Adaptation to stress is a critical strategy for all life. Known strategies involve signaling pathways that induce changes in gene expression. These changes take time and cannot protect against acute assaults. Jin et al. reveal an early stress protection pathway regulated by the vacuole/lysosome.
Abstract
Adaptation to environmental stress is critical for cell survival. Adaptation generally occurs via changes in transcription and translation. However, there is a time lag before changes in gene expression, which suggests that more rapid mechanisms likely exist. In this study, we show that in yeast, the cyclin-dependent kinase Pho85/CDK5 provides protection against hyperosmotic stress and acts before long-term adaptation provided by Hog1. This protection requires the vacuolar/endolysosomal signaling lipid PI3,5P2. We show that Pho85/CDK5 directly phosphorylates and positively regulates the PI3P-5 kinase Fab1/PIKfyve complex and provide evidence that this regulation is conserved in mammalian cells. Moreover, this regulation is particularly crucial in yeast for the stress-induced transient elevation of PI3,5P2. Our study reveals a rapid protection mechanism regulated by Pho85/CDK5 via signaling from the vacuole/lysosome, which is distinct temporally and spatially from the previously discovered long-term adaptation Hog1 pathway, which signals from the nucleus.
Introduction
Organisms adapt to several types of environmental stress, including acute changes in temperature, nutrient limitation, and changes in osmolarity. There are a wide variety of molecular strategies to adjust to these diverse changes. The best-characterized pathways for response to stress involve changes in gene expression that result in the production of proteins that relieve the stress and promote survival. In the budding yeast Saccharomyces cerevisiae, the high-osmolarity glycerol (HOG) pathway is activated by hyperosmotic stress (Saito and Posas, 2012). Upon exposure to hyperosmotic conditions, a signaling cascade is activated that includes a key MAPK, Hog1. Within 1–5 min of exposure, Hog1 is acutely activated and translocates to the nucleus, where it promotes changes in gene expression. However, there is a lag of >30–60 min before achieving these Hog1-dependent changes (O’Rourke and Herskowitz, 2004; Vanacloig-Pedros et al., 2015). This raises the possibility that there are rapid responses that provide protection before changes that establish adaptation.
The vacuole/lysosome is a likely candidate to play roles in response to diverse stresses. The vacuole/lysosome is central for cellular homeostasis and energy metabolism. The vacuole/lysosome is a storage site for several ions and nutrients, which are derived in part via digestion of incoming macromolecules by multiple hydrolases. In addition, in yeast, some types of stress cause changes in the size and morphology of the vacuole (Duex et al., 2006a; Stauffer and Powers, 2015).
The vacuole/lysosome signaling lipid, phosphatidylinositol 3,5-bisphosphate (PI3,5P2) is found in most eukaryotes. Under basal conditions, the levels of PI3,5P2 are exceedingly low, ∼0.05–0.1% of the total phosphatidylinositol lipids. However, signals including insulin and growth factors in mammalian cells and hyperosmotic shock in yeast and cultured adipocytes cause an acute elevation of PI3,5P2 (Dove et al., 1997; Sbrissa et al., 1999; Sbrissa and Shisheva, 2005; Zolov et al., 2012; McCartney et al., 2014). These observations indicate that the synthesis of PI3,5P2 on the vacuole/lysosome is tightly regulated and suggest that PI3,5P2 may function in an early defense pathway that acts before changes in gene expression. However, a requirement for PI3,5P2 in response to stress was unclear because yeast mutants with defects in PI3,5P2 synthesis grew in high salt (Yamamoto et al., 1995).
PI3,5P2 is synthesized by the sole phosphatidylinositol-3-phosphate 5 kinase Fab1 (PIKfyve in mammals; Gary et al., 1998; Sbrissa et al., 1999; Ikonomov et al., 2011; Zolov et al., 2012). Fab1/PIKfyve forms a complex with its regulators, including Fig4, Vac14, Vac7, and Atg18 (Sbrissa et al., 2007; Botelho et al., 2008; Jin et al., 2008). The activation of Fab1 in hyperosmotic shock is likely independent of the HOG signaling pathway (Dove et al., 1997). These observations raise the question of which upstream pathways regulate Fab1 in response to hyperosmotic stress.
In this study, we show that before Hog1-dependent long-term adaptation, PI3,5P2 has critical roles for survival during hyperosmotic shock. Moreover, we find that the cyclin-dependent kinase Pho85 and one of its cyclins, Pho80, are direct activators of the Fab1 complex both under basal conditions and during hyperosmotic stress. Notably, Pho85–Pho80 activation of Fab1 is particularly crucial for yeast survival during stress.
Furthermore, we show that mammalian CDK5-CDK5R1/p35, homologues of yeast Pho85–Pho80, directly phosphorylate human PIKfyve in vitro and regulate the synthesis of PI3,5P2 in mouse fibroblasts. Our study reveals a signaling pathway from the vacuole/lysosome that is regulated by Pho85–Pho80 and promotes survival during hyperosmotic stress, and it also shows that a similar mechanism may exist in mammalian cells.
Results
PI3,5P2 is essential for yeast viability and is particularly crucial during hyperosmotic stress
In yeast, hyperosmotic shock induces an acute, transient elevation of PI3,5P2 (Dove et al., 1997; Duex et al., 2006a). Thus, PI3,5P2 may be part of a signaling pathway that provides rapid and early protection to promote survival during selected environmental stresses. Fab1 is the sole lipid kinase that synthesizes PI3,5P2 from PI3P (Gary et al., 1998). Deletion of FAB1 results in a yeast strain that lacks PI3,5P2. Previous analysis of a fab1Δ mutant revealed that it grew in hyperosmotic media (Yamamoto et al., 1995). Thus, the significance of an acute elevation of PI3,5P2 in hyperosmotic stress was unclear. In yeast, some mutants rapidly acquire additional suppressor mutations that mitigate serious defects. The fab1Δ mutant has a slow growth rate; however, we frequently observed colonies that grew similarly to WT, which we postulated may be caused by suppressor mutations that confer a selective advantage.
To characterize the phenotypes of a fab1Δ mutant in the absence of suppressor mutations, we generated a heterozygous fab1Δ/FAB1 diploid carrying a pFAB1-URA3 plasmid and generated the corresponding haploid strains via sporulation. Viability in the absence of pFAB1-URA3 was tested using 5-fluoroortic acid (5-FOA) to counter select against the URA3-containing plasmid. The fab1Δ mutant showed a dramatic reduction in viability compared with control cells with the pFAB1-URA3 plasmid (Fig. 1 A). The few surviving yeast colonies observed on 5-FOA plates were likely caused by suppressor mutations (Fig. 1 A).
Figure 1.
PI3,5P2 is essential for survival in hyperosmotic stress. (A) fab1Δ and vac7Δ yeast mutants rapidly acquire suppressors, which enable them to grow. WT or fab1Δ yeast carrying pFab1-URA3 and vac7Δ yeast carrying pVac7-URA3 were generated by tetrad dissection. Cells were cultured in SC-Ura liquid, and serial dilutions were spotted onto SC-Ura, 5-FOA, or 5-FOA + 0.9 M NaCl plates and incubated at 24°C for 10 d. (B) Monitoring cell viability by FM4-64 labeling in hyperosmotic stress. From left, WT yeast were heated at 90°C for 5 min or remained at 24°C and then were incubated with FM4-64 for 1 min. WT, or hog1Δ cells treated with 0.9 M NaCl for 21 h, then were incubated with FM4-64 or propidium iodide (PI) for 1 min. (C and D) PI3,5P2 signaling acts before Hog1 for survival during hyperosmotic stress. WT, hog1Δ, or fab1Δ yeast were untreated or treated with 0.9 M NaCl for 4 h and then were incubated with FM4-64 for 1 min. (C) Representative fields of cells. Bars, 10 µm. DIC, differential interference contrast. (D) Quantification of cells with FM4-64 in the cytoplasm. Means ± SD are shown. n = 3. More than 300 cells were counted per condition, per experiment. **, P < 0.001 by Student’s t test.
Vac7 is a critical activator of Fab1 and is defective in the synthesis of PI3,5P2 both under basal conditions and during hyperosmotic shock (Bonangelino et al., 1997; Gary et al., 1998). Notably, the vac7Δ mutant exhibited a growth defect under basal conditions and was not viable under hyperosmotic conditions (Fig. 1 A). These data indicate that PI3,5P2 has roles in cell viability and is particularly important for survival during hyperosmotic stress.
PI3,5P2 signaling functions in an early response to hyperosmotic stress
Both hog1Δ (Brewster et al., 1993) and fab1Δ mutants (Fig. 1 A) die after long-term exposure to high salt. However, the acute elevation of PI3,5P2 occurs within 5 min of exposure to hyperosmotic stress (Duex et al., 2006a). To test whether PI3,5P2 signaling acts before Hog1, we compared rates of hyperosmotic stress–induced cell death in WT, hog1Δ, and fab1Δ cells. To ensure a minimal number of suppressor mutations, we used a slow-growing strain of fab1Δ generated directly after sporulation of a Fab1/fab1Δ diploid. Yeast were treated with 0.9 M NaCl for 4 h, incubated with FM4-64 for 1 min, and then examined by fluorescence microscopy. During this short incubation, FM4-64 labels the plasma membrane of viable cells (Vida and Emr, 1995). However, heating cells at 90°C for 5 min to promote cell death resulted in nonselective permeability to FM4-64 (Fig. 1 B). Thus, monitoring the appearance of cytosolic FM4-64 can be used as a readout for a decrease in cell viability. Note that we tested and found that propidium iodide, which is commonly used to selectively label permeable dead cells, did not label many of the dead cells present in yeast treated with high salt for 21 h (Fig. 1 B).
Under basal conditions, WT and hog1Δ mutants excluded FM4-64 from the cytoplasm, whereas 30% of the fab1Δ cells were permeable, indicating that viability in this strain was decreased. Importantly, after 4 h in 0.9 M NaCl, the differences in cell death between fab1Δ cells and WT or the hog1Δ mutant greatly increased (Fig. 1, C and D). These findings support the hypothesis that PI3,5P2 is protective during hyperosmotic shock and acts before the HOG pathway.
The cyclin-dependent kinase CDK5/Pho85 is an upstream regulator of the synthesis of PI3,5P2
That PI3,5P2 levels transiently change in response to stimuli suggests that upstream pathways modulate Fab1/PIKfyve (Dove et al., 1997; Sbrissa et al., 1999; Zolov et al., 2012; McCartney et al., 2014). Protein kinases regulate several signal transduction pathways during acute environmental changes. Thus, we hypothesized that some protein kinases activate Fab1/PIKfyve in response to hyperosmotic stress. An obvious candidate is Hog1; however, PI3,5P2 levels responded to hyperosmotic stress even in the absence of HOG1 (Fig. S1 A; Dove et al., 1997).
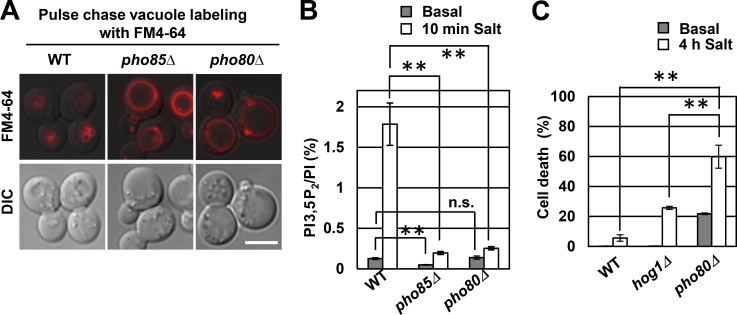
Thus, we sought to identify kinases that positively regulate PI3,5P2, and test whether they function during hyperosmotic stress. To identify protein kinases that positively regulate Fab1, we examined the knockout haploid yeast mutant collection of the 103 nonessential protein kinases. We screened for mutants with enlarged vacuoles because several mutants with defects in the synthesis of PI3,5P2 exhibit this phenotype (Yamamoto et al., 1995; Gary et al., 1998; Bonangelino et al., 2002). Deletion of the PHO85 cyclin-dependent kinase, the pho85Δ mutant, had enlarged vacuoles and was defective in the synthesis of PI3,5P2 both in basal conditions and after hyperosmotic shock (Fig. 2, A and B). Notably, the pho85Δ mutant is not viable in hyperosmotic conditions (Lee et al., 1998), which suggests that Pho85 is critical for survival in hyperosmotic stress. Pho85 partners with 10 cyclins, including Pho80 (Carroll and O’Shea, 2002), and only the pho80Δ mutant had a defect in vacuole size (Figs. 2 A and S1 B). Importantly, the pho80Δ mutant was defective in the hyperosmotic shock–induced elevation of PI3,5P2 (Fig. 2 B) and was not able to grow on high-salt plates (Fig. S1 C). Moreover, after 4 h in 0.9 M NaCl, pho80Δ cells showed early lethality compared with hog1Δ cells (Fig. 2 C).
Figure 2.
Pho85–Pho80 regulates the synthesis of PI3,5P2. (A) Pulse-chase labeling of WT, pho85Δ, or pho80Δ cells with FM4-64 (red). Bar, 5 µm. DIC, differential interference contrast. (B) WT, pho85Δ, or pho80Δ cells were labeled overnight with [3H]inositol (Basal) or were labeled and then treated with 0.9 M NaCl for 10 min (Salt). Corresponding deacylated glycerol-phosphorylated inositol species were analyzed by HPLC. PI, phosphatidylinositol. (C) WT, hog1Δ, or pho80Δ yeast were untreated or treated with 0.9 M NaCl for 4 h and then incubated with FM-64 for 1 min. Quantifications of cells with FM4-64 in the cytoplasm. Means ± SD are shown. n = 3. More than 300 cells were counted per condition, per experiment. **, P < 0.001 by Student’s t test.
These findings suggest that Pho85–Pho80 is (a) an upstream regulator of PI3,5P2 synthesis both under basal conditions and during hyperosmotic stress and (b) during stress, provides an early defense before the HOG pathway. Note that compared with WT yeast, both pho85Δ and pho80Δ mutants had higher levels of PI3P in hyperosmotic stress (Fig. S1 D). This increase in PI3P is likely a result of a defect in the synthesis of PI3,5P2 (Duex et al., 2006a). Importantly, the loss of PI3,5P2 in the pho85Δ and pho80Δ mutants are caused by a loss of activation of Fab1, not a defect in production of PI3P.
CDK5/Pho85 is required for the phosphorylation of the C-terminal region of Fab1 in vivo
To further test whether Pho85 positively regulates PI3,5P2 levels, we acutely inhibited Pho85 using the Pho85-as(F82G) mutant. Pho85-as(F82G) is mutated in the ATP-binding pocket. It retains WT function but is rapidly inactivated when treated with the ATP analogue inhibitor 1-NA-PP1 (Carroll et al., 2001). We treated PHO85 or pho85-as(F82G) with or without 1-NA-PP1 and monitored the levels of PI3,5P2. The pho85-as(F82G) mutant treated with 1-NA-PP1 displayed a defect in vacuole morphology (Fig. 3 A) and was defective in the hyperosmotic shock–induced synthesis of PI3,5P2 (Figs. 3 B and S1 E). This indicates that Pho85 activity is particularly critical for the activation of Fab1 in hyperosmotic stress.
Figure 3.
Activity of Pho85 is required for phosphorylation of Fab1 in hyperosmotic stress. (A) pho85Δ cells expressing pPho85 or pPho85-as(F82G) were labeled with FM4-64 and then treated with DMSO or 20 µM 1-NA-PP1 for 150 min. Bar, 5 µm. DIC, differential interference contrast. (B) pho85Δ cells expressing pPho85 or pPho85-as(F82G) were labeled with [3H]inositol overnight and treated with DMSO or 20 µM 1-NA-PP1 for 150 min (Basal) or with 0.9 M NaCl for 10 min (Salt). PI, phosphatidylinositol. (C) A pho85-as(F82G)/fab1Δ mutant expressing pFab1-TEVr1506-HA6 was treated with DMSO or 20 µM 1-NA-PP1 for 150 min, exposed to 0.9 M NaCl for 5 min, and lysed with 10% TCA. Lysates were treated with TEVp in the presence or absence of PPase. Western blot: anti-HA or anti-Pgk1 (control). Blots are representative of three independent experiments. (D) Quantification of relative mobility from C. Means ± SD are shown. n = 3. *, P < 0.005; **, P < 0.001 by Student’s t test.
Fab1 is a 2,278-aa protein; thus, a simple gel-shift assay to detect in vivo modifications was not feasible. Therefore, to test whether Pho85 is required for phosphorylation of Fab1, we developed a gel-shift assay. We inserted a Tobacco etch virus (TEV) recognition (TEVr) sequence after Fab1-K1505 (Fab1-TEVr1506), added 6×HA to the C terminus, and assessed mobility in an SDS-PAGE gel (Fig. S1, F and G).
We treated yeast expressing Fab1-TEVr1506/Pho85-as(F82G) with 1-NA-PP1 or DMSO and exposed yeast cells to hyperosmotic stress. Then, lysates were treated with protein phosphatase (PPase) and TEV protease (TEVp). PPase treatment resulted in a faster migration of the C terminus of Fab1 (Fig. 3 C, lanes 2 and 4), suggesting that the C-terminal region of Fab1 is phosphorylated in hyperosmotic shock. Moreover, in the absence of PPase, treatment of yeast expressing Fab1-TEVr1506/Pho85-as(F82G) with 1-NA-PP1 resulted in a faster migration of the C terminus of Fab1 compared with the DMSO control (Fig. 3 C, lanes 2 and 3; and Fig. 3 D), suggesting that Pho85 plays a role in the phosphorylation of the C-terminal region of Fab1 in hyperosmotic stress. Note that Pho85-as(F82G) levels were not affected by 1-NA-PP1 or hyperosmotic shock (Fig. S1 H), and Fab1 protein levels were not affected by 1-NA-PP1 (Fig. S1 I).
Fab1 and the cyclin Pho80 interact in vitro and in vivo
Cyclins may determine the specificity of CDK to recognize substrates (Bloom and Cross, 2007). We tested and found that the recombinant Fab1 kinase domain and cyclin-Pho80 directly interact in vitro (Fig. 4 A). Moreover, to test the interaction of Fab1 and Pho80 in vivo, we performed a split YFP assay (Kerppola, 2008). Fab1 was fused with the C-terminal half of the YFP variant Venus (vYFPC) on its N terminus, and Pho80 was fused with N-terminal half of vYFP (vYFPN) on its N terminus (Fig. 4 B). The association of vYFPC-Fab1 and vYFPN-Pho80 was predominantly observed in the nucleus (Fig. 4 B), suggesting that after association with Pho80, Fab1 was dragged from the vacuole membrane to the nucleus, where Pho85 and Pho80 are predominantly localized (Fig. S2 A). Sometimes, split YPF forms an irreversible complex (Kerppola, 2008), and that may have occurred in this study. Notably, inhibition of the pho85-as(F82G) mutant with 1-NA-PP1 resulted in the appearance of some vYFPC-Fab1 and vYFPN-Pho80 on the vacuole (Fig. 4, B and C). This suggests that inhibition of Pho85–Pho80 activity elevates the number of irreversible vYFP formed per cell and facilitates the detection of a fraction on the vacuole.
Figure 4.
The cyclin-Pho80 interacts with Fab1 on the vacuole. (A) MBP or MBP-Fab1(1,941–2,278) was immobilized on amylose resin and then incubated with E. coli lysates containing recombinant His-Pho80. Bound proteins were eluted with SDS sample buffer, separated by SDS-PAGE, and visualized by Coomassie brilliant blue (CBB). Arrows indicate migration positions of indicated proteins. Top: Western blot (WB) with anti-His antibody. Blots are representative of three independent experiments. (B and C) Split YFP assay. A pho85-as(F82G)/fab1Δ mutant expressing vYFPC-Fab1/vYFPN-Pho80 or Fab1-vYFPC/vYFPN-Pho80 was treated with DMSO or 20 µM 1-NA-PP1 for 150 min. (B) Representative fields of cells. Bar, 5 µm. DIC, differential interference contrast. (C) Quantification of percent cells with YFP signal in the nucleus (gray) and in the nucleus plus on the vacuole membrane (white). Means ± SD are shown. n = 3. More than 100 cells were counted per condition, per experiment. **, P < 0.001 by Student’s t test.
The split vYFP association is specific. Note that if the vYFPc tag is moved from the N terminus of Fab1 to the C terminus, an interaction was no longer observed. In support with the finding that both Fab1 constructs are functional, yeast expressing either vYFPC-Fab1 or Fab1-vYFPC have normal-sized vacuoles (Fig. 4 B). Collectively, these observations suggest that Pho85–Pho80 interacts with Fab1 in vivo.
Pho85–Pho80 directly phosphorylates Fab1 and Vac7
We examined whether Pho85–Pho80 directly phosphorylates Fab1 in vitro. We tested eight peptides that span most of Fab1 (Fig. S2 B) and found that immunoprecipitated Pho85–Pho80 from yeast lysates directly phosphorylates two of the eight recombinant Fab1 peptides (Fig. 5, A and B): the kinase domain (1,941–2,278) and a region upstream of the kinase domain (1,501–1,969). These findings suggest that Fab1 is a direct target of Pho85–Pho80.
Figure 5.
Pho85–Pho80 directly phosphorylates Fab1 and Vac7. (A and B) pHA-Pho80 was expressed in a pho80Δ mutant, immunoprecipitated using anti-HA antibody, and then incubated with [γ-32P]ATP and MBP or MBP-Fab1(1,941–2,279) (A) or MBP or MBP-Fab1(1,501–1,969) (B). Proteins were eluted with SDS sample buffer, separated by SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were exposed to x-ray film or stained with Ponceau S. Red arrowheads indicate phosphorylated MBP-Fab1 peptides. Black arrowheads indicate MBP or MBP-Fab1 peptides. (C) Schematic of Fab1 protein representing the FYVE (Fab1, YOTB, Vac1, and EEA1) domain (223–321) that binds PI3P, the CCT domain (814–1,080) that binds Vac14, and the kinase domain (2,022–2,278). Phosphorylation sites were identified by alanine mutagenesis. (D) Schematic of Vac7 protein representing predicted TM domains (TM1, 920–941; TM2, 965–986). Phosphorylation sites are indicated. (E) MBP- or MBP-Vac7–recombinant peptide (394–918) incubated with immunoprecipitated HA-Pho85–Pho80 and [γ-32P]ATP. Proteins were eluted with SDS sample buffer, separated by SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were exposed to x-ray film or stained with Ponceau S. The red arrowhead indicates the phosphorylated MBP-Vac7 peptide. Black arrowheads indicate MBP or MBP-Vac7 peptides. Blots are representative of three independent experiments.
Pho85–Pho80 phosphorylates serine or threonine residues adjacent to proline (SP/TP; Kaffman et al., 1994) and Fab1(1,501–2,278) has 10 SP or TP candidate sites. One of these, T2250, is likely not a Pho85–Pho80 target because fab1-T2250A is a hyperactive mutant (Duex et al., 2006b). We performed alanine scan mutagenesis of the other nine sites and tested the degree of phosphorylation by Pho85–Pho80 in vitro. Peptides carrying a single substitution to alanine at T1569, T1583, T1594, T1691, S1924, T1953, T1963, or S2166 were less phosphorylated by Pho85–Pho80 than the WT control (Fig. S2, C–H), suggesting that these residues are phosphorylated by Pho85–Pho80 (Fig. 5 C). In addition, we raised a phospho-specific antibody to one of these sites, Fab1-T1953, and verified that this site was phosphorylated in vitro and in vivo (Fig. S2, I and J).
In addition to Fab1, we tested and found that Pho85–Pho80 directly phosphorylates Vac7, an activator of Fab1 (Duex et al., 2006b). A recombinant Vac7(394–918) peptide, which includes five putative phosphorylation sites (S593, S607, T858, S903, and S912), was phosphorylated by Pho85–Pho80 in vitro (Fig. 5, D and E), suggesting that Vac7 is also a direct target of Pho85–Pho80.
The Pho85–Pho80 target sites on Fab1 and Vac7 are phosphorylated in vivo
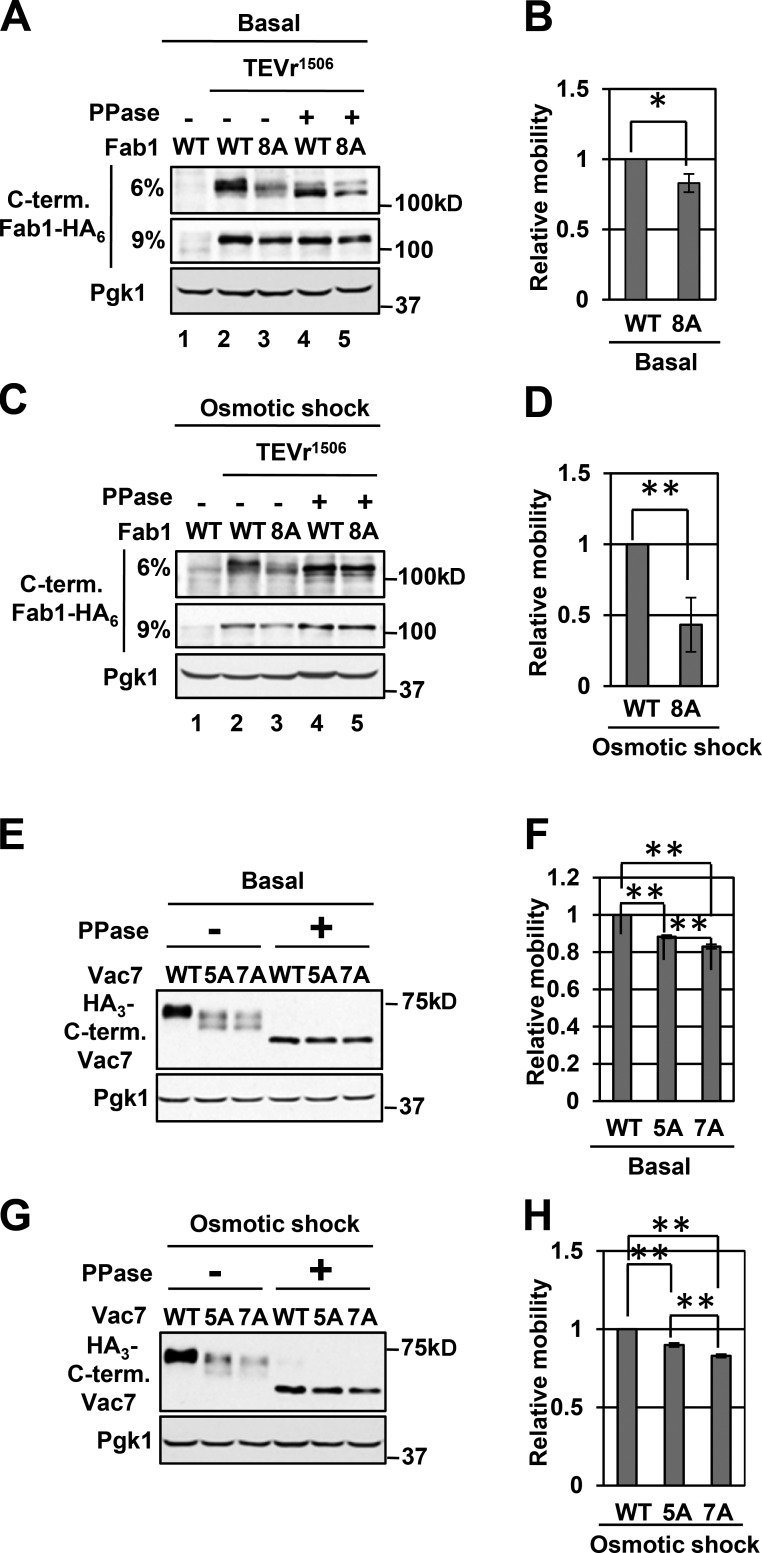
Using a gel shift assay, we tested whether the Pho85–Pho80 sites on Fab1 and Vac7 are phosphorylated in vivo. In the absence of PPase treatment, the Fab18A mutant exhibited a faster migration than WT Fab1 on a 6% SDS-PAGE gel both under basal conditions (Fig. 6 A, lanes 2 and 3; and Fig. 6 B) and in hyperosmotic stress (Fig. 6 C, lanes 2 and 3; and Fig. 6 D), which indicates that some or all sites are phosphorylated under both conditions. Indeed, we observed that Fab1-T1953A was phosphorylated in both conditions (Fig. S2 J). Notably, when compared with basal conditions, during osmotic stress, the degree of the shift in the migration of the Fab18A mutant was greater (Fig. 6, B and D). This raises the possibility that some sites may be specific for hyperosmotic shock. Importantly, when assessed on a 9% SDS-PAGE gel, PPase treatment resulted in single bands of similar intensity, indicating that Fab1 and Fab18A have similar stability (Fig. 6, A and C; lanes 4 and 5). Note that some of these sites may be phosphorylated by other kinases as well. S/P and T/P are also recognition sites for Cdk1 as well as MAPKs.
Figure 6.
Putative Pho85–Pho80 target sites in Fab1 and Vac7 are phosphorylated in vivo. (A and C) A fab1Δ mutant expressing pFab1-TEVr1506-HA6 or pFab18A(T1569A, T1583A, T1594A, T1691A, S1924A, T1953A, T1963A, and S2166A)–TEV1506-HA6 (Basal; A) or incubated with 0.9 M NaCl for 5 min (Osmotic shock; C). Samples were then lysed with 10% TCA. Lysates were treated with TEVp in the presence or absence of PPase. 6% and 9% SDS-PAGE gels were used. Western blot: anti-HA or anti-Pgk1 (control). (B and D) Quantifications of data in A and C, respectively. (E and G) A vac7Δ mutant expressing pVac7-TEVr572-HA3, Vac75A(S593A, S607A, T858A, S903A, and S912A)–TEVr572–HA3 or Vac77A(S593A, S607A, T858A, S903A, S912A, T1050A, and S1074A)–TEVr572-HA3 (Basal; E) or treated with 0.9 M NaCl for 5 min (Osmotic shock; G). Samples were then lysed with 10% TCA. Lysates were treated with TEVp in the presence or absence of PPase. Western blot: anti-HA or anti-Pgk1 (control). Blots are representative of three independent experiments. (F and H) Quantifications of data in E and G, respectively. Means ± SD are shown. n = 3. *, P < 0.005; **, P < 0.001 by Student’s t test.
To determine the phosphorylation status of the C terminus of Vac7, we inserted a TEVr plus 3×HA after Vac7-P571 (Vac7-TEVr572–HA3; Fig. S2 K). TEVp treatment of the lysates generated a C-terminal fragment of Vac7 (Fig. S2 L). In the absence of PPase treatment, the Vac75A mutant exhibited a faster migration than WT Vac7 both under basal conditions and in hyperosmotic shock (Fig. 6, E–H). In addition, we tested T1050 and S1074 adjacent to proline. In the absence of PPase treatment, the Vac77A mutants exhibited a faster migration than the Vac75A mutant both under basal conditions and in hyperosmotic shock (Fig. 6, E–H). If Vac7 has a single transmembrane (TM) domain (Bonangelino et al., 1997), these residues would reside in the vacuole lumen. However, using a relaxed cutoff, we analyzed Vac7 by three independent programs (Topred2 [von Heijne, 1992], TMpred [Hofmann, 1993], and DAS [Cserzö et al., 1997]) and found a second potential TM domain (Figs. 5 D and S3 A). Two TM domains would place T1050 and S1074 on the cytoplasmic side of the vacuole. The C terminus is a relatively conserved region of Vac7 (Fig. S3 A), which supports the possibility that T1050 and S1074 are cytoplasmic regulatory residues. Collectively, these data suggest that the C terminus of Vac7 is phosphorylated and that at least a subset of the Pho85–Pho80 sites are phosphorylated in vivo.
Pho81 is linked to the activation of Fab1 during hyperosmotic shock through the regulation of Pho85–Pho80
Pho81 is a negative regulator of Pho85–Pho80 and is constitutively associated with Pho85-Pho80. During phosphate limitation, conformational changes in Pho81 inhibit Pho85-Pho80 (Lee et al., 2008). Analysis of the dominant mutant pho81-R701S (Wang et al., 1996; Birkeland et al., 2010) suggests that this mutation locks Pho81-R701S into a partially inhibitory conformation. pho81-R701S has a defect in vacuole morphology consistent with a defect in the synthesis of PI3,5P2. Notably, the R701S mutation is in a minimum domain of Pho81, which is necessary and sufficient for the inhibition of Pho85–Pho80 activity (Lee et al., 2008). We found that expression of the dominant pho81-R701S mutant but not of WT or empty vector in a WT strain inhibited the hyperosmotic shock–induced synthesis of PI3,5P2 without impacting basal levels (Fig. 7). These observations suggest that conformational changes in WT Pho81 induce an increase in Pho85–Pho80 activity or association with the Fab1 complex during hyperosmotic stress, and may explain how Pho85–Pho80 is particularly crucial for Fab1 activation during hyperosmotic shock.
Figure 7.

Pho81 regulates Pho85–Pho80 during the activation of Fab1 in the response to hyperosmotic stress. WT strain expressing empty vector, pPho81, or pPho81R701S labeled with [3H]inositol overnight, treated without (Basal) or with 0.9 M NaCl for 10 min (Salt). Means ± SD are shown. n = 3. **, P < 0.001 by Student’s t test. PI, phosphatidylinositol.
Activation of Fab1 by Pho85–Pho80 provides early protection during hyperosmotic stress
Because of modest phenotypes of the fab18A mutant (Fig. S3, B and C), we tested the fab121A allele, which is mutated in all predicted Pho85–Pho80 sites. We found that the fab18A and fab121A mutants had a similar modest effect on vacuole size as well as on basal levels of PI3,5P2 (Fig. S3, B and C). Thus, we tested and monitored vacuole size and levels of PI3,5P2 in the fab121A vac77A double mutant. The fab121A vac77A double mutant exhibited a defect in vacuole size under basal conditions and showed a partial defect in fragmentation of the vacuole in response to salt (Fig. 8 A). To more quantitatively assess defects in the fab121A vac77A double mutant, we directly measured PI3,5P2 levels and found a decrease in the synthesis of PI3,5P2, both under basal conditions (to 68% of WT levels) and during hyperosmotic shock (to 65% of WT levels; Fig. 8 B). That PI3,5P2 levels in the pho80Δ mutant are considerably lower than levels in the fab121A vac77A double mutant suggests that Pho85–Pho80 may phosphorylate (a) additional noncanonical sites within Fab1 or Vac7, (b) additional known proteins in Fab1 complex, and/or (c) unknown proteins that regulate Fab1 activity. Importantly, the >30% drop in PI3,5P2 levels in the fab121A vac77A double mutant strongly suggests that these sites are important for Pho85–Pho80 regulation of PI3,5P2. In further support that these sites are used by Pho85–Pho80 to regulate the Fab1 complex, the fab121A vac77A double mutant showed early lethality compared with WT cells after 4 h of incubation with 0.9 M NaCl (Fig. 8, C and D).
Figure 8.
Pho85–Pho80 activation of Fab1 promotes survival during hyperosmotic stress. (A and B) A diploid fab1Δ vac7Δ mutant expressing pFab1/pVac7 or pFab121A(S75A, T183A, T392A, T526A, T712A, S729A, S855A, S1095, T1208A, T1296A, T1364A, S1447A, T1569A, T1583A, T1594A, T1691A, T1818A, S1924A, T1953A, T1963A, and S2166A)-pVac77A labeled with FM4-64 (Basal) or 5 min of incubation with 0.9 M NaCl (Salt; A). Cultures of the same strains were labeled with [3H]inositol overnight and either not treated or treated with 0.9 M NaCl for 10 min, and PI3,5P2 levels were determined (B). PI, phosphatidylinositol. (C and D) A diploid fab1Δ vac7Δ mutant expressing pFab1/pVac7 or pFab121A/Vac77A yeast untreated or treated with 0.9 M NaCl for 4 h and then incubated with FM4-64 for 1 min. (C) Percent cells with FM4-64 signal in the cytoplasm. (D) Representative fields of cells. (E and F) pho80 or pho80Δ fab1VLA yeast were untreated or treated with 0.9 M NaCl for 4 h and then incubated with FM4-64 for 1 min. The pho80Δ fab1VLA yeast mutant was generated by sporulation of a Pho80/pho80Δ Fab1/fab1VLA diploid. (E) Percent cells with FM4-64 signal in the cytoplasm. (F) Representative fields of cells. Means ± SD are shown. n = 3. *, P < 0.005; **, P < 0.001 by Student’s t test. (C and E) More than 300 cells were counted per condition, per experiment. (G) pho80Δ or pho80Δ/fab1VLA yeast labeled with FM4-64. Bars: (A and G) 5 µm; (D and F) 10 µm. DIC, differential interference contrast.
To further test the hypothesis that Pho85–Pho80 provides protection during stress via activation of Fab1, we introduced a hyperactive FAB1 allele (Duex et al., 2006b), fab1VLA, into a pho80Δ mutant and tested this strain for cell viability in hyperosmotic stress. Importantly, expression of fab1VLA in the pho80Δ mutant rescued early lethality in hyperosmotic stress and rescued vacuole size (Fig. 5, E–G). Collectively, these observations suggest that Pho85-Pho80 provides early protection for survival during hyperosmotic stress via activation of Fab1.
Phosphorylation of the Fab1 complex by Pho85–Pho80 regulates changes in its conformation and/or composition
Phosphorylation of Fab1 could regulate its activity in several ways, including its localization and interaction with its regulators. We tested and found that Fab1 and its regulators, including Vac14, Vac7, and Fig4, properly localized on the vacuole membrane in a pho85Δ mutant (Fig. S4 A). In WT cells, Fab1, Vac14, and Fig4 formed a ternary complex on the vacuole membrane. Similarly, the Fab18A mutant interacted with Vac14 and Fig4 and was properly localized on the vacuole membrane (Fig. S4, B–D), which suggests that the Fab1 ternary complex was not altered.
We postulated that Pho85–Pho80 phosphorylation of the Fab1 complex may cause a change in its conformation and/or composition. To monitor acute changes in Fab1 conformation or association with proteins in vivo, we used limited proteolysis. We engineered a TEVr site after Fab1 K1789 (Fab1-TEVr1790), which is in the middle of the cluster of the Pho85-Pho80 phosphorylation sites (Fig. 9 A). Mild TEVp treatment of Fab1-TEVr1790 or Fab18A-TEVr1790 showed that during hyperosmotic shock, WT Fab1 was more sensitive to TEVp cleavage than the Fab18A mutant, resulting in an increase in the generation of a C-terminal fragment (Fig. 9, B and C). Note that under basal conditions, WT Fab1 and the Fab18A mutant did not show any significant differences to sensitivity to TEVp (Fig. 9, B and D). These observations suggest that during hyperosmotic shock, Pho85–Pho80 likely contributes to an increase in Fab1 activity via conformational changes within the complex and/or the recruitment of additional regulators.
Figure 9.
Phosphorylation of the Fab1 complex by Pho85–Pho80 leads to a change in its conformation and/or composition. (A) Schematic of Fab1-TEVr1790-HA6. Phosphorylation sites identified by alanine mutagenesis and TEVr1790 are indicated. (B) A fab1Δ mutant expressing pFab1-TEVr1790-HA6 or pFab18A-TEV1790-HA6 exposed to 0.9 M NaCl for 0 min (Basal) or 5 min (Salt) and lysed with TEV buffer. After limited digestion with TEVp (++, 0.125 U/µl; +, 0.0625 U/µl) for 10 or 30 min at 4°C, reactions were terminated with an equal amount of 2× SDS sample buffer. Samples were separated by SDS-PAGE. Western blot: anti-HA or Pgk1 (control). Black arrowheads indicate full-length Fab1 or C termini of Fab1 generated by TEV cleavage. Blots are representative of three independent experiments. (C and D) Quantifications of band intensities from osmotic shock (C) and basal (D) experiments from 0.125 U/µl TEVp with 30 min treatment. Means ± SD are shown. n = 3. *, P < 0.005 by Student’s t test.
CDK5–p35 phosphorylates mammalian Fab1/PIKfyve and positively regulates PI3,5P2
CDK5 and its cyclin, p35, are mammalian homologues of yeast Pho85 and Pho80, respectively (Huang et al., 1999; Nishizawa et al., 1999), which suggests that CDK5–p35 may be an upstream regulator of mammalian PIKfyve. The Pho85 phosphorylation sites on Fab1 are not conserved in PIKfyve; however, phosphorylation sites commonly shift in evolution (Holt et al., 2009). Thus, we tested whether CDK5 phosphorylates PIKfyve. We transiently coexpressed N-terminal HA-tagged p35 and N-terminal EGFP-tagged CDK5 in HEK293 cells, immunoprecipitated CDK5–p35 from cell lysates using anti-HA antibody, and then incubated with recombinant PIKfyve peptides. A peptide that is upstream of the kinase domain, PIKfyve(1,214–1,594), was directly phosphorylated in vitro (Fig. 10 A). Although the kinase domain peptide PIKfyve(1,700–2,098) was degraded after elution from the beads, the eluted peptide was phosphorylated by CDK5–p35 (Fig. 10 A). These data suggest that CDK5–p35 directly phosphorylates PIKfyve.
Figure 10.
Cdk5–p35 directly phosphorylates PIKfyve and regulates PI3,5P2 levels. (A) pHA-p35 and pEGFP-Cdk5 were cotransfected into HEK293 cells. The Cdk5–p35 complex was immunoprecipitated using anti-HA antibody. Recombinant proteins: PIKfyve(1,700–2,098) and PIKfyve(1,214–1,594) were used for in vitro kinase assays. Red arrowheads indicate MBP-PIKfyve phosphorylated by Cdk5–p35 in vitro. Black arrowheads indicate MBP or MBP-PIKfyve peptides. (B) MEF labeled with [3H]inositol for 48 h and then treated with DMSO or 20 µM roscovitine for 3 h. Three independent experiments were performed. *, P = 0.014. (C) MEF labeled with [3H]inositol for 48 h, treated with DMSO or 20 µM roscovitine for 3 h, and then treated with high-salt media (final concentration 0.47 M NaCl; Salt) or without (Basal) for 30 min. Means ± SD are shown. n = 3. *, P < 0.005 by Student’s t test. PI, phosphatidylinositol. (D) Model for early protection via PI3,5P2 signaling regulated by CDK5/Pho85 in response to the hyperosmotic stress. In hyperosmotic stress, a protection pathway activated by CDK5/Pho85 is distinct from the HOG pathway both spatially and temporally. Pho85–Pho80 directly activates the Fab1/PIKfyve complex. This activation is particularly crucial for the hyperosmotic stress–induced elevation of PI3,5P2. These changes that signal from the vacuole/lysosome promote survival before the changes regulated by the HOG pathway, which signals from the nucleus.
High NaCl treatment activates CDK5, which regulates the transcription of osmoprotective genes via phosphorylation of TonEBP/OREBP (Gallazzini et al., 2011). To test whether CDK5 regulates the synthesis of PI3,5P2 in cells, we monitored the levels of PI3,5P2 in mouse embryonic fibroblasts (MEFs) treated with roscovitine, an inhibitor of CDK5-CDK2-CDK1 (Meijer et al., 1997). Roscovitine inhibited the synthesis of PI3,5P2 under basal conditions (Fig. 10 B) and caused a defect in a high salt–induced elevation of PI3,5P2 (Fig. 10 C). In MEF cells treated with the PIKfyve inhibitor apilimod, either in the presence or absence of high salt, PI3,5P2 was greatly reduced (Fig. 10 C). Collectively, these experiments suggest that CDK5/Pho85 is an upstream regulator of the synthesis of PI3,5P2 via direct phosphorylation of Fab1/PIKfyve and that this regulation is conserved.
Discussion
Pho85–Pho80 was originally identified as a regulator for response to inorganic phosphate limitation (Carroll and O’Shea, 2002). In this study, we show that Pho85 also plays a role in response to hyperosmotic stress and acts in parallel with and prior to the HOG pathway. We found that Pho85-Pho80 regulates the acute elevation of PI3,5P2 levels. This fits with earlier studies that Pho80 is required for vacuole morphology (Nicolson et al., 1995; Birkeland et al., 2010) and that pho80Δ mutants share similar defects in morphology with mutants defective in the synthesis of PI3,5P2. Our findings indicate that Pho85–Pho80 regulates a pathway that signals from the vacuole and is distinct from the HOG pathway both spatially and temporally (Fig. 10 D).
An early defense for cell survival during stress
PI3,5P2 is key for survival of yeast during stress (Fig. 1 A) and may function as an early defense before achieving changes in gene expression induced via the HOG pathway (Fig. 1, B and C). How might PI3,5P2 function in this capacity? PI3,5P2 may regulate vacuolar/lysosomal ion homeostasis, which is particularly critical during hyperosmotic stress. When yeast are exposed to high salt, the volume of the vacuole transiently decreases and within 30 min returns to its normal size (Duex et al., 2006a). It is tempting to speculate that in the early stages of osmotic stress, water, ions, and osmolytes are released from the vacuole to preserve a balance between cytosolic and extracellular osmolytes. During adaptation, it is possible that ions and osmolytes are generated or pumped into the cell and then transported to the vacuole to restore lumenal conditions optimal for vacuolar function. In support of this hypothesis, the vacuolar ATPase, which is regulated in part by PI3,5P2 (Li et al., 2014), is required for adaptation to osmotic stress and acts in parallel with the HOG pathway (Li et al., 2012). Fab1 plays a role in vacuolar pH during hyperosmotic stress (Ho et al., 2015), and PI3,5P2 also regulates Ca2+ channel currents in the vacuole via regulation of Yvc1 (Dong et al., 2010). Similarly, other transporters and channels may also be regulated by PI3,5P2 to provide a defense against acute environmental change.
Cdk5/Pho85 is a conserved upstream regulator of PI3,5P2
Our study suggests that Pho85–Pho80 activation of Fab1 is conserved in mammalian cells (Fig. 10). We show that CDK5–p35, the mammalian homologue of Pho85–Pho80, directly phosphorylates recombinant PIKfyve. Moreover, we found that treatment of MEF cells with roscovitine lowers the levels of PI3,5P2. Other kinases likely also regulate PIKfyve. Indeed, PIKfyve is phosphorylated by protein kinase B on S318 in vitro, and this site is phosphorylated in cells after treatment with insulin or high sorbitol (Hill et al., 2010). However, cellular consequences for this phosphorylation site are unknown.
It is tempting to speculate that PI3,5P2 signaling regulated by CDK5 has conserved roles in mammalian cells and provides early protection during some types of stress. Indeed, CDK5 is activated during hyperosmotic stress (Gallazzini et al., 2011). Moreover, many stress pathways use signaling cascades, including the p38-MAPK pathway (p38 is a mammalian orthologue of yeast Hog1). These observations suggest that in mammals, CDK5-activated PI3,5P2 signaling may regulate early protection pathways that function before stress-induced pathways that act via changes in gene expression. It is tempting to speculate that activation of PI3,5P2 signaling may more broadly provide early protection for additional types of stress. Furthermore, there may be other types of early protection pathways that act before mechanisms that provide long-term adaption.
In further support that CDK5 is a positive regulator of PIKfyve in mammals, the phenotypes of Vac14−/− mice, which have defects in the levels of PI3,5P2 (Zhang et al., 2007), share some striking similarities with Cdk5−/− mice. Both mouse mutants die perinatally and have cytoplasmic vacuoles in the brain (Ohshima et al., 1996; Zhang et al., 2007). This uncommon phenotype is found in mouse mutants with defects in the levels of PI3,5P2 (Chow et al., 2007; Zhang et al., 2007; Jin et al., 2008; Zolov et al., 2012). There are also similarities in the roles for CDK5 and PIKfyve at the neuronal synapse. Both CDK5 and PIKfyve are required for homeostatic down-regulation (Seeburg et al., 2008; McCartney et al., 2014). Collectively, these studies suggest that CDK5 regulates PIKfyve and may play a critical role in cellular response to stress.
Materials and methods
Strains and media
Yeast strains used are in Table S2. Yeast were grown in yeast extract–peptone-dextrose (1% yeast extract, 2% peptone, 2% dextrose; YEPD) or synthetic complete (SC) minimal medium without a selective amino acid or acids at 24°C. 5-FOA media were made as described previously (Kaiser and Mitchell, 1994). SC medium contained 2% dextrose.
Plasmids
Plasmids used are in Table S1. To generate Fab1 mutants, pRS413 Fab1 was mutagenized by site-directed mutagenesis using the following primers (sequences in Table S3): S75A(FW1 and REV1), T183A(FW2 and REV2), T392A(FW3 and REV3), T526A(FW4 and REV4), T712A(FW5 and REV5), S729A(FW6 and REV6), S855A(FW7 and REV7), S1095A(FW8 and REV8), T1208A(FW9 and REV9), T1296A(FW10 and REV10), T1364A(FW11 and REV11), S1447A(FW12 and REV12), T1569A(FW13 and REV13), T1583A(FW14 and REV14), T1594A(FW15 and REV15), T1691A(FW16 and REV16), T1818A(FW17 and REV17), S1924A(FW18 and REV18), T1953A(FW19 and REV19), T1963A(FW20 and REV20), and S2166A(FW21 and REV21).
To generate HA6-fused Fab1 on the C terminus, pRS413 Fab1 including the 3′UTR was mutagenized for introducing NheI sites by site-directed mutagenesis using the primers FW22 and REV22. An HA6 fragment was amplified by PCR using the primers FW23 and REV23, digested by NheI and SpeI, and inserted the NheI sites of pRS416 Fab1–NheI.
To generate pRS413 Fab1-TEVr1506-HA6, Fab1-HA6 was mutagenized by site-directed mutagenesis using the primers FW24 and REV24. To generate pRS413 Fab1-TEVr1790-HA6, Fab1-HA6 was mutagenized by site-directed mutagenesis using the primers FW25 and REV25.
To insert the AvrII site just after first Met, pRS413 Fab1-HA6 or pRS413 Fab1 was mutagenized by site-directed mutagenesis using the primers FW26 and REV26. To generate pRS413-GST-Fab1-HA6 plasmid, a GST fragment was amplified by PCR using the primers FW27 and REV27, digested by AvrII and SpeI, and inserted to AvrII site of pRS413 AvrII-Fab1-HA6. To generate pRS413-Fab1-vYFPN-Fab1, vYFPN was amplified by PCR using the primers FW28 and REV28, digested by AvrII and SpeI, and inserted to AvrII site of pRS413 AvrII-Fab1.
To generate vac7 mutants, pRS413 Vac7 was mutagenized by site-directed mutagenesis using the following primers, respectively: S593A(FW29 and REV29), S607A(FW30 and REV30), T858A(FW31 and REV31), S903A(FW32 and REV32), S912A(FW33 and REV33), T1050A(FW34 and REV34), and S1074A(FW35 and REV35).
To insert TEVr after Vac7-P571, pRS413 Vac7 was mutagenized by site-directed mutagenesis using the primers FW36 and REV36. To insert the NheI site after TEVr, pRS413 Vac7-TEVr572 was mutagenized by site-directed mutagenesis using the primers FW37 and REV37. To generate pRS413 Vac7-TEVr572-HA3, an HA3 fragment was amplified by PCR using the primers FW38 and REV38, digested by NheI and SpeI, and inserted to NheI site of pRS413 Vac7-TEVr572-NheI.
To generate pVT-Pho85 or HA-Pho85 plasmid, Pho85 or HA-Pho85 was amplified by PCR using the primers FW-Pho85, FW–HA-Pho85, and REV-Pho85. A BamHI–PstI fragment of Pho85 or HA-Pho85 was inserted into the BamHI and PstI sites of pVT102-U. To generate pVT-Pho85F82G or HA-Pho85F82G plasmids, pVT-Pho85 or pVT–HA-Pho85 was mutagenized by site-directed mutagenesis using the primers FW39 and REV39.
To generate the pVT–HA-Pho80 plasmid, Pho80 was amplified by PCR using the primers FW40 and REV40. A BamHI–PstI fragment of HA-Pho80 was inserted into the BamHI and PstI sites of pVT102-U.
To generate pET-Pho80, Pho80 was amplified by PCR using the primers FW41 and REV41, digested by NdeI and XhoI, and inserted to NdeI and XhoI sites of pET28.
To generate pVT-vYFPN, vYFPN was amplified by PCR using the primers FW42 and REV42, digested by BamHI and XhoI, and inserted to BamHI and XhoI sites of pVT-102-U. To generate pVT-vYFPN-Pho80, Pho80 was amplified by PCR using the primers FW43 and REV43, digested by XhoI and PstI, and inserted to XhoI and PstI sites of pVT-vYFPN.
To generate pRS416 Pho81 plasmid, a fragment including the 5′UTR of Pho81 (−1,564 to −5 bp) was amplified by PCR using the primers FW44 and REV44, digested using SacII and SpeI, and inserted to SacII and SpeI sites of pRS416 (pRS416 Pho81-1). A fragment including the 5′UTR (−300 to 2,852 bp) of Pho81 was amplified by PCR using the primers FW45 and REV45, digested using SpeI and EcoRI, and inserted to SpeI and EcoRI sites of pRS416 Pho81-1 (pRS416 Pho81-1 + 2). A fragment of 2,658 bp of Pho81 to the 3′UTR (526 bp) of Pho81 was amplified by PCR using the primers FW46 and REV46, digested using EcoRI and SalI, and inserted to EcoRI and SalI sites of pRS416 Pho81-1 + 2. To generate pRS416 Pho81R701S, pRS416 Pho81 was mutagenized by site-directed mutagenesis using the primers FW47 and REV47.
To generate the pcDNA-HA-p35 plasmid, p35 was amplified by PCR using the primers FW48 and REV48. A p35 fragment was digested by XbaI and XhoI and inserted to XbaI and XhoI sites of pcDNA. To generate pEGFP-Cdk5 plasmid, Cdk5 was amplified by PCR using the primers FW49 and REV49. A fragment pfCdk5 was digested by HindIII and SacII and inserted to HindIII and SacII sites of pEGFP.
To generate the pMAL-Fab1 (1,941–2,278) or pMAL-Fab1 (1,501–1,969) plasmids, a fragment was amplified by PCR using the primers FW50 and REV50 and FW51 and REV51, respectively. A PCR fragment (1,941–2,278) was digested by XbaI and HindIII and inserted to XbaI and HindII sites of pMALc2H10T. A PCR fragment (1,501–1,969) was digested by SalI and PstI and inserted to SalI and PstI sites of pMALc2H10T. To generate pMAL-fab1 mutant plasmids, pMAL-Fab1 was mutagenized by site-directed mutagenesis using the aforementioned primers.
To generate the pMAL-Vac7 (394–918) plasmid, a fragment of Vac7 was amplified by PCR using the primers FW52 and REV52. A fragment was digested by BamHI and PstI and inserted to the BamHI and PstI sites of pMALc2H10T.
To generate pMAL-PIKfyve (1,214–1,594) or pMAL-PIKfyve (1,700–2,098) plasmids, a fragment was amplified by PCR using the primers FW53 and REV53 and FW54 and REV54, respectively. A PCR fragment was digested by BamHI and SalI and inserted to BamHI and SalI sites of pMALc2H10T.
Fluorescence microscopy
Yeast was grown to mid-log phase and labeled with FM4-64 (Vida and Emr, 1995). Images were acquired using a DeltaVision Elite Restoration Microscopy System (Applied Precision Ltd.).
Viability and growth assay in hyperosmotic stress
For plate assays, cells were grown in SC-Ura media to mid-log phase and then resuspended to 0.1 OD/ml, and 10-fold serial dilutions were spotted onto plates and incubated at 24°C for 10 d. For viability assays with FM4-64, cells were grown in YEPD media to mid-log phase and then were treated with or without 0.9 M NaCl for 4 h. Yeast was incubated with 0.012 mg/ml FM4-64 for 1 min, and cells were analyzed by fluorescence microscopy. For growth assays, cells were grown to mid-log phase and then were treated with 0.9 M NaCl for 16 h. Cell numbers were assessed by OD600.
Gel shift assay with PPase and TEVp
Mid-log phase cells were lysed with 10% TCA and resuspended with 2× SDS sample buffer. The dilution buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 µM EDTA, and 0.5% Tween-20) was added to the lysate and incubated with 10,000 U/µl PPase (New England Biolabs, Inc.) and 150 U/ml TEVp (Promega) at 30°C for 1 h. Reactions were terminated with equal volumes of 2× SDS sample buffer.
Protein purification
Proteins were expressed in Escherichia coli BL21 star (DE3) with pRARE. Cells were grown in Luria-Bertani broth with 0.05 mg/ml ampicillin and 0.025 mg/ml spectinomycin at 37°C to an OD600 of 0.3. After 18 h of induction with 0.2 mM IPTG at 16°C, cells were harvested and sonicated in lysis buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM DTT, and 1× EDTA-free protease inhibitor [Roche]). Proteins were purified with amylose resin (New England Biolabs, Inc.), dialyzed in buffer overnight (10 mM Tris-HCl, pH 7.4, 50 mM NaCl, and 1 mM DTT), and then concentrated with an Ultrafree-4 centrifugal filter 50K or 30K membrane (EMD Millipore).
In vitro kinase assay
To test phosphorylation of recombinant Fab1 and Vac7 peptides, pho80Δ mutant cells expressing pHA-Pho80 were grown to mid-log phase, collected, and lysed in 50 mM Tris-HCl, pH 7.5, 2 mM sodium pyrophosphate, 0.1% SDS, 1% sodium deoxycholate, 1% Triton X-100, 1 mM PMSF, and 1× protein inhibitor cocktail (Sigma-Aldrich) and then centrifuged at 13,000 g for 10 min. The supernatant was incubated with anti-HA antibody and immobilized on protein A Sepharose beads (Sigma-Aldrich). Beads were washed three times with lysis buffer, twice with lysis buffer containing 150 mM NaCl, and then twice with kinase buffer (10 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 50 mM NaCl, 2 mM EDTA, and 1 mM DTT). Beads were incubated with 20–40 µg substrate protein, 2.5 mM ATP, and 0.125 µCi γ-[32P]ATP in kinase buffer at 30°C for 30 min. Reactions were terminated with equal volumes 2× SDS sample buffer. For recombinant peptides of human PIKfyve, HEK293 cells expressing pEGFP-Cdk5 and pHA-p35 were lysed in 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1% NP-40, 5 mM sodium pyrophosphate, 1% sodium deoxycholate, 2 mM EDTA, 1 mM DTT, 1 mM PMSF, 1× protein inhibitor cocktail, 10 µg/ml leupeptin, and 10 µg/ml aprotinin and then centrifuged at 13,000 g for 10 min. Supernatants were incubated with anti-HA antibody and immobilized on protein A Sepharose beads. Beads washed three times with lysis buffer, twice with lysis buffer containing 150 mM NaCl, and then twice with kinase buffer (20 mM Hepes, pH 7.4, 20 mM MgCl2, and 1 mM DTT). Beads were incubated with 20–40 µg substrate protein, 2.5 mM ATP, and 0.125 µCi [γ-32P]ATP in kinase buffer at 30°C for 60 min. Reactions were terminated with equal volumes 2× SDS sample buffer.
TEVp protection assay
Mid-log phase cells were lysed in TEV buffer (20 mM Hepes, pH 7.4, 100 mM NaCl, 10 mM EDTA, 1× protease inhibitor [Sigma-Aldrich], and 1 mM DTT), incubated with 0.5% Octyl β-d-glucopyranoside for 30 min, and centrifuged at 13,000 g for 10 min. Supernatants were incubated with TEVp (Promega) at 4°C. Reactions were terminated with an equal volume 2× SDS sample buffer.
Antibodies for Western blot analysis
1:80,000 mouse monoclonal anti-Pgk1 (22C5D8; Thermo Fisher Scientific), 1:10,000 mouse monoclonal anti-HA (901502; BioLegend), 1:10,000 mouse monoclonal anti-GFP (11814460001; Roche), 1:10,000 mouse monoclonal anti-His (51-9000012; BD), 1:10,000 rabbit polyclonal anti-MBP (E8030S; New England Biolabs, Inc.), and 1:10,000 rabbit polyclonal anti-TAP, CAB1001; Thermo Fisher Scientific) were used. 1:3,000 anti–Fab1-T1953-P is a custom rabbit polyclonal antibody raised by 21st Century Biochemicals.
Inositol labeling and measurement of phosphorylated phosphoinositide lipids
Measurements were performed as described previously by Duex et al. (2006a) and Zhang et al. (2007) with the following modification: for MEF cells labeled with myo[2-3H], after 48 h, cultures were rinsed with PBS and treated with 1 ml of 4.5% (vol/vol) perchloric acid for 15 min at room temperature; plates were then rotated gently every 2 min to prevent cells from drying. Cells were scraped off with a cell lifter (3008; Costar) and spun at 12,000 g for 10 min. For yeast labeled with myo[2-3H], after 16 h, cells were rinsed with media, treated with or without high-salt media (0.9 M NaCl), and then lysed in 4.5% (vol/vol) perchloric acid by homogenization using BeadBeater (BIOSPEC PRODUCTS) for 2 min three times. Lysate was spun at 12,000 g for 10 min.
Pellets were washed at room temperature with 1 ml of 0.1 M EDTA and then were resuspended in 50 µl water. To deacylate lipids, samples were transferred to a glass vial, mixed with 1 ml methanol/40% methylamine/l-butanol (45.7% methanol, 10.7% methylamine, and 11.4% l-butanol [vol/vol]), and incubated at 55°C for 1 h. Samples were vacuum dried, resuspended in 0.5 ml water, and extracted twice with an equal volume of butanol/ethyl ether/ethyl formate (20:4:1 [vol/vol]). The aqueous phase was vacuum dried and resuspended in 60 µl water, and 50 µl of each sample was analyzed by HPLC (UFLC CBM-20A Lite; Shimadzu) using an anion exchange 4.6 × 250–mm column (4621-1505; PartiSphere SAX; Whatman).
Online supplemental material
Fig. S1 provides supporting data to Figs. 1, 2, and 3, which provide evidence that Pho85–Pho80 is an upstream regulator of Fab1. Fig. S2 provides supporting data for the Pho85–Pho80 target sites on Fab1 and Vac7. Fig. S3 provides a sequence comparison of Vac7 among some fungal species and provides assays of the Fab121A and Fab18A mutants. Fig. S4 shows that the Fab18A mutant forms a normal complex with Vac14 and Fig4. Fig. S5 provides an explanation of the quantitation of the Western blots. Table S1 is a list of the plasmids used in this study. Table S2 is a list of yeast strains used in this study. Table S3 is a list of the primers used in this study.
Supplementary Material
Acknowledgments
We thank Drs. Sai Srinivas Panapakkam Giridharan, Yanling Zhang, and Sergey Zolov for discussions of methods to measure phosphoinositide lipid levels in MEFs. We thank Dr. Inoki for discussions of methods for in vitro kinase assays. We thank Drs. Daniel Klionsky and Junya Hasegawa and the Weisman Lab for helpful discussions.
This work was supported by National Institutes of Health grants R01-GM050403 and NS064015 and the Protein Folding Diseases FastForward Initiative sponsored by the University of Michigan.
The authors declare no competing financial interests.
Author contributions: N. Jin, Y. Jin, and L.S. Weisman conceived and designed the experiments. N. Jin performed the experiments. N. Jin and L.S. Weisman wrote the manuscript with input from Y. Jin.
Footnotes
Abbreviations used:
- 5-FOA
- 5-fluoroortic acid
- HOG
- high-osmolarity glycerol
- MEF
- mouse embryonic fibroblast
- PPase
- protein phosphatase
- SC
- synthetic complete
- TEV
- Tobacco etch virus
- TEVp
- TEV protease
- TEVr
- TEV recognition
- TM
- transmembrane
References
- Birkeland S.R., Jin N., Ozdemir A.C., Lyons R.H. Jr., Weisman L.S., and Wilson T.E.. 2010. Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole-genome sequencing. Genetics. 186:1127–1137. 10.1534/genetics.110.123232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J., and Cross F.R.. 2007. Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell Biol. 8:149–160. 10.1038/nrm2105 [DOI] [PubMed] [Google Scholar]
- Bonangelino C.J., Catlett N.L., and Weisman L.S.. 1997. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol. Cell. Biol. 17:6847–6858. 10.1128/MCB.17.12.6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino C.J., Nau J.J., Duex J.E., Brinkman M., Wurmser A.E., Gary J.D., Emr S.D., and Weisman L.S.. 2002. Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J. Cell Biol. 156:1015–1028. 10.1083/jcb.200201002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho R.J., Efe J.A., Teis D., and Emr S.D.. 2008. Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol. Biol. Cell. 19:4273–4286. 10.1091/mbc.E08-04-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster J.L., de Valoir T., Dwyer N.D., Winter E., and Gustin M.C.. 1993. An osmosensing signal transduction pathway in yeast. Science. 259:1760–1763. 10.1126/science.7681220 [DOI] [PubMed] [Google Scholar]
- Carroll A.S., and O’Shea E.K.. 2002. Pho85 and signaling environmental conditions. Trends Biochem. Sci. 27:87–93. 10.1016/S0968-0004(01)02040-0 [DOI] [PubMed] [Google Scholar]
- Carroll A.S., Bishop A.C., DeRisi J.L., Shokat K.M., and O’Shea E.K.. 2001. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc. Natl. Acad. Sci. USA. 98:12578–12583. 10.1073/pnas.211195798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C.Y., Zhang Y., Dowling J.J., Jin N., Adamska M., Shiga K., Szigeti K., Shy M.E., Li J., Zhang X., et al. . 2007. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 448:68–72. 10.1038/nature05876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserzö M., Wallin E., Simon I., von Heijne G., and Elofsson A.. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673–676. 10.1093/protein/10.6.673 [DOI] [PubMed] [Google Scholar]
- Dong X.P., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y., Weisman L.S., Delling M., and Xu H.. 2010. PI(3,5)P2 controls membrane trafficking by direct activation of mucolipin Ca2+ release channels in the endolysosome. Nat. Commun. 1:38 10.1038/ncomms1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove S.K., Cooke F.T., Douglas M.R., Sayers L.G., Parker P.J., and Michell R.H.. 1997. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 390:187–192. 10.1038/36613 [DOI] [PubMed] [Google Scholar]
- Duex J.E., Nau J.J., Kauffman E.J., and Weisman L.S.. 2006a Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot. Cell. 5:723–731. 10.1128/EC.5.4.723-731.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex J.E., Tang F., and Weisman L.S.. 2006b The Vac14p–Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J. Cell Biol. 172:693–704. 10.1083/jcb.200512105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallazzini M., Heussler G.E., Kunin M., Izumi Y., Burg M.B., and Ferraris J.D.. 2011. High NaCl-induced activation of CDK5 increases phosphorylation of the osmoprotective transcription factor TonEBP/OREBP at threonine 135, which contributes to its rapid nuclear localization. Mol. Biol. Cell. 22:703–714. 10.1091/mbc.E10-08-0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary J.D., Wurmser A.E., Bonangelino C.J., Weisman L.S., and Emr S.D.. 1998. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143:65–79. 10.1083/jcb.143.1.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E.V., Hudson C.A., Vertommen D., Rider M.H., and Tavaré J.M.. 2010. Regulation of PIKfyve phosphorylation by insulin and osmotic stress. Biochem. Biophys. Res. Commun. 397:650–655. 10.1016/j.bbrc.2010.05.134 [DOI] [PubMed] [Google Scholar]
- Ho C.Y., Choy C.H., Wattson C.A., Johnson D.E., and Botelho R.J.. 2015. The Fab1/PIKfyve phosphoinositide phosphate kinase is not necessary to maintain the pH of lysosomes and of the yeast vacuole. J. Biol. Chem. 290:9919–9928. 10.1074/jbc.M114.613984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K.S.W. 1993. TMbase - A database of membrane spanning proteins segments. Biol. Chem. Hoppe Seyler. 374:166. [Google Scholar]
- Holt L.J., Tuch B.B., Villén J., Johnson A.D., Gygi S.P., and Morgan D.O.. 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 325:1682–1686. 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Patrick G., Moffat J., Tsai L.H., and Andrews B.. 1999. Mammalian Cdk5 is a functional homologue of the budding yeast Pho85 cyclin-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 96:14445–14450. 10.1073/pnas.96.25.14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov O.C., Sbrissa D., Delvecchio K., Xie Y., Jin J.P., Rappolee D., and Shisheva A.. 2011. The phosphoinositide kinase PIKfyve is vital in early embryonic development: Preimplantation lethality of PIKfyve−/− embryos but normality of PIKfyve+/− mice. J. Biol. Chem. 286:13404–13413. 10.1074/jbc.M111.222364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N., Chow C.Y., Liu L., Zolov S.N., Bronson R., Davisson M., Petersen J.L., Zhang Y., Park S., Duex J.E., et al. . 2008. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P2 in yeast and mouse. EMBO J. 27:3221–3234. 10.1038/emboj.2008.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A., Herskowitz I., Tjian R., and O’Shea E.K.. 1994. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 263:1153–1156. 10.1126/science.8108735 [DOI] [PubMed] [Google Scholar]
- Kaiser C.M.S., and Mitchell A.. 1994. Methods in yeast genetics and genomics. Cold Spring Harbor Laboratory Press, New York. 256 pp. [Google Scholar]
- Kerppola T.K. 2008. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37:465–487. 10.1146/annurev.biophys.37.032807.125842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Colwill K., Aneliunas V., Tennyson C., Moore L., Ho Y., and Andrews B.. 1998. Interaction of yeast Rvs167 and Pho85 cyclin-dependent kinase complexes may link the cell cycle to the actin cytoskeleton. Curr. Biol. 8:1310–1321. 10.1016/S0960-9822(07)00561-1 [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Huang K., Quiocho F.A., and O’Shea E.K.. 2008. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 4:25–32. 10.1038/nchembio.2007.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.C., Diakov T.T., Rizzo J.M., and Kane P.M.. 2012. Vacuolar H+-ATPase works in parallel with the HOG pathway to adapt Saccharomyces cerevisiae cells to osmotic stress. Eukaryot. Cell. 11:282–291. 10.1128/EC.05198-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.C., Diakov T.T., Xu T., Tarsio M., Zhu W., Couoh-Cardel S., Weisman L.S., and Kane P.M.. 2014. The signaling lipid PI(3,5)P2 stabilizes V1-V(o) sector interactions and activates the V-ATPase. Mol. Biol. Cell. 25:1251–1262. 10.1091/mbc.E13-10-0563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney A.J., Zolov S.N., Kauffman E.J., Zhang Y., Strunk B.S., Weisman L.S., and Sutton M.A.. 2014. Activity-dependent PI(3,5)P2 synthesis controls AMPA receptor trafficking during synaptic depression. Proc. Natl. Acad. Sci. USA. 111:E4896–E4905. 10.1073/pnas.1411117111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer L., Borgne A., Mulner O., Chong J.P., Blow J.J., Inagaki N., Inagaki M., Delcros J.G., and Moulinoux J.P.. 1997. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 243:527–536. 10.1111/j.1432-1033.1997.t01-2-00527.x [DOI] [PubMed] [Google Scholar]
- Nicolson T.A., Weisman L.S., Payne G.S., and Wickner W.T.. 1995. A truncated form of the Pho80 cyclin redirects the Pho85 kinase to disrupt vacuole inheritance in S. cerevisiae. J. Cell Biol. 130:835–845. 10.1083/jcb.130.4.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M., Kanaya Y., and Toh-E A.. 1999. Mouse cyclin-dependent kinase (Cdk) 5 is a functional homologue of a yeast Cdk, pho85 kinase. J. Biol. Chem. 274:33859–33862. 10.1074/jbc.274.48.33859 [DOI] [PubMed] [Google Scholar]
- O’Rourke S.M., and Herskowitz I.. 2004. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell. 15:532–542. 10.1091/mbc.E03-07-0521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T., Ward J.M., Huh C.G., Longenecker G., Veeranna H.C. Pant R.O. Brady L.J. Martin, and Kulkarni A.B.. 1996. Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc. Natl. Acad. Sci. USA. 93:11173–11178. 10.1073/pnas.93.20.11173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., and Posas F.. 2012. Response to hyperosmotic stress. Genetics. 192:289–318. 10.1534/genetics.112.140863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D., and Shisheva A.. 2005. Acquisition of unprecedented phosphatidylinositol 3,5-bisphosphate rise in hyperosmotically stressed 3T3-L1 adipocytes, mediated by ArPIKfyve-PIKfyve pathway. J. Biol. Chem. 280:7883–7889. 10.1074/jbc.M412729200 [DOI] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O.C., and Shisheva A.. 1999. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J. Biol. Chem. 274:21589–21597. 10.1074/jbc.274.31.21589 [DOI] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O.C., Fu Z., Ijuin T., Gruenberg J., Takenawa T., and Shisheva A.. 2007. Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J. Biol. Chem. 282:23878–23891. 10.1074/jbc.M611678200 [DOI] [PubMed] [Google Scholar]
- Seeburg D.P., Feliu-Mojer M., Gaiottino J., Pak D.T., and Sheng M.. 2008. Critical role of CDK5 and Polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron. 58:571–583. 10.1016/j.neuron.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer B., and Powers T.. 2015. Target of rapamycin signaling mediates vacuolar fission caused by endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Biol. Cell. 26:4618–4630. 10.1091/mbc.E15-06-0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacloig-Pedros E., Bets-Plasencia C., Pascual-Ahuir A., and Proft M.. 2015. Coordinated gene regulation in the initial phase of salt stress adaptation. J. Biol. Chem. 290:10163–10175. 10.1074/jbc.M115.637264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T.A., and Emr S.D.. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792. 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. 1992. Membrane protein structure prediction: Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487–494. 10.1016/0022-2836(92)90934-C [DOI] [PubMed] [Google Scholar]
- Wang Y.X., Zhao H., Harding T.M., Gomes de Mesquita D.S., Woldringh C.L., Klionsky D.J., Munn A.L., and Weisman L.S.. 1996. Multiple classes of yeast mutants are defective in vacuole partitioning yet target vacuole proteins correctly. Mol. Biol. Cell. 7:1375–1389. 10.1091/mbc.7.9.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., DeWald D.B., Boronenkov I.V., Anderson R.A., Emr S.D., and Koshland D.. 1995. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol. Biol. Cell. 6:525–539. 10.1091/mbc.6.5.525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zolov S.N., Chow C.Y., Slutsky S.G., Richardson S.C., Piper R.C., Yang B., Nau J.J., Westrick R.J., Morrison S.J., et al. . 2007. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc. Natl. Acad. Sci. USA. 104:17518–17523. 10.1073/pnas.0702275104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolov S.N., Bridges D., Zhang Y., Lee W.W., Riehle E., Verma R., Lenk G.M., Converso-Baran K., Weide T., Albin R.L., et al. . 2012. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl. Acad. Sci. USA. 109:17472–17477. 10.1073/pnas.1203106109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.