferase: a positive selection marker for mammalian cells. PLoS One. 8:e68509 10.1371/journal.pone.0068509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto N., Matsuzaki S., Inoue K., Hattori T., Shimizu S., Hashimoto R., Yamatodani A., Katayama T., and Tohyama M.. 2006. Hyperactivation of midbrain dopaminergic system in schizophrenia could be attributed to the down-regulation of dysbindin. Biochem. Biophys. Res. Commun. 345:904–909. 10.1016/j.bbrc.2006.04.163 [DOI] [PubMed] [Google Scholar]
- Lapierre L.A., Kumar R., Hales C.M., Navarre J., Bhartur S.G., Burnette J.O., Provance D.W. Jr., Mercer J.A., Bähler M., and Goldenring J.R.. 2001. Myosin vb is associated with plasma membrane recycling systems. Mol. Biol. Cell. 12:1843–1857. 10.1091/mbc.12.6.1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimore J., Tornieri K., Ryder P.V., Gokhale A., Zlatic S.A., Craige B., Lee J.D., Talbot K., Pare J.F., Smith Y., and Faundez V.. 2011. The schizophrenia susceptibility factor dysbindin and its associated complex sort cargoes from cell bodies to the synapse. Mol. Biol. Cell. 22:4854–4867. 10.1091/mbc.E11-07-0592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimore J., Zlatic S.A., Gokhale A., Tornieri K., Singleton K.S., Mullin A.P., Tang J., Talbot K., and Faundez V.. 2014. Mutations in the BLOC-1 subunits dysbindin and muted generate divergent and dosage-dependent phenotypes. J. Biol. Chem. 289:14291–14300. 10.1074/jbc.M114.553750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K., Zhang X., Ding X., Guo X., Chen M., Zhu B., Xu T., Zhuang Y., Xu R., and Han M.. 2009. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc. Natl. Acad. Sci. U S A. 106:10207–10212. 10.1073/pnas.0812037106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang Q., Oiso N., Novak E.K., Gautam R., O’Brien E.P., Tinsley C.L., Blake D.J., Spritz R.A., Copeland N.G., et al. 2003. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat. Genet. 35:84–89. 10.1038/ng1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wei Q., Zhang Y., Ling K., and Hu J.. 2010. The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J. Cell Biol. 189:1039–1051. 10.1083/jcb.200912001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Zhang J., and Guo W.. 2014. The role of Sec3p in secretory vesicle targeting and exocyst complex assembly. Mol. Biol. Cell. 25:3813–3822. 10.1091/mbc.E14-04-0907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marley A., and von Zastrow M.. 2010. Dysbindin promotes the post-endocytic sorting of G protein-coupled receptors to lysosomes. PLoS One. 5:e9325 10.1371/journal.pone.0009325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazelova J., Ransom N., Astuto-Gribble L., Wilson M.C., and Deretic D.. 2009. Syntaxin 3 and SNAP-25 pairing, regulated by omega-3 docosahexaenoic acid, controls the delivery of rhodopsin for the biogenesis of cilia-derived sensory organelles, the rod outer segments. J. Cell Sci. 122:2003–2013. 10.1242/jcs.039982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry M.P., Borchers M., Novak E.K., Lee N.A., Ohtake P.J., Lee J.J., and Swank R.T.. 2002. Pulmonary pathologies in pallid mice result from nonhematopoietic defects. Exp. Mol. Pathol. 72:213–220. 10.1006/exmp.2002.2431 [DOI] [PubMed] [Google Scholar]
- Milenkovic L., Scott M.P., and Rohatgi R.. 2009. Lateral transport of smoothened from the plasma membrane to the membrane of the cilium. J. Cell Biol. 187:365–374. 10.1083/jcb.200907126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki T., Wu G., Hayashi T., Xenophontos S.L., Veldhuisen B., Saris J.J., Reynolds D.M., Cai Y., Gabow P.A., Pierides A., et al. 1996. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 272:1339–1342. 10.1126/science.272.5266.1339 [DOI] [PubMed] [Google Scholar]
- Morgan N.V., Pasha S., Johnson C.A., Ainsworth J.R., Eady R.A., Dawood B., McKeown C., Trembath R.C., Wilde J., Watson S.P., and Maher E.R.. 2006. A germline mutation in BLOC1S3/reduced pigmentation causes a novel variant of Hermansky-Pudlak syndrome (HPS8). Am. J. Hum. Genet. 78:160–166. 10.1086/499338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K., and Bonifacino J.S.. 2002. Pallidin is a component of a multi-protein complex involved in the biogenesis of lysosome-related organelles. Traffic. 3:666–677. 10.1034/j.1600-0854.2002.30908.x [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Chih B., Nelson C.D., Lane W.S., Scales S.J., and Jackson P.K.. 2010. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24:2180–2193. 10.1101/gad.1966210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin A.P., Gokhale A., Larimore J., and Faundez V.. 2011. Cell biology of the BLOC-1 complex subunit dysbindin, a schizophrenia susceptibility gene. Mol. Neurobiol. 44:53–64. 10.1007/s12035-011-8183-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peränen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., and Jackson P.K.. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213. 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Nachury M.V., Seeley E.S., and Jin H.. 2010. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu. Rev. Cell Dev. Biol. 26:59–87. 10.1146/annurev.cellbio.042308.113337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R., Starcevic M., Spencer M.J., and Dell’Angelica E.C.. 2006. Reinvestigation of the dysbindin subunit of BLOC-1 (biogenesis of lysosome-related organelles complex-1) as a dystrobrevin-binding protein. Biochem. J. 395:587–598. 10.1042/BJ20051965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa K., Salazar G., Smith Y., and Faundez V.. 2009. Roles of BLOC-1 and adaptor protein-3 complexes in cargo sorting to synaptic vesicles. Mol. Biol. Cell. 20:1441–1453. 10.1091/mbc.E08-05-0456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak E.K., and Swank R.T.. 1979. Lysosomal dysfunctions associated with mutations at mouse pigment genes. Genetics. 92:189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numakawa T., Yagasaki Y., Ishimoto T., Okada T., Suzuki T., Iwata N., Ozaki N., Taguchi T., Tatsumi M., Kamijima K., et al. 2004. Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Hum. Mol. Genet. 13:2699–2708. 10.1093/hmg/ddh280 [DOI] [PubMed] [Google Scholar]
- Papermaster D.S., Schneider B.G., and Besharse J.C.. 1985. Vesicular transport of newly synthesized opsin from the Golgi apparatus toward the rod outer segment. Ultrastructural immunocytochemical and autoradiographic evidence in Xenopus retinas. Invest. Ophthalmol. Vis. Sci. 26:1386–1404. [PubMed] [Google Scholar]
- Pazour G.J., Wilkerson C.G., and Witman G.B.. 1998. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141:979–992. 10.1083/jcb.141.4.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Baker S.A., Deane J.A., Cole D.G., Dickert B.L., Rosenbaum J.L., Witman G.B., and Besharse J.C.. 2002a The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 157:103–113. 10.1083/jcb.200107108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., San Agustin J.T., Follit J.A., Rosenbaum J.L., and Witman G.B.. 2002b Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr. Biol. 12:R378–R380. 10.1016/S0960-9822(02)00877-1 [DOI] [PubMed] [Google Scholar]
- Rauchman M.I., Nigam S.K., Delpire E., and Gullans S.R.. 1993. An osmotically tolerant inner medullary collecting duct cell line from an SV40 transgenic mouse. Am. J. Physiol. 265:F416–F424. [DOI] [PubMed] [Google Scholar]
- Rogers K.K., Wilson P.D., Snyder R.W., Zhang X., Guo W., Burrow C.R., and Lipschutz J.H.. 2004. The exocyst localizes to the primary cilium in MDCK cells. Biochem. Biophys. Res. Commun. 319:138–143. 10.1016/j.bbrc.2004.04.165 [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L., and Scott M.P.. 2007. Patched1 regulates hedgehog signaling at the primary cilium. Science. 317:372–376. 10.1126/science.1139740 [DOI] [PubMed] [Google Scholar]
- Rosenbaum J.L., and Witman G.B.. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3:813–825. 10.1038/nrm952 [DOI] [PubMed] [Google Scholar]
- Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. 2005. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 437:1173–1178. 10.1038/nature04209 [DOI] [PubMed] [Google Scholar]
- Ryder P.V., Vistein R., Gokhale A., Seaman M.N., Puthenveedu M.A., and Faundez V.. 2013. The WASH complex, an endosomal Arp2/3 activator, interacts with the Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type IIα. Mol. Biol. Cell. 24:2269–2284. 10.1091/mbc.E13-02-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G., Craige B., Styers M.L., Newell-Litwa K.A., Doucette M.M., Wainer B.H., Falcon-Perez J.M., Dell’Angelica E.C., Peden A.A., Werner E., and Faundez V.. 2006. BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 cargoes. Mol. Biol. Cell. 17:4014–4026. 10.1091/mbc.E06-02-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Agustin J.T., Klena N., Granath K., Panigrahy A., Stewart E., Devine W., Strittmatter L., Jonassen J.A., Liu X., Lo C.W., and Pazour G.J.. 2016. Genetic link between renal birth defects and congenital heart disease. Nat. Commun. 7:11103 10.1038/ncomms11103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattar S., and Gleeson J.G.. 2011. The ciliopathies in neuronal development: a clinical approach to investigation of Joubert syndrome and Joubert syndrome-related disorders. Dev. Med. Child Neurol. 53:793–798. 10.1111/j.1469-8749.2011.04021.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seixas C., Choi S.Y., Polgar N., Umberger N.L., East M.P., Zuo X., Moreiras H., Ghossoub R., Benmerah A., Kahn R.A., et al. 2016. Arl13b and the exocyst interact synergistically in ciliogenesis. Mol. Biol. Cell. 27:308–320. 10.1091/mbc.E15-02-0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty S.R., Tenza D., Truschel S.T., Chou E., Sviderskaya E.V., Theos A.C., Lamoreux M.L., Di Pietro S.M., Starcevic M., Bennett D.C., et al. 2007. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol. Biol. Cell. 18:768–780. 10.1091/mbc.E06-12-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty S.R., Tenza D., Sviderskaya E.V., Bennett D.C., Raposo G., and Marks M.S.. 2008. Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature. 454:1142–1146. 10.1038/nature07163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram A., Dennis M.K., Chaudhuri R., De Jesus-Rojas W., Tenza D., Setty S.R., Wood C.S., Sviderskaya E.V., Bennett D.C., Raposo G., et al. 2012. Differential recognition of a dileucine-based sorting signal by AP-1 and AP-3 reveals a requirement for both BLOC-1 and AP-3 in delivery of OCA2 to melanosomes. Mol. Biol. Cell. 23:3178–3192. 10.1091/mbc.E11-06-0509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic M., and Dell’Angelica E.C.. 2004. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 279:28393–28401. 10.1074/jbc.M402513200 [DOI] [PubMed] [Google Scholar]
- Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., and Burke B.. 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147:913–920. 10.1083/jcb.147.5.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Zhang A., Baker B., Sun L., Howard A., Buswell J., Maurel D., Masharina A., Johnsson K., Noren C.J., et al. 2011. Development of SNAP-tag fluorogenic probes for wash-free fluorescence imaging. ChemBioChem. 12:2217–2226. 10.1002/cbic.201100173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Amsterdam A., Pazour G.J., Cole D.G., Miller M.S., and Hopkins N.. 2004. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 131:4085–4093. 10.1242/dev.01240 [DOI] [PubMed] [Google Scholar]
- Talbot K., Eidem W.L., Tinsley C.L., Benson M.A., Thompson E.W., Smith R.J., Hahn C.G., Siegel S.J., Trojanowski J.Q., Gur R.E., et al. 2004. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J. Clin. Invest. 113:1353–1363. 10.1172/JCI200420425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush D.R., and Novick P.. 1995. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J. Cell Biol. 130:299–312. 10.1083/jcb.130.2.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush D.R., Maurice T., Roth D., and Novick P.. 1996. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M., and Ingham P.W.. 1996. smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 382:547–551. 10.1038/382547a0 [DOI] [PubMed] [Google Scholar]
- Volpicelli L.A., Lah J.J., Fang G., Goldenring J.R., and Levey A.I.. 2002. Rab11a and myosin Vb regulate recycling of the M4 muscarinic acetylcholine receptor. J. Neurosci. 22:9776–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Huo K., Ma L., Tang L., Li D., Huang X., Yuan Y., Li C., Wang W., Guan W., et al. 2011. Toward an understanding of the protein interaction network of the human liver. Mol. Syst. Biol. 7:536 10.1038/msb.2011.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wei H., Yu Y., Sun J., Yang Y., Xing G., Wu S., Zhou Y., Zhu Y., Zhang C., et al. 2004. Characterization of Ceap-11 and Ceap-16, two novel splicing-variant-proteins, associated with centrosome, microtubule aggregation and cell proliferation. J. Mol. Biol. 343:71–82. 10.1016/j.jmb.2004.08.034 [DOI] [PubMed] [Google Scholar]
- Ward C.J., Hogan M.C., Rossetti S., Walker D., Sneddon T., Wang X., Kubly V., Cunningham J.M., Bacallao R., Ishibashi M., et al. 2002. The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat. Genet. 30:259–269. 10.1038/ng833 [DOI] [PubMed] [Google Scholar]
- Ward C.J., Yuan D., Masyuk T.V., Wang X., Punyashthiti R., Whelan S., Bacallao R., Torra R., LaRusso N.F., Torres V.E., and Harris P.C.. 2003. Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum. Mol. Genet. 12:2703–2710. 10.1093/hmg/ddg274 [DOI] [PubMed] [Google Scholar]
- Weickert C.S., Rothmond D.A., Hyde T.M., Kleinman J.E., and Straub R.E.. 2008. Reduced DTNBP1 (dysbindin-1) mRNA in the hippocampal formation of schizophrenia patients. Schizophr. Res. 98:105–110. 10.1016/j.schres.2007.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz O.A., and Rodriguez-Boulan E.. 2009. Apical trafficking in epithelial cells: signals, clusters and motors. J. Cell Sci. 122:4253–4266. 10.1242/jcs.032615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake C.J., Baye L.M., Nachury M.V., Wright K.J., Ervin K.E., Phu L., Chalouni C., Beck J.S., Kirkpatrick D.S., Slusarski D.C., et al. 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. U S A. 108:2759–2764. 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D.S. 2002. Transport to the photoreceptor outer segment by myosin VIIa and kinesin II. Vision Res. 42:455–462. 10.1016/S0042-6989(01)00228-0 [DOI] [PubMed] [Google Scholar]
- Wu G., D’Agati V., Cai Y., Markowitz G., Park J.H., Reynolds D.M., Maeda Y., Le T.C., Hou H. Jr., Kucherlapati R., et al. 1998. Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell. 93:177–188. 10.1016/S0092-8674(00)81570-6 [DOI] [PubMed] [Google Scholar]
- Wu H., Turner C., Gardner J., Temple B., and Brennwald P.. 2010. The Exo70 subunit of the exocyst is an effector for both Cdc42 and Rho3 function in polarized exocytosis. Mol. Biol. Cell. 21:430–442. 10.1091/mbc.E09-06-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., He X., Yang L., Zhou Z., Cullinane A.R., Wei A., Zhang Z., Hao Z., Zhang A., He M., et al. 2012. The BLOS1-interacting protein KXD1 is involved in the biogenesis of lysosome-related organelles. Traffic. 13:1160–1169. 10.1111/j.1600-0854.2012.01375.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder B.K., Hou X., and Guay-Woodford L.M.. 2002. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J. Am. Soc. Nephrol. 13:2508–2516. 10.1097/01.ASN.0000029587.47950.25 [DOI] [PubMed] [Google Scholar]
- Young R.W. 1967. The renewal of photoreceptor cell outer segments. J. Cell Biol. 33:61–72. 10.1083/jcb.33.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Sharma S., Skowronek A., and Erdmann K.S.. 2016. The serologically defined colon cancer antigen-3 (SDCCAG3) is involved in the regulation of ciliogenesis. Sci. Rep. 6:35399 10.1038/srep35399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Xu R., Zhu B., Yang X., Ding X., Duan S., Xu T., Zhuang Y., and Han M.. 2007. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 134:901–908. 10.1242/dev.02783 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wong M.L., Alberts B., and Mitchison T.. 1995. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 378:578–583. 10.1038/378578a0 [DOI] [PubMed] [Google Scholar]
- Zuo X., Zhang J., Zhang Y., Hsu S.C., Zhou D., and Guo W.. 2006. Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat. Cell Biol. 8:1383–1388. 10.1038/ncb1505 [DOI] [PubMed] [Google Scholar]
- Zuo X., Guo W., and Lipschutz J.H.. 2009. The exocyst protein Sec10 is necessary for primary ciliogenesis and cystogenesis in vitro. Mol. Biol. Cell. 20:2522–2529. 10.1091/mbc.E08-07-0772 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.