Mitochondrial stress activates a concerted mitonuclear response to safeguard and repair mitochondrial function. Quirós et al. assessed the transcriptome, proteome, and metabolome of mammalian cells under four types of stressors and combine population genetic analyses and in vivo studies to show that ATF4 coordinates the mitochondrial stress response.
Abstract
Mitochondrial stress activates a mitonuclear response to safeguard and repair mitochondrial function and to adapt cellular metabolism to stress. Using a multiomics approach in mammalian cells treated with four types of mitochondrial stressors, we identify activating transcription factor 4 (ATF4) as the main regulator of the stress response. Surprisingly, canonical mitochondrial unfolded protein response genes mediated by ATF5 are not activated. Instead, ATF4 activates the expression of cytoprotective genes, which reprogram cellular metabolism through activation of the integrated stress response (ISR). Mitochondrial stress promotes a local proteostatic response by reducing mitochondrial ribosomal proteins, inhibiting mitochondrial translation, and coupling the activation of the ISR with the attenuation of mitochondrial function. Through a trans–expression quantitative trait locus analysis, we provide genetic evidence supporting a role for Fh1 in the control of Atf4 expression in mammals. Using gene expression data from mice and humans with mitochondrial diseases, we show that the ATF4 pathway is activated in vivo upon mitochondrial stress. Our data illustrate the value of a multiomics approach to characterize complex cellular networks and provide a versatile resource to identify new regulators of mitochondrial-related diseases.
Introduction
Mitochondria are key for energy metabolism within the cell, as they produce most of the cellular ATP through oxidative phosphorylation (OXPHOS) and are essential for regulating intermediate metabolism (Nunnari and Suomalainen, 2012). Changes in mitochondrial function impact not only cellular metabolism but also whole-body metabolism, thus affecting healthspan and lifespan. Nearly 300 human diseases are caused by mutations or defects in mitochondrial proteins, half of which are consequence of defects in OXPHOS, for which treatment options remain scarce (Koopman et al., 2012). Additionally, a decline in mitochondrial function is also at the basis of many other common disorders, including diseases that affect the metabolic, muscular, neurological, and immune systems (Andreux et al., 2013), that are often linked with aging (Houtkooper et al., 2010; López-Otín et al., 2013, 2016). Mitochondrial function is also affected in cancer. However, unlike in other diseases, here they are not at the origin of tumorigenesis; cancer cells modify cellular metabolism and mitochondrial function for their own purposes, using them as anabolic machines (Zong et al., 2016). Understanding how mitochondria respond to stress and how mitonuclear communication pathways adapt cellular metabolism to environmental changes is thus not only critical from a basic point of view but also has great translational value given the link between mitochondria and a variety of diseases.
Mitochondria are signaling organelles that continuously communicate with the nucleus (Chandel, 2015; Quirós et al., 2016; Matilainen et al., 2017). This coordination is extremely important for energy harvesting, because OXPHOS complex components are encoded in both nuclear and mitochondrial genome, and their expression needs to be tightly regulated to ensure proper assembly and function (Couvillion et al., 2016). Although mitochondria contain an intricate and intrinsic mitochondrial protein quality control system, mainly mediated by proteases (Quirós et al., 2015), upon mitochondrial stress, mitonuclear signals also instruct the nucleus to activate an adaptive response. Mitochondrial stress stimulates the synthesis of specific mitochondrial proteins by activating mitochondrial biogenesis and counteracting the impact of the mitochondrial stress on cellular homeostasis (Yoneda et al., 2004; Houtkooper et al., 2013; Palikaras et al., 2015). One of the best-characterized retrograde stress responses is the mitochondrial unfolded protein response (UPRmt), which has been studied mainly in invertebrates, such as Caenorhabditis elegans and Drosophila melanogaster. In these organisms, mitochondrial proteotoxic stress activates the UPRmt, which results in the induction of the transcription of a set of genes encoding proteases, chaperones, and metabolic enzymes aimed at restoring mitochondrial function and cellular homeostasis (Yoneda et al., 2004; Durieux et al., 2011; Nargund et al., 2012; Houtkooper et al., 2013; Owusu-Ansah et al., 2013). However, in mammals, it is largely unknown how the UPRmt is activated, executed, and integrated with other general cellular stress responses.
To shed light on these mitochondrial stress pathways, we used a multiomics approach in mammalian cells challenged by four different compounds that alter mitochondrial function. Of note, the canonical UPRmt response that was defined in invertebrates was not activated by these stressors. Instead, under the conditions used, they activate an integrated stress response (ISR) in which the activating transcription factor 4 (ATF4) is the main effector. ATF4 promotes the expression of particular cytoprotective genes that reprogram and rewire cellular metabolism toward the synthesis of key metabolites, in particular serine. Mitochondrial stress also promotes a local proteostatic response by specifically decreasing mitochondrial ribosomal proteins (MRPs), which attenuate mitochondrial translation. In conclusion, our data assign a primordial role to ATF4 and the MRPs as main effectors of mitochondrial stress signaling and define a critical role of both inputs in regulating mitochondrial function. In addition our data provide a useful resource that can be exploited to identify new regulators of mitochondrial homeostasis and actors in mitochondrial-related diseases.
Results
Mitochondrial stressors effectively alter mitochondrial function
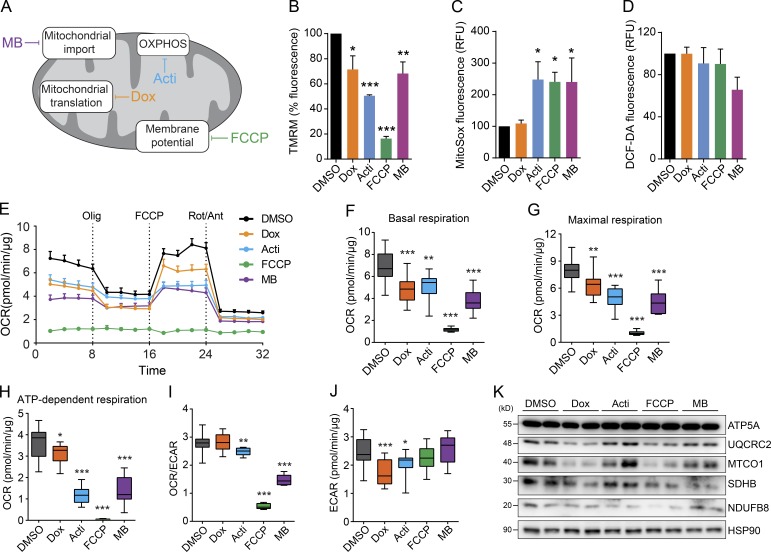
To identify a putative common mitochondrial stress response, we treated cells with four drugs that alter mitochondrial proteostasis in a different way: doxycycline, which inhibits mitochondrial translation (Moullan et al., 2015); actinonin, which alters the stability of OXPHOS proteins (Richter et al., 2015); the ionophore carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), which disrupts mitochondrial membrane potential; and MitoBloCK-6 (MB), a small molecule that impairs mitochondrial protein import of both Mia40/Erv1 and TIM22 pathways (Fig. 1 A; Dabir et al., 2013). We used HeLa cells because of their ease of handling, high metabolic plasticity, and previous success to study mitochondrial function (Lanning et al., 2014; Münch and Harper, 2016). First, we validated our experimental system by characterizing different parameters of mitochondrial function affected by each treatment. The compounds decreased mitochondrial membrane potential to a different extent, with FCCP being the most effective (Fig. 1 B). Further, they all increased mitochondrial reactive oxygen species (ROS), except for doxycycline, without increasing the general cellular ROS levels (Fig. 1, C and D). The different stressors also decreased both basal and maximal mitochondrial respiration (Fig. 1, E–G), as well as ATP-dependent respiration (Fig. 1 H). Except in the case of doxycycline, we also observed a decrease in the ratio of oxidative versus glycolytic metabolism (oxygen consumption rate [OCR]/extracellular acidification rate [ECAR]); ECAR levels were also decreased (or showed a tendency toward a decrease), suggesting that mitochondrial stress promotes a general decline of cellular metabolism instead of a shift to glycolysis to compensate for the mitochondrial defects (Fig. 1, I and J). Accordingly, expression of various OXPHOS subunits was also decreased with these compounds, with the exception for actinonin (Fig. 1 K). Together, these results validate our system to map mitochondrial stress pathways that are activated by distinct compounds that alter mitochondrial proteostasis in different ways and to different degrees.
Figure 1.
Mitochondrial stress alters mitochondrial function. (A) Schematic representation of the mechanism of action of the compounds selected for the analysis. Acti, actinonin; Dox, doxycycline; MB, MitoBloCK-6. (B) Mitochondrial membrane potential after 24 h of treatment with the selected chemicals. Tetramethylrhodamine, methyl ester (TMRM) was used to determine mitochondrial membrane potential (n = 4 independent experiments; mean values ± SEM). (C) Mitochondrial and (D) total ROS levels after 24 h of treatment with the selected chemicals. Dichlorofluorescin diacetate (DCF-DA) reflects total cellular ROS levels, whereas MitoSox measure mitochondrial superoxide level. RFU, relative fluorescence units (n = 4 independent experiments; mean values ± SEM). (E) Oxygen consumption rate (OCR) of cells treated with the different compounds. Dashed vertical lines indicate the subsequent addition of the ATPase inhibitor oligomycin (Olig.), the uncoupling reagent FCCP and the inhibitors of the electron transport chain rotenone/antimycin A (Rot/Ant). (F and G) Boxplots representing OCR (F) in basal conditions and (G) after treatment with the uncoupler FCCP (maximal respiration). (H) Boxplot representing the ATP-dependent respiration (oligomycin-sensitive respiration) calculated as the difference in OCR before and after the addition of oligomycin. (I) Ratio of OCR and extracellular acidification rate (ECAR) as an indicator of the relation between mitochondrial respiration and glycolysis. (J) ECAR in basal conditions as indication of glycolytic rate. For E–J, n = 2 independent experiments, using 10 replicates per experiment; mean values ± SEM of a representative experiment. (K) Inmunoblot analysis showing the effects of the compounds on different mitochondrial OXPHOS subunits (ATPA5, complex V; UCQRC2, complex III; MTCO1, complex IV; SDHB, complex II; and NDUFB8, complex I). HSP90 was used as loading control. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Differential changes in transcriptome and proteome upon mitochondrial stress
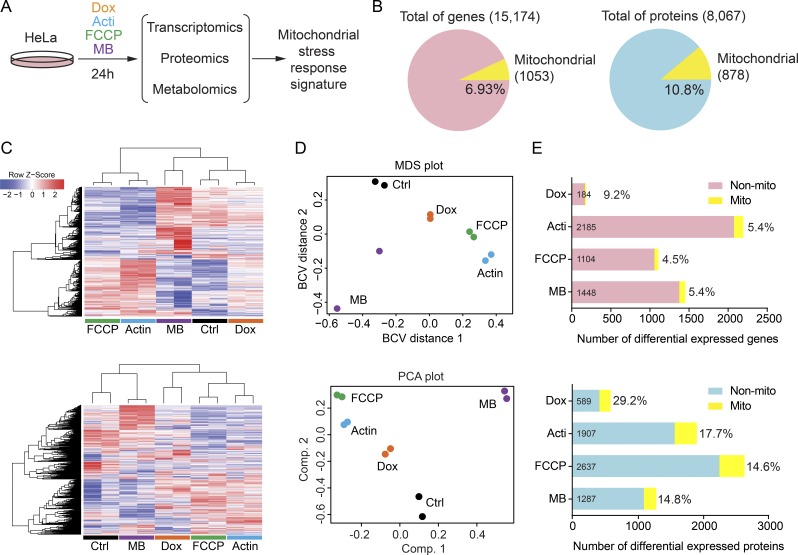
Changes in the transcriptome, proteome, and metabolome of HeLa cells treated for 24 h with the four different compounds (doxycycline, actinonin, FCCP, and MB) were determined by RNA sequencing, quantitative proteomic analysis using tandem mass tag (TMT) technology, and nontargeted metabolomics analysis (Fig. 2 A). We initially focused on changes in transcripts and proteins, and we identified 15,174 unique transcripts and 8,067 unique proteins in all conditions, of which 1,053 transcripts and 878 proteins were mitochondrial, based on MitoCarta2.0 (Calvo et al., 2016), representing 6.9% and 10.8% of the transcriptome and proteome, respectively (Fig. 2 B and Tables S3 and S4). Hierarchical clustering and heatmap analysis of the duplicate samples indicated the reproducible nature of our analysis, as exemplified by the similarity between replicates, but also highlighted differential expression changes between the treatments (Fig. 2 C). Multidimensional scaling and principal-component analysis also confirmed a good accordance between biological duplicates, as well as the close clustering of transcripts and proteins upon different treatments (Fig. 2 D). In fact, these clustering methods suggested that doxycycline induces a similar but less pronounced response as compared with actinonin and FCCP, whereas MB displayed a completely different transcriptional and proteomic response (Fig. 2, C and D). Using a significance level of false discovery rate (FDR) <0.05, we identified the differentially expressed genes and proteins upon each stress (Fig. 2 E; and Fig. S1, A and B). For each compound, the observed response was different, showing in some conditions more changes at the transcript level, as for actinonin and MB, whereas upon doxycycline and FCCP treatment, the protein changes were more pronounced (Fig. 2 E and Fig. S1, A and B). Importantly, in all conditions, changes affecting mitochondria were much higher in the proteome than in the transcriptome, with the percentage of differentially expressed mitochondrial proteins being approximately three times higher than differential genes (Figs. 2 E and S1 C).
Figure 2.
RNA sequencing and proteomic analysis upon mitochondrial stress. (A) Schematic representation of the omics experiments. HeLa cells were treated with compounds for 24 h, and the transcriptome, proteome, and metabolome were analyzed. Control cells were treated with DMSO. (B) Pie charts representing the total number of genes and proteins identified in all conditions by RNA sequencing and TMT quantitative proteomic analysis. Number of mitochondrial genes and proteins are represented in both charts, as well as the percentage relative to the total. (C) Heatmap analysis of the transcriptome (top) and proteome (bottom). Hierarchical clustering of samples (columns) and genes/proteins (rows) was based on Pearson’s correlation coefficient to measure the distance and the mean to cluster the samples. (D) Multidimensional scaling (MDS) using the distances between transcript expression profiles (top) and principal-component analysis (PCA) of the proteomic profiles (bottom) show a similar clustering between the duplicates and treatments. BCV, biological coefficient of variation; Comp, principal component. (E) Bar plots representing the number of differentially expressed (DE) genes (top) and proteins (bottom) in each condition (FDR 5%). Percentage of DE mitochondrial genes and proteins is shown, observing a greater impact on the mitochondrial proteome than on the mitochondrial transcriptome. Acti, actinonin; Dox, doxycycline; MB, MitoBloCK-6.
Next, we compared the changes in the levels of 7,885 transcripts and proteins that were commonly detected in each analysis (Fig. S1 D). Also for the common genes, changes in the proteome were also more pronounced than those observed in the transcriptome, and only a limited overlap between the changes in transcripts and proteins was detected, demonstrating that stress response differentially affects the transcriptome and proteome (Fig. S1 E). Nevertheless, we observed a positive and highly significant correlation between the transcriptomic and proteomic changes in all conditions (Fig. S1 F). However, despite the extreme significance owing to the high number of observations, it is evident that the higher correlations (rho of ∼0.4) were associated with compounds that induced major changes, such as actinonin and FCCP (Fig. S1 F). This is in agreement with the fact that each stress generates a different response and that mRNA and protein levels are not perfectly correlated (Chick et al., 2016; Williams et al., 2016). Altogether, these results demonstrate that mitochondrial stress differentially alters the cellular transcriptome and proteome, with the changes at the protein level in mitochondria being more pronounced.
Mitochondrial stress decreases MRPs, inhibits mitochondrial translation, and alters epigenetic regulators
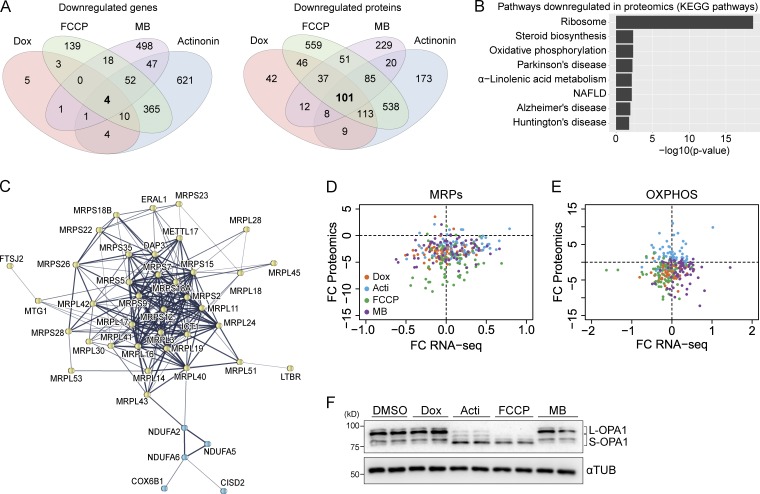
We next explored the down-regulated transcripts and proteins that were common to all stress conditions. Using a FDR of 5%, we identified only four genes down-regulated in RNA sequencing, whereas 101 proteins were identified by proteomic analysis (Fig. 3 A and Table S5). These differences are in agreement with the global changes in the transcriptome and proteome described above (Fig. 2 E). Notably, among the 101 down-regulated proteins, 44 were mitochondrial (Table S5). Enrichment analysis of these down-regulated proteins identified “Ribosome” as the top down-regulated pathway, as well as “Oxidative phosphorylation” and “Mitochondrial translation” (Figs. 3 B and S2 A). Network analysis of down-regulated proteins pointed out the specific enrichment in MRPs, proteins related to mitochondrial translation, and OXPHOS subunits (Fig. 3 C). Interestingly, the decrease in MRPs was only observed in the proteome; no changes were observed at the transcript level (less than onefold change; Figs. 3 D and S2 B). OXPHOS proteins were also decreased at the protein level, except for actinonin, where an increase in these proteins was observed (Figs. 3 E and S2 C). This result demonstrates that actinonin causes an accumulation of nonfunctional OXPHOS proteins, because mitochondrial respiration was decreased (Fig. 1, E and F), validating the proposed mechanism of action of actinonin, which was reported to impair the turnover of de novo mitochondrial protein synthesis (Richter et al., 2015). Similarly to MRPs, mitochondrial stress did not change OXPHOS transcripts (Figs. 3 E and S2 C). Notably, individual analysis of each stressor also evidenced changes in mitochondrial translation and OXPHOS as the main enriched pathways (Tables S6 and S7).
Figure 3.
Mitochondrial stress inhibits mitochondrial translation and decreases mitochondrial ribosomal proteins and OXPHOS complexes. (A) Venn diagrams showing commonly down-regulated genes (top) and proteins (bottom) with an FDR < 0.05. (B) Enrichment analysis of 101 proteins down-regulated was performed using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and represented as negative of log10 of p-value after Bonferroni correction. (C) A STRING network showing the enrichment in mitochondrial ribosomal proteins (MRPs; yellow) and OXPHOS proteins (blue). Line thickness indicates the strength of the association. (D and E) Correlation of transcript and protein data for the (D) MRPs and (E) OXPHOS components with each stressor. Values are represented as log2-fold changes (FC) relative to control. Dashes lines depict the 0 values to indicate the up- or down-regulation in each analysis. (F) Western blot analysis of OPA1 upon treatment for 24 h with each stressor. Long OPA1 isoforms (L-OPA1) and short OPA1 isoforms (S-OPA1) are indicated. Acti, actinonin; Dox, doxycycline; MB, MitoBloCK-6; NAFLD, non-alcoholic fatty liver disease.
Of note, mitochondrial stress also decreased the protein levels of OPA1 (Table S5), a mitochondrial inner membrane GTPase that participates in the inner membrane fusion and cristae maintenance (Frezza et al., 2006; Cogliati et al., 2013). Upon different mitochondrial insults, long OPA1 isoforms are proteolytically processed by the stress-dependent mitochondrial protease OMA1, inhibiting mitochondrial fusion and promoting mitochondrial fragmentation (Quirós et al., 2012; Baker et al., 2014). However, despite the decrease in total protein levels, only actinonin and FCCP triggered the processing of the long OPA1 isoforms (Figs. 3 F and S2 D; showing different times).
Besides mitochondrial proteins, some epigenetic regulators were also down-regulated upon mitochondrial stress. These include the histone methyl transferases SETD2, SUV39H1, and ATF7IP, as well as components of some histone acetyltransferase complexes, such as MORF4L1, ZZZ3, and MRGBP (Table S5). These data confirm recent findings that mitochondrial activity is required for histone acetylation (Martínez-Reyes et al., 2016) and that mitochondrial stress activates different histone methyl transferases and demethylases to regulate stress adaptation (Schroeder et al., 2013; Merkwirth et al., 2016; Tian et al., 2016).
Mitochondrial stress activates metabolic pathways of amino acid biosynthesis
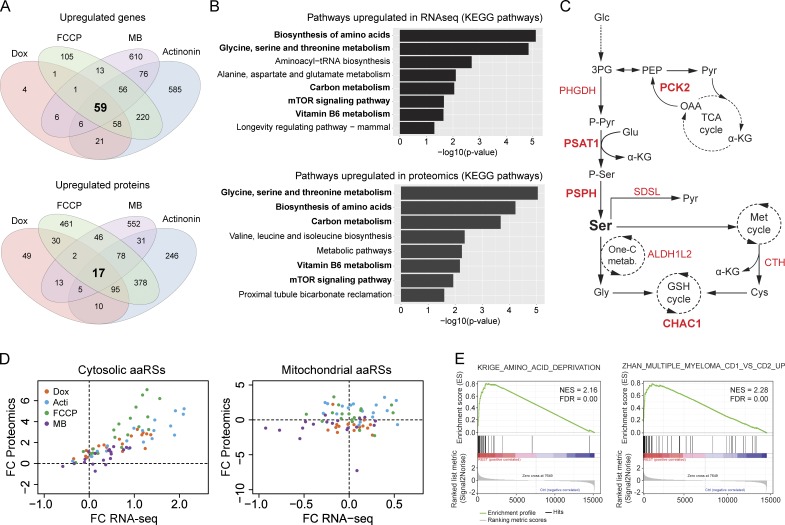
Next, we analyzed the common up-regulated genes and proteins in mitochondrial stress conditions. Unlike down-regulated changes, here we found that the major increase was observed at the transcript level, with 59 genes and only 17 proteins commonly up-regulated (Fig. 4 A and Table S5). Notably, among those changes, only four genes and two proteins were mitochondrial (Table S5), illustrating that the main response upon mitochondrial stress does not involve the induction of mitochondrial genes. Enrichment analysis of both RNA sequencing and proteomic analysis identified pathways related to biosynthesis of amino acids (Fig. 4 B), in particular biosynthesis of serine (Figs. 4 B and S3 A). Specifically, pathways of “Biosynthesis of amino acids,” “Glycine, serine and threonine metabolism,” “Carbon metabolism,” “Vitamin B6 metabolism,” and “mTOR signaling pathway” were enriched in both transcriptome and proteome (Fig. 4 B). Analysis of individual genes and proteins also showed the up-regulation of the key enzymes of serine biosynthesis (PSAT1, PSPH, and PHGDH), as well as serine dehydratase, SDSL (Fig. 4 C and Table S5). These changes suggest that serine is generated de novo from 3-phosphoglycerate and converted into pyruvate, also replenishing intermediates of the tricarboxylic acid cycle, such as α-ketoglutarate (Fig. 4 C). In addition, we observed an induction of the mitochondrial enzyme phosphoenolpyruvate carboxykinase 2 (PCK2), which allows the cells to use alternative cataplerotic pathways to convert tricarboxylic acid cycle intermediates into glycolytic intermediates (Fig. 4 C and Table S5). Furthermore, the up-regulation of some enzymes of the one-carbon metabolism (ALDH1L2), cysteine biosynthesis (CTH), and glutathione (GSH) cycle (CHAC1) also suggested an increase in the flux toward the transsulfuration pathway to synthesize GSH, supported by the methionine cycle (Fig. 4 C and Table S5). In addition, several amino acid transporters (SLC1A4, SLC1A5, SLC6A9, and SLC7A11), as well as key enzymes for the synthesis of asparagine (ASNS) and arginine (ASS1), were also induced (Table S5).
Figure 4.
Mitochondrial stress increases biosynthesis of amino acids. (A) Venn diagrams showing commonly up-regulated transcripts (top) and proteins (bottom) with an FDR < 0.05. (B) Enrichment analysis of 59 transcripts (top) and 17 proteins (bottom) up-regulated were performed using KEGG pathways and are represented as negative of log10 of p-value after Bonferroni correction. Common enriched pathways in RNA sequencing and proteomics are represented in bold. (C) Graphical representation of enzymes and metabolites of de novo serine biosynthesis and their integration in cellular metabolism. Enzymes highlighted in red are up-regulated in RNA sequencing and in red bold in both RNA sequencing and proteomics. (D) Correlation of cytosolic and mitochondrial amino acid-tRNA synthetase (aaRS) genes and proteins in each stress condition. Values are represented as log2-fold changes (FC) relative to control. Dashes lines depict the 0 values to indicate the up- or down-regulation in each analysis. (E) Enrichment score plots from GSEA using a combine mitochondrial stress expression extracted from the RNA seq. 3PG, 3-phosphoglycerate; Acti, actinonin; Dox, doxycycline; FDR, false discovery rate; Glc, glucose; MB, MitoBloCK-6; NES, normalized enrichment score; PEP, phosphoenolpyruvate; PHGDH, phosphoglycerate dehydrogenase; PSAT1, phosphoserine aminotransferase 1; PSPH, phosphoserine phosphatase; Pyr, pyruvate; SDSL, serine dehydratase; Ser, serine; TCA, tricarboxylic acid.
Moreover, “Aminoacyl−tRNA biosynthesis” was also enriched in RNA sequencing results (Fig. 4 B). However, only cytosolic amino acid–tRNA synthetases (aaRSs) were consistently induced at both the mRNA and protein level; mitochondrial aaRSs showed variable regulation depending of each stressor (Fig. 4 D). Gene set enrichment analysis (GSEA) of the transcriptomes of all stressors evidenced similarity in gene expression changes with those seen after amino acid deprivation (Krige et al., 2008) and with several cancer studies (Figs. 4 E and S3 B). Notably, despite the differential changes induced by each stressor, individual enrichment analysis of transcripts modulated by each compound also showed the activation of those metabolic pathways as the main stress responses (Tables S6 and S7).
We also observed the up-regulation of two proteins associated with the ubiquitin-proteasome system, UBA5 and UHRF1BP1 (Table S5), which might suggest an increase in protein degradation. In fact, in other species like yeast, mitochondrial stress can trigger the activation of the proteasome to degrade mitochondrial proteins accumulated in the cytosol (Wrobel et al., 2015). However, proteasome activity was decreased in all stress conditions, with the exception of doxycycline (Fig. S3 C), which can be explained by the increase in mitochondrial ROS (Fig. 1 C), which is known to inhibit proteasome activity (Segref et al., 2014).
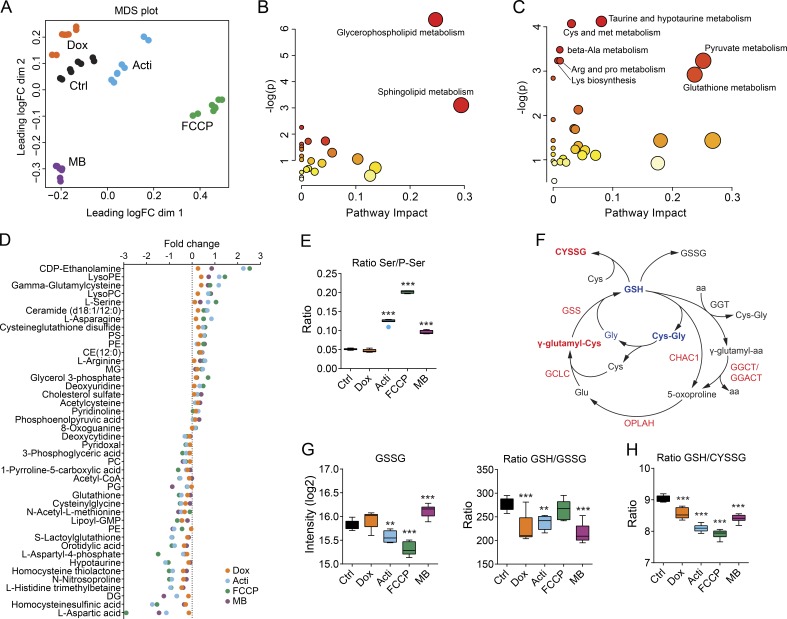
Mitochondrial stress alters cellular metabolism
To study the metabolic footprint of mitochondrial stress, we analyzed the changes in the metabolome induced by exposure to mitochondrial stressors. Approximately 300 detectable peaks were putatively annotated as deprotonated metabolites by accurate mass and used to explore the differences between conditions. Both hierarchical clustering with heatmap analysis and multidimensional scaling segregated the samples by compound, tightly grouping the replicates (Figs. 5 A and S3 D). These results were similar to those observed in the transcriptome and proteome analysis (Fig. 2, C and D); however, in this case, the moderate impact of doxycycline was even more evident (Figs. 5 A and S3 D). Analysis of differentially expressed metabolites and pathway enrichment analysis with the unique metabolites showed that pathways of glycerophospholipid and sphingolipid metabolism were the most highly induced upon mitochondrial stress (Fig. 5 B). These pathways were also indicated by analyzing the top up-regulated and down-regulated metabolites in common between the stressors, where we found increased levels of ceramide, CDP-ethanolamine, and several species of lysophosphatidycholine and lysophosphatidylethanolamine (Fig. 5 D). These changes reveal that the increase in serine might be used to promote the synthesis of lipids and phospholipids through ceramide, which has a key role during mitochondrial stress in C. elegans (Liu et al., 2014; Kim et al., 2016). Of note, we also found several metabolites related to cholesterol and prostaglandin metabolism to be up-regulated (Fig. 5 D), in line with previous studies (Rauthan et al., 2013; Andreux et al., 2014; Liu et al., 2014). Importantly, these findings suggest a high degree of conservation in the link between mitochondrial stress and lipid metabolism across different organisms.
Figure 5.
Metabolomic analysis of mitochondrial stress. (A) Multidimensional scaling (MDS) using the unique metabolites identified. Metabolome pathway enrichment of (B) up-regulated and (C) down-regulated metabolites upon mitochondrial stress. Scatterplots represents p-values from integrated enrichment analysis and impact values from pathway topology analysis. The node color is based on the p-values and the node radius represents the pathway impact values. (D) Dot plot representing the fold change of the top changed metabolites in each mitochondrial stress condition. Fold change is represented in log scale and all metabolites show significant differences with an FDR <0.05. (E) Box-whisker plots representing the ratio of serine and phosphoserine levels in each mitochondrial stress condition. (F) Graphical representation of enzymes and metabolites of the glutathione (GSH) cycle. Enzymes and metabolites highlighted in red are up-regulated upon mitochondrial stress, whereas those in blue are down-regulated. (G and H) Box-whisker plots representing the levels of (G) oxidized glutathione (GSSG) and the ratios of GSH to GSSG (GSH/GSSG) and (H) GSH to cysteine glutathione disulfide (GSH/CYSSG). **, P < 0.01; ***, P < 0.001. Acti, actinonin; CE, cholesterol ester; DG, diacylglycerol; Dox, doxycycline; MB, MitoBloCK-6; MG, monoacylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PS, phosphatidylserine.
On the other hand, analysis of the top down-regulated pathways showed alterations in several amino acid–related pathways, such as taurine and hypotaurine, cysteine and methionine, and GSH metabolism, among others (Fig. 5 C). Notably, many amino acids and derivatives were also significantly altered, but our resolution could not completely distinguish them from other metabolites, although in general an increase in almost all amino acids was observed (Table S8). Analysis of individual metabolites confirmed the up-regulation of serine, asparagine, and arginine (Fig. 5 D), consistent with the increase in their biosynthetic enzymes (Fig. 4 C and Table S5). The increase in serine, which was one of the most highly induced metabolites, was also indicated by the increase in the ratio serine/phosphoserine, providing further support for the up-regulation of the activity of the serine biosynthetic pathway (Fig. 5, D and E). Conversely, one of the metabolites most decreased in all conditions was aspartic acid (Fig. 5 D), a finding that is consistent with the up-regulation of ASNS and the consequent increase in asparagine (Fig. 5 D and Table S5).
Moreover, one of the most up-regulated metabolites was γ-glutamylcysteine (Fig. 5 D), which is a precursor of GSH (Fig. 5 F). However, GSH levels were decreased (Fig. 5, D and F), suggesting a decrease in its synthesis, an increase in its degradation, or an increase in the oxidized forms (Fig. 5 F). To assess the origin of this reduction in GSH, we ascertained the levels of the different enzymes and metabolites implicated in its cycle. Oxidized glutathione (GSSG) showed differential levels depending on the stressor; however, the GSH/GSSG ratio was decreased with almost all stressors (Fig. 5 G), indicating that the oxidative damage may explain the GSH depletion. Further, almost all the enzymes of the GSH cycle were increased at the protein level, and gamma-glutamylcyclotransferase CHAC1 was extremely up-regulated (Fig. S3 E), suggesting an increase in GSH cycle metabolism. Intriguingly, we observed an increase in the levels of cysteine-GSH disulfide (Fig. 5, D and F) and a decrease in the GSH/cysteine-GSH disulfide ratio (Fig. 5 H), which further explains the decline in GSH levels.
Besides amino acid related metabolism, other important metabolites were also changed, reflective of global metabolic changes induced upon mitochondrial stress. We found increased levels of phosphoenolpyruvic acid, which was consistent with the up-regulation of PCK2 and the activation of the cataplerotic reaction from OAA (Figs. 4 C and 5 D). On the other hand, the decrease in orotidylic acid (Fig. 5 D), which is implicated in the synthesis of uridine, was remarkable and likely attributed to the decreased activity of dihydroorotate dehydrogenase, which requires the electron transport chain for its normal function (Grégoire et al., 1984). Further, acetyl coenzyme A was also down-regulated (Fig. 5 D), agreeing with the starvation-like status and the decrease in mitochondrial function (Mariño et al., 2014; Martínez-Reyes et al., 2016). Altogether, these changes demonstrate that upon mitochondrial dysfunction, a compensatory response is triggered, aimed at synthesizing key metabolites, which maintain cellular metabolism, promote the synthesis of certain lipids, and regulate the pool of GSH.
ATF4 coordinates the mitochondrial stress response
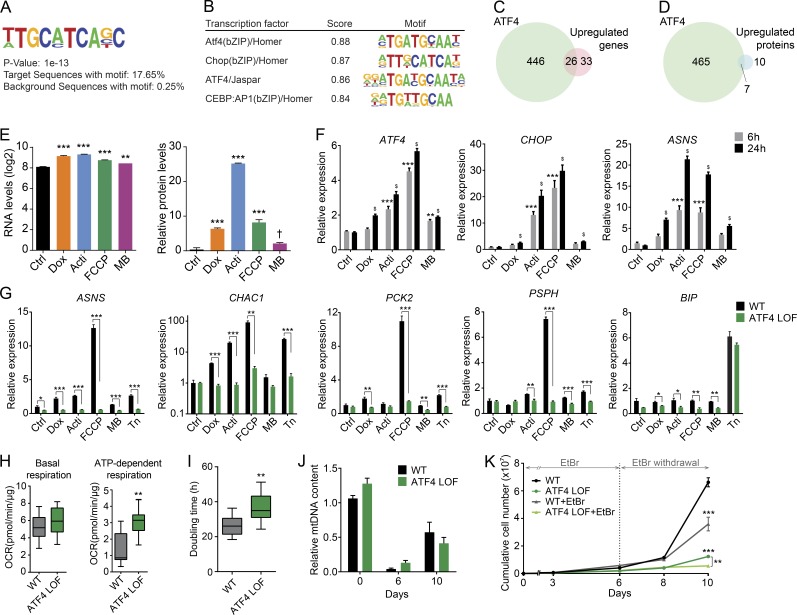
To identify the putative factors that control this transcriptional stress response, we performed a de novo motif analysis in the common up-regulated genes using HOMER (Heinz et al., 2010). Among all possible motifs, the top-scoring motif, and the only one that was not assigned as a possible false positive, was 5′-TTGCATGACG-3′, which was found in 17.65% of the target genes (Fig. 6 A). This target sequence was similar to the motif bound by bZIP-containing proteins, such as ATF4, DDIT3/CHOP, and CEBPB, with ATF4 being the most likely and highest scoring transcription factor associated with this sequence (Fig. 6 B). We indeed confirmed the likely involvement of ATF4 by comparing the chromatin immunoprecipitation and sequence data available for this factor, as well as for CHOP (Han et al., 2013), with the changes in transcript level observed in our RNA sequencing data. Remarkably, approximately half of the genes (26/59) that were induced in common by the different mitochondrial stressors were also ATF4 targets (Fig. 6 C). This overlap was also evident when comparing up-regulated proteins (Fig. 6 D). Of note, some of the up-regulated genes (13/59) also were potential CHOP targets; however, because all those genes were also ATF4 targets and CHOP only overlapped with half of them, we hypothesized that CHOP activation is a downstream event in this context (Fig. S4 A). Accordingly, ATF4 transcript and protein levels were induced upon mitochondrial stress (Fig. 6 E), being activated also at early time points (Fig. 6 F).
Figure 6.
ATF4 mediates the mitochondrial stress response. (A) Consensus de novo motif identified using HOMER software by selecting the common up-regulated genes in all stress conditions. (B) Representation of the common transcription factors that match with the identified motif. (C and D) Venn diagram showing the similarity between the ATF4 targets and the common induced (C) transcripts and (D) proteins reflecting mitochondrial stress in RNA sequencing and proteomics analysis, respectively. ATF4 chromatin immunoprecipitation and sequence: GSE35681. (E) Relative mRNA, represented as normalized counts, and relative protein levels of ATF4. Data were extracted from RNA sequencing and proteomics, respectively. †, P < 0.1; **, P < 0.01; ***, P < 0.001. (F) Transcript levels of ATF4, CHOP (DDIT3), and ASNS upon 6 and 24 h of exposure to the different mitochondrial stressors. Expression of ATF4 and its targets genes showed time-dependence activation. **, P < 0.01; ***, P < 0.001 relative to control at 6 h; $, P < 0.001 relative to control at 24 h. (G) mRNA expression analysis of ATF4 targets, ASNS, CHAC1, PCK2, PSPH, and the ER stress marker BIP upon 6 h of treatment with the different mitochondrial stressors and the ER stressor tunicamycin (Tn at 2.5 µg/ml) in WT and ATF4 LOF HeLa cells. (H) Boxplots showing the basal and ATP-dependent respiration of ATF4 LOF HeLa cells. OCR, oxygen consumption rate. (I) Doubling time in hours of WT and ATF4 LOF HeLa cells. (J) Relative mtDNA levels of WT and ATF4 LOF HeLa cells after 6 d of 50 ng/ml EtBr treatment and 4 d of recovery (10 d). (K) Cumulative cell number of WT and ATF4 LOF HeLa cells treated or untreated with EtBr for 10 d (6 d with EtBr and 4 d of recovery). All experiments were independently performed at least two times, using triplicates for each condition; data are presented as mean ± SEM of a representative experiment, *, P < 0.05; **, P < 0.01; ***, P < 0.001. Acti, actinonin; Dox, doxycycline; MB, MitoBloCK-6.
To further consolidate the role of ATF4 in mitochondrial stress response, we generated HeLa cells deficient in ATF4 using CRISPR-Cas9 technology (Fig. S4 B). ATF4 loss of function (LOF) was confirmed by treating these cells with the mitochondrial stressor actinonin and the ER stressor tunicamycin (Fig. S4 C), which also induces ATF4 during the unfolded protein response in the ER (UPRer; Hetz, 2012). ATF4 LOF abrogated the mitochondrial stress response, illustrated by the decreased levels of several of its targets, including ASNS, CHAC1, PCK2, and PSHP (Fig. 6 G). Treatment with tunicamycin also activated the same ATF4-dependent response, which indeed was suppressed in the ATF4-deficient cells (Fig. 6 G). Mitochondrial stress did not, however, activate the expression of the ER chaperone BIP (also known as HSP5A or GRP78), a classical UPRer marker, and therefore, ATF4 LOF did not abolish the tunicamycin-mediated BIP induction (Fig. 6 G). Similarly, other UPRer branches, including those governed by the ATF6 or IRE1/XBP1 pathways, were not activated upon mitochondrial stress (Fig. S4 D).
ATF4 LOF increased ATP-dependent respiration (Fig. 6 H), suggesting a role in regulating mitochondrial function; however, it also promoted a decrease in cellular proliferation, exemplified by an increase in cellular doubling time (Fig. 6 I). We then explored the role of ATF4 during the recovery from mitochondrial stress induced by mitochondrial DNA (mtDNA) depletion. WT and ATF4 LOF HeLa cells were treated with ethidium bromide (EtBr) for 6 d, showing a comparable depletion in mtDNA (Fig. 6 J). After EtBr removal, both cell types recovered mtDNA levels at similar rates (Fig. 6 J), indicating that ATF4 is not involved in mtDNA metabolism. Strikingly, ATF4 LOF cells proliferated significantly less than control cells, with and without EtBr treatment; this decrease in proliferation was even more enhanced upon recovery from mtDNA depletion (Fig. 6 K), indicating that ATF4 is essential to maintain cell proliferation and to protect against mitochondrial stress.
ATF4 is activated through the ISR
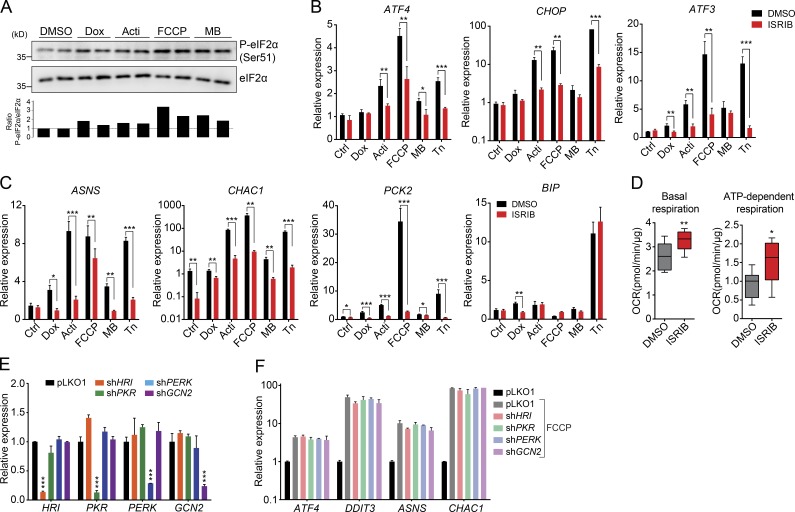
ATF4 is one of the prototypical transcription factors activated during the ISR. The ISR is activated by different cellular stressors and culminates in the phosphorylation of the translation initiation factor eIF2α, which reduces total protein synthesis while allowing preferential translation of some transcription factors that have upstream ORFs in their 5′ UTRs, like ATF4 and ATF5 (Ron, 2002; Pakos-Zebrucka et al., 2016). eIF2α can be phosphorylated by four kinases, each of which is activated by different forms of cellular stress: PERK (ER stress), GCN2 (amino acid starvation), PKR (double-stranded RNA), and HRI (reduced heme levels; Balachandran et al., 2000; Harding et al., 2000, 2003; Han et al., 2001). Although ATF4 acts as a common downstream target that integrates signals from those eIF2α kinases, it has also been reported to be activated by an independent mechanism upon mitochondrial stress (Münch and Harper, 2016) or even in an mTOR-dependent way (Ben-Sahra et al., 2016). All mitochondrial stressors that we used stimulated the phosphorylation of eIF2α (Fig. 7 A). Furthermore, treatment with ISRIB, an ISR inhibitor that blocks eIF2α phosphorylation (Sidrauski et al., 2015), reduced the induction of ATF4 and several of its target genes (Fig. 7, B and C). As observed upon ATF4 LOF (Fig. 6 G), ISRIB treatment also abolished the activation of the ATF4 pathway mediated by tunicamycin (Fig. 7, B and C) but did not suppress the activation of BIP (Fig. 7 C). Further, ISRIB also increased basal and ATP-dependent respiration (Fig. 7, D), supporting that the activation of the ISR and the attenuation in cytosolic translation also affect mitochondrial function in nonstress conditions.
Figure 7.
ATF4 is activated through the ISR. (A) Western blot analysis showing the increased phosphorylation of eIF2α (Ser51) upon 6 h of treatment with the different mitochondrial stressors. Bottom, ratio between P-eIF2α and eIF2α total levels. (B and C) mRNA expression analysis of ATF4 and its target genes, CHOP (DDIT3), ASNS, CHAC1, PCK2, and the ER stress marker BIP, upon 6 h of treatment with the different mitochondrial stressors and the ER stressor tunicamycin (Tn at 2.5 µg/ml) in HeLa cells, together with the inhibitor of the integrated stress response (ISRIB at 500 nM). (D) Boxplots showing an increase in basal and ATP-dependent respiration of HeLa cells treated with 500 nM of ISRIB for 24 h. OCR: oxygen consumption rate. (E) mRNA expression analysis of eIF2α kinases upon knock down with specific shRNAs. Data are presented as mean ± SEM of two independent shRNAs for each gene. Statistical differences were calculated compared with pLKO1. (F) mRNA expression analysis of ATF4 and some of its target genes upon knock down of the eIF2α kinases and 6 h of treatment with FCCP. Data are presented as mean ± SEM of two independent shRNAs for each gene. No statistical differences were found between the FCCP treated conditions. All experiments were independently performed at least two times, using triplicates for each condition; data are presented as mean ± SEM of a representative experiment; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Acti, actinonin; Dox, doxycycline; MB, MitoBloCK-6.
Finally, to identify which kinase acts upstream of the ISR activation upon mitochondrial stress, we knocked down the four known eIF2α kinases individually in HeLa cells using shRNAs (Fig. 7 E) and thereafter treated the cells with FCCP, which activates the ATF4 pathway most robustly. Individual knockdown of the kinases, however, did not abolish the induction of ATF4 and its target genes (Fig. 7 F). These results demonstrate that mitochondrial stress activates the ATF4 pathway through the ISR but independently of the activation of an individual eIF2α kinase.
Mitochondrial stress activates the ATF4 pathway in vitro and in vivo
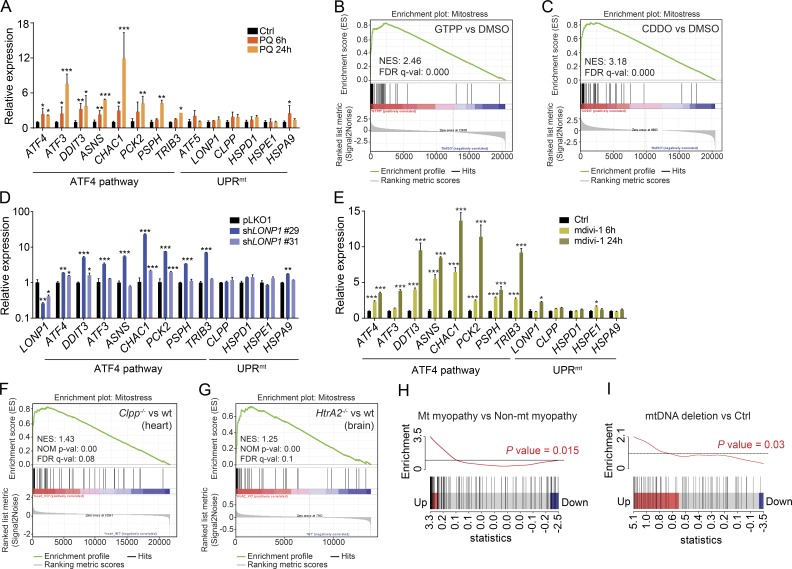
It has been previously reported that alterations in mitochondrial proteostasis activate the UPRmt (Yoneda et al., 2004; Durieux et al., 2011; Houtkooper et al., 2013; Münch and Harper, 2016). However, in our study, we did not detect consistently the activation of the prototypical UPRmt, as reflected by HSPD1, HSPE1, or CLPP expression (Fig. S4 E). To unravel if these differences were based on the type of stress, we tried to reproduce the original conditions used to trigger the UPRmt in mammalian cells by transfecting a mutant form of ornithine transcarbamylase (ΔOTC), which was reported to induce protein misfolding in mitochondria (Zhao et al., 2002). However, we could not detect the activation of the UPRmt, as previously reported (Fig. S4 F; Zhao et al., 2002). Next, we used paraquat, an inhibitor of mitochondrial complex I that increases mitochondrial ROS levels, which has been also described as potent UPRmt inducer through the activation of ATF5 (Fiorese et al., 2016). However, paraquat treatment consistently activated the ATF4 pathway, having no effect on ATF5 and the UPRmt genes (Fig. 8 A). Inhibition of the mitochondrial chaperone TRAP1 by GTPP or of the Lon protease (LONP1) by CDDO has also been reported to activate the UPRmt, as well as the ATF4 pathway in vitro (Münch and Harper, 2016). We then compared the transcripts induced by both inhibitors with the changes observed in our stress conditions by performing a GSEA using the core set of genes commonly up-regulated with our four stressors, hereafter referred as mt-stress genes (Table S5). Expression of mt-stress genes was significantly enriched in HeLa cells treated with GTPP and CDDO (Münch and Harper, 2016; Fig. 8, B and C), indicating that inhibition of TRAP1 and LONP1 also activated a similar stress response pathway mediated by ATF4. Likewise, knockdown of LONP1 expression, which promotes the accumulation of misfolded proteins and induces a robust mitochondrial proteotoxic stress (Bernstein et al., 2012; Quirós et al., 2014), also activated the ATF4 pathway but did not consistently activate the classical UPRmt markers (Fig. 8 D). Importantly, the ATF4 pathway was also induced by the same mitochondrial stressors in mouse embryonic fibroblasts (MEFs; Fig. S4 G), demonstrating that this stress response is not restricted to a specific cellular model. In combination, these results suggest that under the conditions tested, mitochondrial stress in mammalian cells does not activate the classical prototypical UPRmt, defined in worms (Yoneda et al., 2004; Haynes et al., 2007; Durieux et al., 2011; Houtkooper et al., 2013) or in mammals (Martinus et al., 1996; Zhao et al., 2002; Münch and Harper, 2016), but rather induces the ATF4 pathway via the activation of the ISR.
Figure 8.
Mitochondrial stress activates the ATF4 pathway in vitro and in vivo independent of the UPRmt. (A) mRNA expression analysis of the ATF4 pathway and UPRmt genes in HeLa cells 6 and 24 h after exposure to paraquat (PQ) at a final concentration of 400 µM. (B and C) Enrichment score plots from GSEA using RNA sequencing data of cells treated with (B) the TRAP1 inhibitor GTPP and (C) the LONP1 inhibitor CDDO (GSE75247). List of mitochondrial stress genes (mt-stress genes; Table S5) was used as the gene set of interest. FDR q-val, false discovery rate adjusted p-value; NES, normalized enrichment score. (D and E) mRNA expression analysis of the ATF4 pathway and UPRmt genes in HeLa cells after (D) knockdown of LONP1 using two different shRNAs and (E) 6 and 24 h of exposure to mdivi-1 at a final concentration of 50 µM. (F and G) Enrichment score plots from GSEA using microarray data (F) from hearts of mice deficient in ClpP (GSE40207) and (G) from brains of mice deficient in Htra2 (GSE13035). NOM p-val, nominal p-value. (H and I) Barcode plot representing the enrichment in the core mt-stress genes in (H) mitochondrial myopathies versus nonmitochondrial myopathies (GSE43698) and (I) myopathies caused by mtDNA deletions (GSE1462). Enrichment was analyzed using the fold-change relative to control. p-value was calculated using the ROAST test, which confirmed that gene expression changes induced by mitochondrial stress significantly correlate with gene expression changes caused by mitochondrial myopathies. Black vertical lines represent the genes, and the red and blue rectangles the cutoff for the up-regulated and down-regulated genes, respectively. The black horizontal line indicates neutral enrichment, whereas the red line shows the enrichment of up-regulated genes. All experiments were independently performed at least two times, using triplicates for each condition; data are presented as mean ± SEM of a representative experiment; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We next studied the relationship between ATF4 activation and other mitochondrial stress responses, such as the regulation of mitochondrial dynamics. As we previously described, stressors that promote OPA1 processing and mitochondrial fragmentation, like FCCP and actinonin (Figs. 3 F and S2 D) or knockdown of LONP1 (Quirós et al., 2014), also activated the ATF4 pathway. However, because OPA1 was not processed in all stress conditions analyzed (Figs. 3 F and S2 D), the transcriptional stress response mediated by ATF4 and mitochondrial fragmentation are probably two noninterdependent stress pathways. Further, treatment with mdivi-1, an inhibitor of the dynamin GTPase DRP1 that blocks mitochondrial division (Cassidy-Stone et al., 2008), also activated the ATF4 pathway in a time-dependent way (Fig. 8 E), demonstrating that different mitochondrial stress conditions can activate the ATF4 pathway independent of mitochondrial fragmentation. Notably, mdivi-1 treatment also did not consistently activate the UPRmt (Fig. 8 E).
Finally, we explored whether the activation of mt-stress genes can also arise in mitochondrial dysfunction in vivo or in human mitochondrial diseases. For this, we analyzed the expression of the mt-stress genes in tissues derived from mice deficient in the mitochondrial quality control proteases Clpp (Gispert et al., 2013) and HtrA2 (Moisoi et al., 2009). Deficiency in both mitochondrial proteases, which trigger mitochondrial proteotoxic stress and promote severe physiological alterations, activated the same ATF4 response in heart and brain, respectively, as illustrated in the enrichment plots obtained from GSEA (Fig. 8, F and G). Importantly, we also found that expression of the mt-stress genes was significantly correlated with gene expression changes observed in human mitochondrial myopathies (Fig. 8 H), especially in those with mtDNA deletions (Fig. 8 I). Collectively, these data demonstrate that mitochondrial stress activates the ATF4 pathway both in vitro and in vivo and is also implicated in human disease with a mitochondrial origin.
Genetic link between ATF4 pathway and mitochondrial stress in human and mouse populations
To further explore the link between the activation of the ATF4 pathway and mitochondrial function, we analyzed the expression of mt-stress genes in the GTEx data (GTEx Consortium, 2013) and the BXD mouse genetic reference population (Andreux et al., 2012). Network analysis using multiple tissue correlations showed a tight clustering of the mt-stress genes, supporting the idea of a common in vivo coregulation across 49 human tissues (Fig. 9 A) and 16 mouse tissues (Fig. 9 B) in a nonstressed basal situation. Next, we correlated the expression of different transcription factors, including ATF4, DDIT3/CHOP, and CEBPB, with all the other genes in the 49 human tissues from GTEx data, which show a tighter clustering for the mt-stress genes compared with the BXD data. Enrichment analysis of the top negative-correlated genes identified several metabolic and mitochondrial pathways as linked with ATF4, but not with DDIT3/CHOP, befitting with the predominant role of ATF4 in mitochondrial homeostasis (Fig. 9 C). Importantly, OXPHOS correlated negatively with the expression of both ATF4 and CEBPB (Fig. 9 C). As expected, NCOR1, a well-known mitochondrial repressor (Yamamoto et al., 2011), also showed a robust negative correlation with OXPHOS (Fig. 9 C), validating our experimental approach. These results indicate that the activation of the ATF4 pathway seems to oppose mitochondrial function, in agreement with our previous results in cells upon ATF4 LOF or treatment with ISRIB (Figs. 6 H and 7 D). Importantly, the fact that the relationship of ATF4 and mitochondrial function occurs in basal nonstressed conditions in vivo, in which the mt-stress genes showed a transcriptional coregulation, extends the ATF4 function from a pure stress response to a role in homeostasis.
Figure 9.
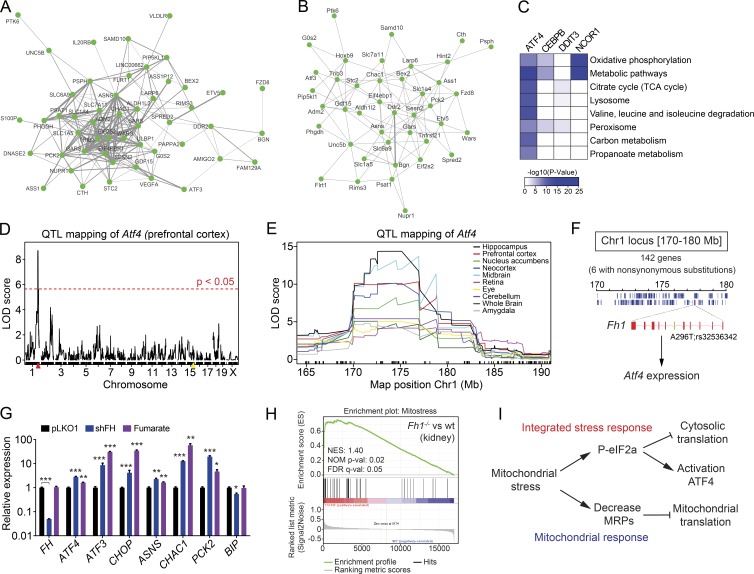
Genetic link of ATF4 and mitochondrial stress in human and mouse populations. (A and B) Multitissue correlation network analysis of transcript levels across (A) 49 human tissues in GTEx and (B) 16 mouse tissues in the BXD genetic reference population. Nodes represent the transcripts and the width of the ties among nodes indicate the probability to show a significant positive correlation in all tissues analyzed. The human network shows a tighter clustering likely caused by the higher number of tissues and samples per tissues. (C) Heatmap representing the KEGG pathway analysis of the top negative-correlated genes across 49 tissues using GTEx data for ATF4, CEBPB, DDIT3/CHOP, and NCOR1 (used as positive control). Color key represents the negative logarithm base 10 of the p-value of each pathway obtained in the analysis. (D) Expression quantitative trait locus (eQTL) analysis of Atf4 transcript level in the prefrontal cortex. The yellow mark represents the Atf4 gene locus on chromosome 15, whereas the red mark represent the position of a trans-eQTL for Atf4 expression levels on chromosome 1 (Chr1). (E) eQTL mapping of Atf4 transcript levels across several tissues identifies a common and strong trans-eQTL on chromosome 1 (170–180 Mb). (F) Representation of the chromosome 1 locus (170–180 Mb) containing 142 genes, 6 of which have nonsynonymous substitutions, and only one gene, Fh1, encodes a mitochondrial protein. Fh1 contains a nonsynonymous sequence variant (A296T; rs32536342) that segregates in the BXDs. This sequence variant regulates the expression levels of Fh1, which in turn regulates Atf4 expression. (G) mRNA expression analysis of HeLa cells after knockdown of fumarate hydratase (shFH) and treatment with monomethyl fumarate at 2.5 mM for 24 h. Data are presented as mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (H) Enrichment score plot from GSEA using microarray data from renal cysts of mice with renal tubule specific inactivation of Fh1 (Fh1−/−; GSE29988). The list of mitochondrial stress genes (mt-stress genes; Table S5) was used as the gene set of interest. FDR q-val: false discovery rate adjusted p-value; NES, normalized enrichment score; NOM p-val, nominal p-value. (I) Scheme summarizing our working hypothesis. Mitochondrial stress stimulates the phosphorylation of the eIF2α, which inhibits cytosolic translation and activates the ATF4 pathway. At the same time, mitochondrial stress also reduces the expression of MRPs to inhibit mitochondrial translation and protect mitochondrial function.
To deepen into this in vivo coregulation of ATF4 and its targets genes, we explored the expression of Atf4 in the BXD mouse genetic reference population. We mapped the Atf4 transcript levels to a trans–expression quantitative trait locus on chromosome 1 (170–180 Mb) in multiple mouse tissues, especially those related to the central nervous system (Fig. 9, D and E). Notably, several Atf4 target genes, including Asns and most of the cytosolic aaRS, also have a trans–expression quantitative trait locus in the same locus at chromosome 1 (Mozhui et al., 2008), suggesting that this locus can control their expression through the regulation of Atf4 expression. By exploring the genes under this quantitative trait locus (QTL), we identified six genes (Copa, Atp1a4, Fh1, Adamts4, Usp21, and Olfr248) that contain nonsynonymous substitutions, including nonsense, splicing site, and frameshift mutations, which segregate in the BXDs. One of these genes, and the only mitochondrial protein, was fumarate hydratase 1 (Fh1; Fig. 9 F), a tricarboxylic acid cycle enzyme that catalyzes the reversible hydration/dehydration of fumarate to l-malate. In the BXD population, a missense variant in Fh1 gene modulates its expression level in several tissues (Wang et al., 2016), whereas the LOF of the C. elegans orthologue of Fh1, fum-1, activates the UPRmt in worms (Wang et al., 2016). In combination, these data suggest a conserved role of Fh1 in the mitochondrial stress response from C. elegans to mammals.
To validate Fh1 as a possible candidate gene regulating Atf4 expression, we hence knocked down FH expression and treated HeLa cells with fumarate, the FH substrate. Both LOF of FH and treatment with fumarate activated the ATF4 pathway in vitro (Fig. 9 G). Similarly, analysis of kidney microarray extracted from Fh1-deficient mice (Adam et al., 2011) also evidenced the activation of the ATF4 pathway, as illustrated by the GSEA enrichment plot of the mt-stress genes (Fig. 9 H). These results demonstrate that changes in the Fh1 expression levels activate Atf4 expression, which in turn would activate the expression of its targets genes, including Asns, the cytosolic aaRS and the rest of the mt-stress genes. Collectively, these results hence reveal that modulation of mitochondrial function in both basal and stress conditions regulates the expression of ATF4 and its target genes in both human and mouse populations.
Altogether, our results demonstrate that the activation of ATF4 through the ISR is a general retrograde response that regulates both cellular metabolism and mitochondrial function to allow a global cellular adaptation to mitochondrial stress.
Discussion
By using a combined analysis of the transcriptome, proteome, and metabolome of HeLa cells exposed to four different mitochondrial stressors, we obtained a multilayered footprint/map of mitochondrial stress response pathways. By analyzing the common and global changes in our multiomics data, we identified that, despite the profound changes observed in the mitochondrial proteome, the stress response is mainly performed by a network of genes/proteins and metabolites that regulates cytosolic translation and rewires cellular metabolism. The use of four different stressors has enabled us to discern specific responses obtained by the use of individual compounds or genetic interventions from the overall and common response that occurs upon mitochondrial stress.
Despite some differences in response observed with each stressor, we identified that the general retrograde response is coordinated by ATF4, which acts downstream of the ISR. The activation of this response aims to decrease global translation, similarly to what is observed in a yeast model of mitochondrial stress (Wang and Chen, 2015), and enable the translation of specific cytoprotective genes. These results are in agreement with previous data that have shown the activation of the ISR under different mitochondrial stress conditions in mammals (Tyynismaa et al., 2010; Martínez-Reyes et al., 2012; Michel et al., 2015; Bao et al., 2016; Münch and Harper, 2016) and in worms (Baker et al., 2012). In addition to the mitonuclear response regulated by ATF4, mitochondrial stress also triggers a decrease in MRP levels. It is of note that variation in the expression levels of the MRPs is both at the origin of a mitochondrial stress response that regulates lifespan (Houtkooper et al., 2013) as well as a key target of this feedback regulation to adapt mitochondrial proteostasis. This makes the MRPs nodal points in mitonuclear stress communication pathways. Interestingly, the activation of the ISR can also couple the decrease in cytosolic protein translation with the attenuation of mitochondrial import (Rainbolt et al., 2013), which would collaborate in reducing mitochondrial function under stress conditions.
Surprisingly, under the conditions used, we did not detect activation of the prototypical UPRmt, which is generally characterized by the induction of the chaperones HSP60 and HSP10 and the protease CLPP, among others, and which has been mainly described in invertebrates and cellular models as the primary transcriptional response to mitochondrial proteotoxic stress (Zhao et al., 2002; Haynes et al., 2007; Durieux et al., 2011; Houtkooper et al., 2013). Some recent evidence might help to explain this apparent contradiction, such as the requirement of additional regulatory components, like epigenetic modifications or non–cell-autonomous factors (Durieux et al., 2011; Merkwirth et al., 2016; Tian et al., 2016), as well as particular specificities of given cells, tissues, or stressors to activate the UPRmt (Dogan et al., 2014; Münch and Harper, 2016).
Overall, our multiomics analysis enabled us to identify an ATF4-dependent mitochondrial stress signature that is shared between many mitochondrial stress conditions in cellular and animal models and that is characterized primarily by the activation of amino acid biosynthesis pathways. A similar response was also observed in mitochondrial myopathies in vivo, extending its relevance to human pathology. Perhaps most importantly, the observation of a tight coregulated network of mt-stress genes in different human (GTEx) and mouse (BXD) tissues under physiological conditions assigns a key role of ATF4 and its upstream mediator fumarate hydratase in the regulation of cellular metabolism in equilibrium in the absence of stress. ATF4 and its coregulated gene circuit could hence be important both to adapt mitochondrial and cellular function to genetic variation, metabolic activity, and environmental cues in homeostasis (health), as well as provide a mechanistic basis to explain mitochondrial dysfunction in stress and disease.
Conceptually, our work extends our understanding of mitonuclear communication in stress and homeostasis in mammals. We demonstrate that ATF4 is a main player in the mitonuclear stress response, being activated by the ISR and controlling cytosolic translation and homeostasis. This ATF4-driven response is coupled with an ATF4-independent local mitochondrial proteostatic response, which attenuates mitochondrial translation by decreasing the levels of the MRPs (Fig. 9 I). The activation of these two events exemplifies the important role of mitonuclear communication to respond to stress, synchronizing nuclear and mitochondrial genomes by concurrently decreasing cytosolic and mitochondrial translation. We furthermore anticipate that our comprehensive approach to study mitochondrial stress in cells will provide a platform that will contribute to the discovery of essential new players in the pathogenesis of diseases linked with mitochondrial dysfunction.
Materials and methods
Cell lines and reagents
HeLa, 293T, and COS7 cells were obtained from ATCC. MEFs were supplied by C. López-Otín (University of Oviedo, Oviedo, Spain). All cell lines were cultured in DMEM plus l-glutamine and pyruvate with 10% inactivated FBS and penicillin and streptomycin (Thermo Fisher Scientific) and maintained at 37°C and 5% CO2. Reagents were obtained from the following sources: doxycycline hyclate (D9891; Sigma-Aldrich), actinonin (A6671; Sigma-Aldrich), FCCP (C2920; Sigma-Aldrich), MB (10–1472-5; Tebu.bio), Rotenone (R8875; Sigma-Aldrich), antimycin A (A8674; Sigma-Aldrich), oligomycin (495455; EMD Millipore), tunicamycin (T7765; Sigma-Aldrich), ISRIB (SML0843; Sigma-Aldrich), monomethyl fumarate (651419; Sigma-Aldrich), Mdivi-1 (M0199; Sigma-Aldrich), and paraquat dichloride hydrate (36541; Sigma-Aldrich). pCAGGS vectors containing WT (pCAGGS-OTC) and mutant (pCAGGS-ΔOTC) forms of ornithine-transcarbamylase were a gift from N. Hoogenraad (La Trobe University, Melbourne, Australia). Cells were treated during 6 h or 24 h with 30 µg/ml doxycycline, 50 µM actinonin, 10 µM FCCP, or 50 µM MB. Induction of ER stress was performed by treating the cells with 2.5 µg/ml (6 h) or 1 µg/ml (24 h) tunicamycin. ISRIB was used at 500 nM, monomethyl fumarate at 2.5 mM, Mdivi-1 at 50 µM, and paraquat at 400 µM.
RNA sequencing library preparation and analysis
Total RNA from HeLa cells was extracted using TRIzol (Thermo Fisher Scientific) and purified by column using RNeasy Mini kit (QIAGEN). 500 ng total RNA was enriched for poly(A) RNA, and then barcoded libraries for sequencing were prepared using the Lexogen mRNA Sense kit for Ion Torrent according to manufacturer’s standard protocol. Libraries were amplified 13 cycles, and the quality of each library was checked on an Agilent Technologies Bioanalyzer DNA High Sensitivity chip. The libraries were pooled based on the concentration of each sample between 200 and 350 bp, purified on a Pippin Prep gel, quantified by the Agilent Technologies Bioanalyzer, and sequenced on an Ion Torrent Proton sequencer at 30x coverage depth. The reads obtained were first trimmed out to remove adaptor sequences and then mapped using a combined pipeline of TopHat2 (Kim et al., 2013) and bowtie2 (Langmead and Salzberg, 2012) against the human genome (GRCh37). HTSeq-count (Anders et al., 2015) was then used to obtain the counts of each gene. Differential expressed genes in each condition were identified using R/Bioconductor package edgeR (Robinson et al., 2010). The accession number for the RNA sequencing data reported in this paper is GEO GSE84631.
Proteomic analysis
HeLa cells treated with all described compounds were washed twice with ice-cold PBS and scraped in PBS. Cell pellets were lysed with urea buffer (8 M urea, 75 mM NaCl, 50 mM EPPS, pH 8.5, with added cOmplete protease inhibitors [Roche] and PhosSTOP phosphatase inhibitors [Roche]). Cell debris was spun down for 15 min at 13,000 g at 4°C. Protein concentration was determined using the BCA assay (Thermo Fisher Scientific). 100 µg lysate was reduced with 5 mM TCEP for 30 min and then alkylated with 14 mM iodoacetamide for 30 min in the dark, and all reactions were incubated at room temperature. Then, proteins were precipitated by methanol/chloroform precipitation and resuspended in 200 mM EPPS, pH 8.5. Protein digestion was performed for 16 h at room temperature with LysC at a 1:100 (LysC/protein) ratio. The day after, trypsin was added at a 1:75 (trypsin/protein) ratio and incubated at 37°C for 5 h. Then, isobaric labeling of the peptides was performed using the 10-plex TMT reagents (Thermo Fisher Scientific). In brief, 100 µg peptides was labeled with 0.2 mg TMT reagent (previously dissolved in acetonitrile [ACN]), final concentration of ACN ∼30% (vol/vol), for 1 h at room temperature. Finally, the reactions were quenched for 15 min with hydroxylamine to a final concentration of 0.3% (vol/vol). The TMT-labeled peptides were combined equally and dried via vacuum centrifugation. Dried peptides were resuspended in 3% FA, 3% ACN, desalted using a Sep-Pak C18 solid-phase extraction cartridge (Waters), and dried down again via vacuum centrifugation. The pooled TMT-labeled peptides were then subjected to a basic pH reversed-phase HPLC fractionation as described (Paulo et al., 2015) and fractionated into 24 fractions. Half of them were vacuum centrifuged to dryness, dissolved in 3% FA, 3% ACN, desalted via StageTip, dried again via vacuum centrifugation and resuspended in 10 µl of 3% FA, 3% ACN for mass spectrometry analysis. All samples were run in an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) coupled to a Proxeon EASY-nLC II LC pump (Thermo Fisher Scientific). Peptides were fractionated on a microcapillary column packed with GP-18 resin (1.8 µm, 200 Å; Sepax). Peptides were separated using a 135-min gradient of 6–26% ACN in 0.125% formic acid at a flow rate of ∼350 nl/min. Each analysis used the multinotch MS3-based TMT method (McAlister et al., 2014). For each FTMS1 spectrum detected in the Orbitrap (resolution of 120k; mass range 400–1,400 m/z; automatic gain control [AGC] target 5 × 105, maximum injection time of 100 ms), precursors for MS2/MS3 analysis were selected using the TopSpeed parameter of 2 s. MS2 analysis consisted of collision-induced dissociation (quadrupole ion-trap analysis; AGC 8 × 103; normalized collision energy = 35; maximum injection time 150 ms). To create TMT reporter ions, a synchronous-precursor-selection MS3 scan was collected on the top 10 most intense ions in the MS2 spectrum. Synchronous-precursor-selection MS3 precursors were fragmented by high-energy collision–induced dissociation and analyzed in the Orbitrap (normalized collision energy = 55%, AGC = 105, maximum injection time = 150 ms, and resolution = 60 K). The data analysis was performed with a SEQUEST-based in-house software pipeline (Huttlin et al., 2010). In short, mass spectra were searched against the human Uniprot database (February 2014) and a reverse decoy database. A cysteine carbamidomethylation (+57.0215 D) and TMT tag (+229.1629 D) on lysine residues and peptide N termini were added as static modifications, and methionine oxidation (+15.9949 D) was set as a variable modification. Precursor ion tolerance was set at 20 ppm and product ion tolerance at 0.9 D. Peptide-spectrum matches were adjusted to a 1% FDR using a linear discriminant analysis (Huttlin et al., 2010), quantified by extracting the signal-to-noise ratio, and proteins were further collapsed to a final protein-level FDR of 1% (McAlister et al., 2014; Paulo et al., 2015). Differential expressed proteins were identified by fitting the data to a linear model and performing an empirical Bayes moderated t-statistics test, comparing each stress condition with control cells using the R/Bioconductor package limma (Ritchie et al., 2015). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD006293.
Metabolome analysis
Cells were seeded at density of 3 × 105 cells per well in six-well plates and treated the next day with selected drugs in quadruplicate. 24 h later, cells were washed twice with 75 mM ammonium carbonate buffer, pH 7.4 (37°C), and cell metabolism was quenched by shock freezing of plates in liquid N2. Metabolites were extracted by adding ice-cold acetonitrile/methanol/water (40:40:20, vol/vol). The extraction procedure was repeated twice. Scraped cells and supernatants were pooled in the same tube. To remove cell debris, tubes were centrifuged (4°C, 13,000 rpm, 2 min), and the supernatants collected were assayed by flow injection analysis using time-of-flight mass spectrometry (6550 QTOF; Agilent Technologies) operated in the negative ionization mode. High-resolution mass spectra were recorded from 50–1,000 m/z and analyzed as described previously (Fuhrer et al., 2011). Detected ions were putatively annotated by as searching matching metabolites in the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2006) using an m/z tolerance of 0.001 and including all common electrospray derivatives. Differences between treatments and control cells were determined using the R/Bioconductor package limma (Ritchie et al., 2015).
Bioinformatics and statistical analysis
For RNA sequencing and quantitative proteomic analysis, Gene Ontology and KEGG pathway enrichment analysis were performed using goanna and kegga, respectively, from limma and edgeR packages. Pathways, genes and proteins selected in each condition were filtered after Benjamini-Hochberg correction at an adjusted P < 0.05 (FDR 5%). Network analysis of common down-regulated proteins was performed using STRING (version 10.0; http://string-db.org). De novo motif analysis was performed using HOMER (http://homer.salk.edu/homer) by selecting the common up-regulated genes in RNA sequencing analysis. Metabolic pathway analysis was performed using MetaboAnalyst 3.0 (http://www.metaboanalyst.ca; Xia et al., 2015) using a hypergeometric test for overrepresentation analysis and relative-betweenness centrality for pathway topology analysis. Correlation analysis was performed using the GTEx v6 dataset (http://www.gtexportal.org) for human data and the datasets available in GeneNetwork (http://www.genenetwork.org) for the BXD mouse genetic reference population. For GTEx data, reads per kilobase per million mapped reads (RPKM) of each transcript were filtered (selecting genes with more than 0.1 RPKM in at least 10 individuals), log2 transformed, and quantile normalized across all samples for each tissue; only tissues with more than 30 individuals were used. Genes with a positive (Person’s rho > 0) or negative correlation (Pearson’s rho < 0) in each case and a significant correlation (P < 0.05) in each tissue were filtered. Common genes in 75% of the GTEx data (49 tissues) or BXD data (16 tissues) were selected for the analysis. Multitissue correlation networks were generated with Cytoscape (http://www.cytoscape.org), using edge width as an indication the percentage of tissues with positive correlation between genes (nodes). Enrichment analysis of top correlated genes was conducted using kegga function from the limma package. QTL analysis was performed using R/qtl package (Broman et al., 2003) selecting the following traits obtained from GeneNetwork: INIA Amygdala Cohort Affy MoGene 1 0 ST Mar11 RMA, UTHSC Mouse BXD Whole Brain RNA Sequence Nov12 RPKM Untrimmed, GE NIAAA Cerebellum mRNA M430v2 May05 RMA, Hippocampus Consortium M430v2 Jun06 PDNN, VU BXD Midbrain Agilent Technologies SurePrint G3 Mouse GE May12 Quantile, HQF BXD Neocortex ILM6v1 1 Dec10v2 RankInv, VCU BXD NAc Sal M430 2 0 Oct07 RMA, VCU BXD PFC Sal M430 2 0 Dec06 RMA, and Full HEI Retina Illumina V6 2 Apr10 RankInv. All data analysis and plots were generated using R and RStudio (R Core Team and RStudio Team) and modified using Adobe Illustrator CC. For the rest of the analysis, data were expressed as mean ± SEM, and p-values were calculated using two-tailed Student’s t test for pairwise comparisons, one-way ANOVA for multiple comparisons, and two-way ANOVA for multiple comparisons involving two independent variables. Statistical tests were performed using RStudio and GraphPad Prism 6.0 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; n ≥ 3).
Seahorse analysis
The OCR and ECAR were measured using Seahorse XF96 equipment (Seahorse Bioscience Inc.) as previously described (Andreux et al., 2014). In brief, cells were seeded at 10,000 cells per well and treated with different medium conditions or compounds for 24 h in 100 µl medium. Before the measurements, plates were equilibrated in a CO2-free incubator at 37°C for 1 h. Analysis were performed using 1 µM oligomycin, 10 µM FCCP, and 1 µM rotenone/antimycin as indicated. Data were normalized to protein levels. ATP-dependent respiration (or oligomycin-sensitive respiration) was calculated as the difference in OCR before and after the addition of oligomycin.
Mitochondrial membrane potential and ROS measurements
Mitochondrial membrane potential (Δψm) was measured using the fluorescent tetramethylrhodamine methyl ester probe (Sigma-Aldrich), and ROS levels were measured using 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA; Abcam) and MitoSox (Thermo Fisher Scientific). Cells were stained with selected probes and fluorescence was analyzed using Gallios Flow Cytometer (Beckman Coulter). In brief, for tetramethylrhodamine methyl ester, cells were stained with 200 nM for 15 min at 37°C, washed twice and resuspended in PBS. For H2DCFDA and MitoSox, cells were stained with 20 and 5 µM, respectively, and fluorescence was analyzed without washing.
26S proteasome assay
The in vitro assay of the 26S proteasome activity was performed as previously described (Vilchez et al., 2012) with minor changes. In brief, cells were lysed in proteasome activity assay buffer (50 mM Tris-HCl, pH 7.5, 250 mM sucrose, 5 mM MgCl2, 0.5 mM EDTA, 2 mM ATP, and 1 mM DTT) by passing 10 times through a 29G syringe. Lysates were centrifuged at 10,000 g for 10 min at 4°C, and protein concentration of cell homogenates was determined by Lowry method. 10 µg total protein was incubated in proteasome activity assay buffer with 100 µM Suc-Leu-Leu-Val-Tyr-AMC (Bachem) to measure chymotrypsin-like proteasome activity. Peptide cleavage was monitored by measuring fluorescence (380 nm excitation, 460 nm emission) every 5 min for 1 h at room temperature on a VICTOR X4 plate reader (PerkinElmer). Proteasome activity was determined by calculating the slope of the curves.
Quantitative RT-PCR
Total RNA from cells was extracted using TRIzol (Thermo Fisher Scientific). 1–2 µg RNA was used for reverse transcription using the QuantiTect Reverse Transcription kit (QIAGEN). 10× diluted cDNA was used for quantitative RT-PCR reactions using the LightCycler 480 System (Roche) and a LightCycler 480 SYBR Green I Master Mix (Roche). All data were normalized to ACTB and GAPDH expression. Primer sets for quantitative real-time PCR analyses are shown in Table S1.
mtDNA levels
mtDNA levels were analyzed as previously described (Quiros et al., 2017) using tRNA-Leu primers for mtDNA and POLG primers for nDNA. In brief, 10–50 ng total DNA extracted from cells was used for qPCR using the LightCycler 480 System (Roche) and a LightCycler 480 SYBR Green I Master Mix (Roche). Relative mtDNA copy number was calculated by normalizing the amplification of tRNA-Leu (mtDNA) with the expression of POLG (nuclear DNA).
Immunoblot analysis
Cultured cells were washed with PBS and lysed in RIPA buffer containing 50 mM Tris buffer, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 10 mM EDTA, with cOmplete protease inhibitors (Roche) and PhosSTOP phosphatase inhibitors (Roche). Once homogenized, lysed cells were sonicated and centrifuged at 13,000 g at 4°C for 15 min, and supernatants were collected. The protein concentration of the supernatant was evaluated using the Lowry technique. Equal protein amounts were boiled in Laemmli buffer and run in SDS-polyacrylamide. After electrophoresis, gels were electrotransferred onto polyvinylidene fluoride membranes (EMD Millipore), blocked with 5% nonfat dried milk in TBS-T (TBS with 0.05% Tween-20), and incubated with primary antibodies following the commercial instructions. The following primary antibodies were used: mouse anti–β-actin (sc-47778; Santa Cruz Biotechnology, Inc.); mouse anti-HSP90 (BD Biosciences, 610418); OXPHOS antibody cocktail (mouse mAbs, ab110413; Abcam); mouse anti-HSP60 (ADI-SPA-806; Enzo Life Sciences); mouse anti-CLPP (WH0008192M1-100; Sigma-Aldrich); mouse anti-HSPA9 (ABIN361739; Antibodies online); rabbit anti-LONP1 (HPA002192; Sigma-Aldrich); rabbit anti-OTC (sc-102051; Santa Cruz Biotechnology, Inc.); mouse anti-OPA1 (BD, 612606); rabbit anti-CREB-2 (ATF4, sc-200; Santa Cruz Biotechnology, Inc.); phospho-eIF2α (Ser51) rabbit mAb (9721; Cell Signaling Technology); eIF2α rabbit mAb (9722; Cell Signaling Technology); and α tubulin (T5168; Sigma-Aldrich). Secondary antibodies used were goat anti–rabbit HRP (AC2114; Azure Biosystems) and goat anti–mouse HRP secondary antibody (AC2115; Azure Biosystems). All primary antibodies were used at 1:1,000, and secondary antibodies were used at 1:10,000. Images were scanned on an Azure Imager c300 (Azure Biosystems).
Viral package and cell infection
Lentiviruses were produced by cotransfecting HEK293T cells with lentiviral and packaging (pCMV-dR8.2 dvpr) and envelope (pCMV-VSV-G) plasmids in a ratio of 4:3:1, respectively, using FUGENE 6 (Promega) following the manufacturer’s instructions. The packaging and envelope vectors were created by Robert Weinberg (Whitehead Institute for Biomedical Research, Cambridge, MA) and obtained through Addgene (plasmids 8455 and 8454). Transfection medium was removed 24 h after transfection, and fresh medium was added to the plate. Cell supernatants were collected at 24 and 48 h and filtered through a 0.45-µm sterile filter. Cells were seeded in six-well plates at 20–30% confluence 24 h before infection. The following day, 1 ml viral supernatant supplemented with 5 mg/ml polybrene (EMD Millipore) was added to growing cells. This step was repeated twice, and cells were left to recover for 24 h in growth media before 2 µg/ml puromycin selection. pLKO.1 vector was used as a control. The shRNAs used are listed in Table S2.
CRISPR-Cas9–mediated gene knockout
Gene knockout of ATF4 in HeLa cells was conducted via CRISPR-Cas-9–mediated genome editing as previously described (Sanjana et al., 2014). gRNAs against exon 1 of ATF4 were designed using Benchling (https://benchling.com) and cloned into the Lenti CRISPR v2 plasmid, a gift from F. Zhang (Broad Institute of MIT and Harvard, Cambridge, MA; (plasmid 52961; Addgene). Cells were infected in an analogous way as the previously described for shRNA and selected for 48 h with 2 µg/ml puromycin. After selection, cells were single-cell sorted with a flow cytometer into 96-well plates. Clones were grown for 2 wk, and the resultant colonies were trypsinized and expanded. Gene knockout was confirmed by Sanger sequencing and Western blot analysis. The sgRNA sequence for ATF4 was 5′-TCTCTTAGATGATTACCTGG-3′ (plus strand).
Online supplemental material
Fig. S1 shows RNA sequencing and proteomic analysis upon mitochondrial stress. Fig. S2 shows an extended data from the analysis of the commonly down-regulated proteins upon mitochondrial stress. Fig. S3 shows extended data from the analysis of the commonly up-regulated transcripts and proteins and from the metabolomics analysis. Fig. S4 shows the generation of ATF4 LOF cells and the exclusion of UPRer and UPRmt pathways upon mitochondrial stress. Table S1 shows a list of primer sets used for quantitative RT-PCR analyses. Table S2 shows the list of shRNAs used. Tables S3 and S4 contain the normalized RNA sequencing and proteomics data. Table S5 contains the list of the common differentially expressed genes and proteins in mitochondrial stress. Tables S6 and S7 contain gene enrichment analysis of each stressor in RNA sequencing and proteomics, respectively. Table S8 contains nontargeted metabolomics raw data.
Supplementary Material
Acknowledgments
We thank A. Mottis, O. Matilainen, V. Sorrentino, P. Xu, P. Jha, and C. Bárcena for comments and discussions and C. López-Otín for reagents and support. We also thank C. Costelle for processing RNA samples and J.A. Paulo for setting up the mass spectrometers for the proteomics analysis.
P.M. Quirós was supported by a long-term fellowship from the European Molecular Biology Organization (ALTF 480–2014). D. D’Amico was supported by a fellowship funded by Associazione Italiana per la Ricerca sul Cancro and Marie Curie Actions. D. Finley is supported by National Institutes of Health grant R01GM095526. S.P. Gygi is supported by National Institutes of Health grant GM67945. J. Auwerx is supported by grants from the École Polytechnique Fédérale de Lausanne, the Swiss National Science Foundation (31003A-140780), the AgingX program of the Swiss Initiative for Systems Biology (51RTP0-151019), the Krebsliga Schweiz (KFS-3082-02-2013), and the National Institutes of Health (R01AG043930).
The authors declare no competing financial interests.
Author contributions: P.M. Quirós and J. Auwerx conceived and designed the project. P.M. Quirós performed the experiments and analyzed all data. M.A. Prado, D. Finley, and S.P. Gygi performed proteomic analysis. N. Zamboni measured the metabolites. D. D’Amico helped perform qPCR experiments. R.W. Williams contributed to the RNA sequencing and QTL mapping. P.M. Quirós and J. Auwerx wrote the manuscript.
Footnotes
Abbreviations used:
- aaRS
- aa–tRNA synthetase
- ACN
- acetonitrile
- AGC
- automatic gain control
- ATF
- activating transcription factor
- ECAR
- extracellular acidification rate
- EtBr
- ethidium bromide
- FCCP
- carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone
- FDR
- false discovery rate
- GSEA
- gene set enrichment analysis
- GSH
- glutathione
- GSSG
- oxidized glutathione
- ISR
- integrated stress response
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- LOF
- loss of function
- MB
- MitoBloCK-6
- MEF
- mouse embryonic fibroblast
- MRP
- mitochondrial ribosomal protein
- mtDNA
- mitochondrial DNA
- OCR
- oxygen consumption rate
- OXPHOS
- oxidative phosphorylation
- QTL
- quantitative trait locus
- ROS
- reactive oxygen species
- TMT
- tandem mass tag
- UPRer
- unfolded protein response in the ER
- UPRmt
- mitochondrial unfolded protein response
References
- Adam J., Hatipoglu E., O’Flaherty L., Ternette N., Sahgal N., Lockstone H., Baban D., Nye E., Stamp G.W., Wolhuter K., et al. . 2011. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: Roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 20:524–537. 10.1016/j.ccr.2011.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreux P.A., Williams E.G., Koutnikova H., Houtkooper R.H., Champy M.F., Henry H., Schoonjans K., Williams R.W., and Auwerx J.. 2012. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 150:1287–1299. 10.1016/j.cell.2012.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreux P.A., Houtkooper R.H., and Auwerx J.. 2013. Pharmacological approaches to restore mitochondrial function. Nat. Rev. Drug Discov. 12:465–483. 10.1038/nrd4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreux P.A., Mouchiroud L., Wang X., Jovaisaite V., Mottis A., Bichet S., Moullan N., Houtkooper R.H., and Auwerx J.. 2014. A method to identify and validate mitochondrial modulators using mammalian cells and the worm C. elegans. Sci. Rep. 4:5285 10.1038/srep05285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.M., Nargund A.M., Sun T., and Haynes C.M.. 2012. Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLoS Genet. 8:e1002760 10.1371/journal.pgen.1002760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.J., Lampe P.A., Stojanovski D., Korwitz A., Anand R., Tatsuta T., and Langer T.. 2014. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 33:578–593. 10.1002/embj.201386474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S., Roberts P.C., Brown L.E., Truong H., Pattnaik A.K., Archer D.R., and Barber G.N.. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 13:129–141. 10.1016/S1074-7613(00)00014-5 [DOI] [PubMed] [Google Scholar]
- Bao X.R., Ong S.E., Goldberger O., Peng J., Sharma R., Thompson D.A., Vafai S.B., Cox A.G., Marutani E., Ichinose F., et al. . 2016. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife. 5:5 10.7554/eLife.10575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I., Hoxhaj G., Ricoult S.J., Asara J.M., and Manning B.D.. 2016. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 351:728–733. 10.1126/science.aad0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein S.H., Venkatesh S., Li M., Lee J., Lu B., Hilchey S.P., Morse K.M., Metcalfe H.M., Skalska J., Andreeff M., et al. . 2012. The mitochondrial ATP-dependent Lon protease: A novel target in lymphoma death mediated by the synthetic triterpenoid CDDO and its derivatives. Blood. 119:3321–3329. 10.1182/blood-2011-02-340075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K.W., Wu H., Sen S., and Churchill G.A.. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 19:889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- Calvo S.E., Clauser K.R., and Mootha V.K.. 2016. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44(D1):D1251–D1257. 10.1093/nar/gkv1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Stone A., Chipuk J.E., Ingerman E., Song C., Yoo C., Kuwana T., Kurth M.J., Shaw J.T., Hinshaw J.E., Green D.R., and Nunnari J.. 2008. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 14:193–204. 10.1016/j.devcel.2007.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel N.S. 2015. Evolution of mitochondria as signaling organelles. Cell Metab. 22:204–206. 10.1016/j.cmet.2015.05.013 [DOI] [PubMed] [Google Scholar]
- Chick J.M., Munger S.C., Simecek P., Huttlin E.L., Choi K., Gatti D.M., Raghupathy N., Svenson K.L., Churchill G.A., and Gygi S.P.. 2016. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 534:500–505. 10.1038/nature18270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S., Frezza C., Soriano M.E., Varanita T., Quintana-Cabrera R., Corrado M., Cipolat S., Costa V., Casarin A., Gomes L.C., et al. . 2013. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 155:160–171. 10.1016/j.cell.2013.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion M.T., Soto I.C., Shipkovenska G., and Churchman L.S.. 2016. Synchronized mitochondrial and cytosolic translation programs. Nature. 533:499–503. 10.1038/nature18015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabir D.V., Hasson S.A., Setoguchi K., Johnson M.E., Wongkongkathep P., Douglas C.J., Zimmerman J., Damoiseaux R., Teitell M.A., and Koehler C.M.. 2013. A small molecule inhibitor of redox-regulated protein translocation into mitochondria. Dev. Cell. 25:81–92. 10.1016/j.devcel.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan S.A., Pujol C., Maiti P., Kukat A., Wang S., Hermans S., Senft K., Wibom R., Rugarli E.I., and Trifunovic A.. 2014. Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell Metab. 19:458–469. 10.1016/j.cmet.2014.02.004 [DOI] [PubMed] [Google Scholar]
- Durieux J., Wolff S., and Dillin A.. 2011. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 144:79–91. 10.1016/j.cell.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorese C.J., Schulz A.M., Lin Y.F., Rosin N., Pellegrino M.W., and Haynes C.M.. 2016. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr. Biol. 26:2037–2043. 10.1016/j.cub.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B., and Scorrano L.. 2006. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 126:177–189. 10.1016/j.cell.2006.06.025 [DOI] [PubMed] [Google Scholar]
- Fuhrer T., Heer D., Begemann B., and Zamboni N.. 2011. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 83:7074–7080. 10.1021/ac201267k [DOI] [PubMed] [Google Scholar]
- Gispert S., Parganlija D., Klinkenberg M., Dröse S., Wittig I., Mittelbronn M., Grzmil P., Koob S., Hamann A., Walter M., et al. . 2013. Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum. Mol. Genet. 22:4871–4887. 10.1093/hmg/ddt338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grégoire M., Morais R., Quilliam M.A., and Gravel D.. 1984. On auxotrophy for pyrimidines of respiration-deficient chick embryo cells. Eur. J. Biochem. 142:49–55. 10.1111/j.1432-1033.1984.tb08249.x [DOI] [PubMed] [Google Scholar]
- GTEx Consortium 2013. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45:580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A.P., Yu C., Lu L., Fujiwara Y., Browne C., Chin G., Fleming M., Leboulch P., Orkin S.H., and Chen J.J.. 2001. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20:6909–6918. 10.1093/emboj/20.23.6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Back S.H., Hur J., Lin Y.H., Gildersleeve R., Shan J., Yuan C.L., Krokowski D., Wang S., Hatzoglou M., et al. . 2013. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15:481–490. 10.1038/ncb2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Bertolotti A., Zeng H., and Ron D.. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 5:897–904. 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. . 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 11:619–633. 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- Haynes C.M., Petrova K., Benedetti C., Yang Y., and Ron D.. 2007. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell. 13:467–480. 10.1016/j.devcel.2007.07.016 [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C. 2012. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13:89–102. [DOI] [PubMed] [Google Scholar]
- Houtkooper R.H., Williams R.W., and Auwerx J.. 2010. Metabolic networks of longevity. Cell. 142:9–14. 10.1016/j.cell.2010.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R.W., and Auwerx J.. 2013. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 497:451–457. 10.1038/nature12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., and Gygi S.P.. 2010. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 143:1174–1189. 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., and Hirakawa M.. 2006. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 34:D354–D357. 10.1093/nar/gkj102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.E., Grant A.R., Simic M.S., Kohnz R.A., Nomura D.K., Durieux J., Riera C.E., Sanchez M., Kapernick E., Wolff S., and Dillin A.. 2016. Lipid biosynthesis coordinates a mitochondrial-to-cytosolic stress response. Cell. 166:1539–1552.e16. 10.1016/j.cell.2016.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman W.J., Willems P.H., and Smeitink J.A.. 2012. Monogenic mitochondrial disorders. N. Engl. J. Med. 366:1132–1141. 10.1056/NEJMra1012478 [DOI] [PubMed] [Google Scholar]
- Krige D., Needham L.A., Bawden L.J., Flores N., Farmer H., Miles L.E., Stone E., Callaghan J., Chandler S., Clark V.L., et al. . 2008. CHR-2797: an antiproliferative aminopeptidase inhibitor that leads to amino acid deprivation in human leukemic cells. Cancer Res. 68:6669–6679. 10.1158/0008-5472.CAN-07-6627 [DOI] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanning N.J., Looyenga B.D., Kauffman A.L., Niemi N.M., Sudderth J., DeBerardinis R.J., and MacKeigan J.P.. 2014. A mitochondrial RNAi screen defines cellular bioenergetic determinants and identifies an adenylate kinase as a key regulator of ATP levels. Cell Reports. 7:907–917. 10.1016/j.celrep.2014.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Samuel B.S., Breen P.C., and Ruvkun G.. 2014. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 508:406–410. 10.1038/nature13204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C., Blasco M.A., Partridge L., Serrano M., and Kroemer G.. 2013. The hallmarks of aging. Cell. 153:1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C., Galluzzi L., Freije J.M., Madeo F., and Kroemer G.. 2016. Metabolic control of longevity. Cell. 166:802–821. 10.1016/j.cell.2016.07.031 [DOI] [PubMed] [Google Scholar]
- Mariño G., Pietrocola F., Eisenberg T., Kong Y., Malik S.A., Andryushkova A., Schroeder S., Pendl T., Harger A., Niso-Santano M., et al. . 2014. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol. Cell. 53:710–725. 10.1016/j.molcel.2014.01.016 [DOI] [PubMed] [Google Scholar]
- Martínez-Reyes I., Sánchez-Aragó M., and Cuezva J.M.. 2012. AMPK and GCN2-ATF4 signal the repression of mitochondria in colon cancer cells. Biochem. J. 444:249–259. 10.1042/BJ20111829 [DOI] [PubMed] [Google Scholar]
- Martínez-Reyes I., Diebold L.P., Kong H., Schieber M., Huang H., Hensley C.T., Mehta M.M., Wang T., Santos J.H., Woychik R., et al. . 2016. TCA cycle and mitochondrial membrane potential are necessary for diverse biological functions. Mol. Cell. 61:199–209. 10.1016/j.molcel.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinus R.D., Garth G.P., Webster T.L., Cartwright P., Naylor D.J., Høj P.B., and Hoogenraad N.J.. 1996. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur. J. Biochem. 240:98–103. 10.1111/j.1432-1033.1996.0098h.x [DOI] [PubMed] [Google Scholar]
- Matilainen O., Quirós P.M., and Auwerx J.. 2017. Mitochondria and epigenetics: Crosstalk in homeostasis and stress. Trends Cell Biol. https://doi.org 10.1016/j.tcb.2017.02.004. [DOI] [PubMed] [Google Scholar]
- McAlister G.C., Nusinow D.P., Jedrychowski M.P., Wühr M., Huttlin E.L., Erickson B.K., Rad R., Haas W., and Gygi S.P.. 2014. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86:7150–7158. 10.1021/ac502040v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth C., Jovaisaite V., Durieux J., Matilainen O., Jordan S.D., Quiros P.M., Steffen K.K., Williams E.G., Mouchiroud L., Tronnes S.U., et al. . 2016. Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell. 165:1209–1223. 10.1016/j.cell.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel S., Canonne M., Arnould T., and Renard P.. 2015. Inhibition of mitochondrial genome expression triggers the activation of CHOP-10 by a cell signaling dependent on the integrated stress response but not the mitochondrial unfolded protein response. Mitochondrion. 21:58–68. 10.1016/j.mito.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Moisoi N., Klupsch K., Fedele V., East P., Sharma S., Renton A., Plun-Favreau H., Edwards R.E., Teismann P., Esposti M.D., et al. . 2009. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 16:449–464. 10.1038/cdd.2008.166 [DOI] [PubMed] [Google Scholar]
- Moullan N., Mouchiroud L., Wang X., Ryu D., Williams E.G., Mottis A., Jovaisaite V., Frochaux M.V., Quiros P.M., Deplancke B., et al. . 2015. Tetracyclines disturb mitochondrial function across eukaryotic models: A call for caution in biomedical research. Cell Reports. 10:1681–1691. 10.1016/j.celrep.2015.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozhui K., Ciobanu D.C., Schikorski T., Wang X., Lu L., and Williams R.W.. 2008. Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. PLoS Genet. 4:e1000260 10.1371/journal.pgen.1000260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch C., and Harper J.W.. 2016. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature. 534:710–713. 10.1038/nature18302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund A.M., Pellegrino M.W., Fiorese C.J., Baker B.M., and Haynes C.M.. 2012. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 337:587–590. 10.1126/science.1223560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., and Suomalainen A.. 2012. Mitochondria: In sickness and in health. Cell. 148:1145–1159. 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E., Song W., and Perrimon N.. 2013. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 155:699–712. 10.1016/j.cell.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., and Gorman A.M.. 2016. The integrated stress response. EMBO Rep. 17:1374–1395. 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K., Lionaki E., and Tavernarakis N.. 2015. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 521:525–528. 10.1038/nature14300 [DOI] [PubMed] [Google Scholar]
- Paulo J.A., O’Connell J.D., Gaun A., and Gygi S.P.. 2015. Proteome-wide quantitative multiplexed profiling of protein expression: carbon-source dependency in Saccharomyces cerevisiae. Mol. Biol. Cell. 26:4063–4074. 10.1091/mbc.E15-07-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros P.M., Goyal A., Jha P., and Auwerx J.. 2017. Analysis of mtDNA/nDNA ratio in mice. Curr. Protoc. Mouse Biol. 7:47–54. 10.1002/cpmo.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós P.M., Ramsay A.J., Sala D., Fernández-Vizarra E., Rodríguez F., Peinado J.R., Fernández-García M.S., Vega J.A., Enríquez J.A., Zorzano A., and López-Otín C.. 2012. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J. 31:2117–2133. 10.1038/emboj.2012.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós P.M., Español Y., Acín-Pérez R., Rodríguez F., Bárcena C., Watanabe K., Calvo E., Loureiro M., Fernández-García M.S., Fueyo A., et al. . 2014. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Reports. 8:542–556. 10.1016/j.celrep.2014.06.018 [DOI] [PubMed] [Google Scholar]
- Quirós P.M., Langer T., and López-Otín C.. 2015. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 16:345–359. 10.1038/nrm3984 [DOI] [PubMed] [Google Scholar]
- Quirós P.M., Mottis A., and Auwerx J.. 2016. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 17:213–226. 10.1038/nrm.2016.23 [DOI] [PubMed] [Google Scholar]
- Rainbolt T.K., Atanassova N., Genereux J.C., and Wiseman R.L.. 2013. Stress-regulated translational attenuation adapts mitochondrial protein import through Tim17A degradation. Cell Metab. 18:908–919. 10.1016/j.cmet.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauthan M., Ranji P., Aguilera Pradenas N., Pitot C., and Pilon M.. 2013. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proc. Natl. Acad. Sci. USA. 110:5981–5986. 10.1073/pnas.1218778110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U., Lahtinen T., Marttinen P., Suomi F., and Battersby B.J.. 2015. Quality control of mitochondrial protein synthesis is required for membrane integrity and cell fitness. J. Cell Biol. 211:373–389. 10.1083/jcb.201504062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Invest. 110:1383–1388. 10.1172/JCI0216784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder E.A., Raimundo N., and Shadel G.S.. 2013. Epigenetic silencing mediates mitochondria stress-induced longevity. Cell Metab. 17:954–964. 10.1016/j.cmet.2013.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Kevei É., Pokrzywa W., Schmeisser K., Mansfeld J., Livnat-Levanon N., Ensenauer R., Glickman M.H., Ristow M., and Hoppe T.. 2014. Pathogenesis of human mitochondrial diseases is modulated by reduced activity of the ubiquitin/proteasome system. Cell Metab. 19:642–652. 10.1016/j.cmet.2014.01.016 [DOI] [PubMed] [Google Scholar]
- Sidrauski C., Tsai J.C., Kampmann M., Hearn B.R., Vedantham P., Jaishankar P., Sokabe M., Mendez A.S., Newton B.W., Tang E.L., et al. . 2015. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife. 4:e07314 10.7554/eLife.07314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Garcia G., Bian Q., Steffen K.K., Joe L., Wolff S., Meyer B.J., and Dillin A.. 2016. Mitochondrial stress induces chromatin reorganization to promote longevity and UPR(mt). Cell. 165:1197–1208. 10.1016/j.cell.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H., Carroll C.J., Raimundo N., Ahola-Erkkilä S., Wenz T., Ruhanen H., Guse K., Hemminki A., Peltola-Mjøsund K.E., Tulkki V., et al. . 2010. Mitochondrial myopathy induces a starvation-like response. Hum. Mol. Genet. 19:3948–3958. 10.1093/hmg/ddq310 [DOI] [PubMed] [Google Scholar]
- Vilchez D., Boyer L., Morantte I., Lutz M., Merkwirth C., Joyce D., Spencer B., Page L., Masliah E., Berggren W.T., et al. . 2012. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 489:304–308. 10.1038/nature11468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., and Chen X.J.. 2015. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature. 524:481–484. 10.1038/nature14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Pandey A.K., Mulligan M.K., Williams E.G., Mozhui K., Li Z., Jovaisaite V., Quarles L.D., Xiao Z., Huang J., et al. . 2016. Joint mouse-human phenome-wide association to test gene function and disease risk. Nat. Commun. 7:10464 10.1038/ncomms10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E.G., Wu Y., Jha P., Dubuis S., Blattmann P., Argmann C.A., Houten S.M., Amariuta T., Wolski W., Zamboni N., et al. . 2016. Systems proteomics of liver mitochondria function. Science. 352:aad0189 10.1126/science.aad0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L., Topf U., Bragoszewski P., Wiese S., Sztolsztener M.E., Oeljeklaus S., Varabyova A., Lirski M., Chroscicki P., Mroczek S., et al. . 2015. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 524:485–488. 10.1038/nature14951 [DOI] [PubMed] [Google Scholar]
- Xia J., Sinelnikov I.V., Han B., and Wishart D.S.. 2015. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 43(W1):W251–W257. 10.1093/nar/gkv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Williams E.G., Mouchiroud L., Cantó C., Fan W., Downes M., Héligon C., Barish G.D., Desvergne B., Evans R.M., et al. . 2011. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 147:827–839. 10.1016/j.cell.2011.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T., Benedetti C., Urano F., Clark S.G., Harding H.P., and Ron D.. 2004. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 117:4055–4066. 10.1242/jcs.01275 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Wang J., Levichkin I.V., Stasinopoulos S., Ryan M.T., and Hoogenraad N.J.. 2002. A mitochondrial specific stress response in mammalian cells. EMBO J. 21:4411–4419. 10.1093/emboj/cdf445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W.X., Rabinowitz J.D., and White E.. 2016. Mitochondria and cancer. Mol. Cell. 61:667–676. 10.1016/j.molcel.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.