Hammarskjold and Rekosh preview new results from the Müller-McNicoll laboratory suggesting that the differentiation state modulates SR protein nucleocytoplasmic shuttling and function.
Abstract
Serine- and arginine-rich proteins play important roles in splicing, nuclear export, and translation. In this issue, Botti et al. (2017. J. Cell Biol. https://doi.org/10.1083/jcb.201610051) show that SRSF2 and SRSF5, previously thought to be nuclear, shuttle with messenger RNA to the cytoplasm in pluripotent P19 cells, but not in differentiated cells.
RNA-binding proteins (RBPs) play major roles in development and cellular gene regulation in all eukaryotic organisms. In humans, >1,500 genes are predicted to code for RBPs (7.5% of all human genes), illustrating the complexity of RNA regulation in cellular processes such as transcription, splicing, RNA trafficking, and translation (Gerstberger et al., 2014). Many of the RBPs show tissue-specific expression, and many are highly expressed only in early development. In addition, most of the genes are subject to alternative splicing, leading to expression of multiple protein isoforms. RBPs are subject to different posttranslational modifications, such as arginine methylation and serine and tyrosine phosphorylation. These can affect and regulate protein–RNA, protein–protein interactions, cellular localization, and other properties. Although many RBPs have been the subject of in-depth studies, we still know little about many of them and about their roles in normal function and disease.
The serine- and arginine-rich (SR) family RBPs were some of the first proteins to be identified as crucial regulators of both constitutive and alternative splicing (Cáceres et al., 1997). Several of the SR proteins have also been shown to be important connectors between splicing and nuclear export (Änkö, 2014). In addition, some of the members are known to shuttle between the nucleus and cytoplasm in human cell lines (such as HeLa cells and 293T cells) and are also present in polysomal fractions, linking mRNA export to translation (Sanford et al., 2004; Swartz et al., 2007). Based on these and other studies, the 12 canonical SR proteins were divided into shuttling (SRSF1, SRSF3, SRSF4, SRSF6, SRSF7, and SRSF10) and nonshuttling (SRSF2, SRSF5, SRSF8, SRSF9, SRSF11, and SRSF12). In this issue, Botti et al. (2017) unexpectedly show that although the nonshuttling proteins SRSF2 and SRSF5 indeed shuttle poorly in HeLa cells, they display a considerable amount of shuttling in pluripotent P19 mouse embryonic carcinoma cells (ECCs) (Fig. 1).
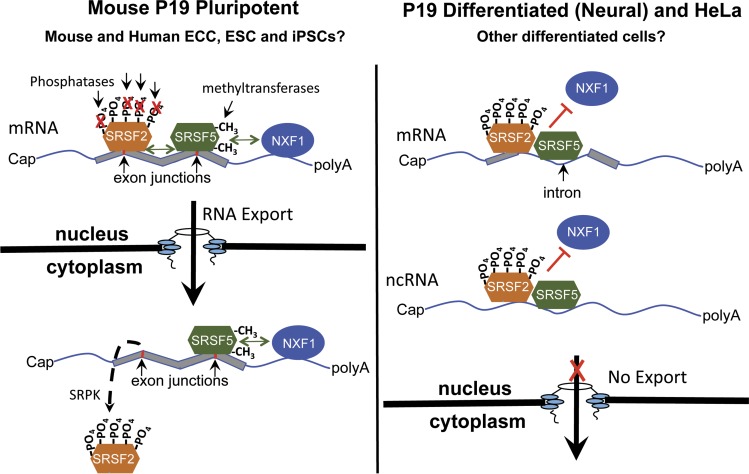
Figure 1.
SRSF5 and SRSF2 shuttle in pluripotent mouse P19 cells and cooperate with NXF1 in the export of mRNAs (model suggested by the data in Botti et al., 2017). (Left) In pluripotent P19 cells, increased arginine methylation of SRSF5 and hypophosphorylation of SRSF2 allow these two SR proteins to interact with NXF1 and mRNA in the export of specific mRNAs to the cytoplasm. NXF1 and SRSF5 stay bound to mRNAs in the cytoplasm, whereas SRSF2 is removed from the complex before translation. (Right) In P19 differentiated neural cells (and HeLa cells), SRSF2 and SRSF5 shuttle poorly and bind mainly to introns and noncoding RNAs. This is accompanied by reduced arginine methylation of SRSF5 and hyperphosphorylation of SRSF2.
Botti et al. (2017) used a novel quantitative heterokaryon assay that compares the shuttling behavior of seven canonical SR protein family members (SRSF1–7). This assay uses pluripotent mouse P19 cells stably expressing GFP-tagged SR proteins from genomic loci as donor cells and HeLa cells stably expressing the membrane marker CAAX-mCherry as recipients. To identify true heterokaryons after cell fusion, the authors screened for cells with two nuclei showing GFP expression (P19) within a plasma membrane labeled with CAAX-mCherry (HeLa). Using this assay, they made the observation that SRSF2 and SRSF5 show different shuttling behavior in the pluripotent P19 cells versus nonpluripotent HeLa cells. This finding led them to further investigate the factors that influenced nucleocytoplasmic shuttling of these proteins. They assessed SR protein interaction with nuclear export factor 1 (NXF1), SR presence in polysomal fractions, and phosphorylation sites. Using individual-nucleotide resolution cross-linking and immunoprecipitation (iCLIP) and mass spectrometry, Botti et al. (2017) show that differences in shuttling are accompanied by hypophosphorylation of SRSF2 and elevated arginine methylation of SRSF5 in P19 cells (Fig. 1). This facilitated binding of SRSF5 to NXF1 and enhanced shuttling of SRSF2 through cobound SRSF5 mRNA. Interestingly, results are also presented showing that perturbation of SRSF5 levels affects the cytoplasmic mRNA levels of the pluripotency factors Lin28a and Pou5f1/Oct4 in P19 cells and that SRSF5 is bound to these mRNAs in the cytoplasm. Finally, the study shows that neural differentiation of the P19 cells in vitro leads to a significant reduction in shuttling of SRSF5 (Fig. 1). In summary, the results of this study demonstrate that differential posttranslational modification of SR proteins influences shuttling and expression of pluripotency factors. Thus, this is the first demonstration that the cellular differentiation state can modulate mRNA export activity in P19 cells, and the work also suggests that posttranslational modifications of SR proteins influence their behavior.
The P19 cell line, established in 1982, was one of the first pluripotent cell lines (McBurney, 1993). P19 cells are easy to maintain and propagate compared with most embryonic lines and can be differentiated into different lineages using appropriate growth factors. In the Botti et al. (2017) study, mouse P19 cells were the only pluripotent cells studied, and the cells were only differentiated into neural cells. In addition, P19 cells are not “normal” cells, but cancer cells. Although there are many shared properties between the genetic programs of noncancer and cancer stem cells, such as the properties of self-renewal, there are also several differences. It is therefore not possible to know whether the findings from Botti et al. (2017) represent a general distinction between pluripotent and differentiated cells. It will thus be of considerable interest to study shuttling of SRSF2/SRSF5 in mouse embryonic stem cells (ESCs) and in stem cell progeny along lineages other than the neural protocol used by Botti et al. (2017). In this context, it should be mentioned that NXF1 was previously reported to be down-regulated in neuronal cells, potentially signifying differences in mRNA export between different lineages (Zhang et al., 2007). How SRSF2 and SRSF5 behave in pluripotent and differentiated human cells, including ESCs and induced pluripotent stem cells, also remains to be tested.
In addition, it would seem important to study SRSF2 and SRSF5 and their mRNA targets in a human ECC cell line, such as NCCIT. These cells were part of a recent study from the Wysocka laboratory that reported that the endogenous retrovirus HERV-K is induced in early human embryonic development, leading to expression of the HERV-K (HML-2) Rec RBP (Grow et al., 2015). Rec is involved in the nucleocytoplasmic export of HERV-K mRNA through binding to the XPO1 (CRM1) export receptor and serves an analogous function to the HIV Rev protein. The experiments presented in the paper included iCLIP and analysis of mRNA in polysomes in NCCIT cells expressing FLAG-tagged Rec, and it was shown that several mRNAs were specifically bound to Rec in the pluripotent cells. Interestingly, one of the identified targets was Lin28a mRNA, one of the mRNAs also identified by Botti et al. (2017) as interacting with SR proteins. It will thus be interesting to investigate how cytoplasmic levels of this mRNA are influenced by perturbations of SRSF5 in NCCIT cells.
In addition to a role as a general mRNA export receptor, NXF1 is also involved in the export of mRNAs with retained introns through direct binding of NXF1 and its cofactor NXT1 to a cis-acting element in the mRNA known as the constitutive transport element (CTE; Li et al., 2006). Although this element was first identified in the retrovirus Mason-Pfizer monkey virus (MPMV), it was subsequently shown that the Nxf1 gene itself contains a CTE in an intron that is retained in an alternatively spliced mRNA isoform (Li et al., 2006). The CTE is present in the Nxf1 gene in the same intron in most mammalian species and is also present in the Nxf1 gene in teleost fish, demonstrating that NXF1/NXT1 interaction with CTEs is a conserved mechanism (Wang et al., 2015). The Nxf1 mRNA with the retained intron is stably expressed in many cells, but the short protein that can be translated from this mRNA isoform is only present in some tissues. It was recently shown to be highly expressed in hippocampal and other neurons in rodents (Li et al., 2016). Whether the short NXF1 protein is expressed when P19 cells are differentiated into neural cells and whether the long isoform of NXF1 is down-regulated, as was previously reported (Zhang et al., 2007), remains to be determined. If this is the case, this could have general effects on mRNA export and potentially contribute to the nonshuttling behavior of SRSF5 in the neural cells.
Interestingly, it was previously demonstrated that at least two of the shuttling SR proteins enhanced expression mediated by CTEs and NXF1/NXT, even though SR proteins should not be essential for recruiting NXF1 to the mRNA in this case (Swartz et al., 2007). Specifically, it was shown that the SR proteins promoted polysome association and translation of CTE-containing mRNA. It would thus be of interest to analyze whether the Nxf1 CTE is active in pluripotent cells and whether SRSF2 and/or SRSF5 plays a role in Nxf1 CTE regulation in these cells. The exact role of NXT1 in NXF1-mediated mRNA export also remains to be better investigated, but this was not part of the present study (Botti et al., 2017).
SR proteins were first described in the early 1990s, a few years before the identification of the mRNA export receptors NXF1 and XPO1 (CRM1). Even though numerous studies throughout the years have highlighted the connections between splicing, export, and translation in eukaryotic cells and the important roles of SR proteins in all of these processes, we still have much to discover and elucidate. With the rapid advance of novel tools for genetic studies in mice and other animals, stem cell technologies, and techniques such as RNaseq, we have now finally entered an era where we can begin to uncover the mechanisms by which SR proteins and other RBPs operate together to fine-tune gene regulation in development and differentiation. This will lead to much needed novel insights into both normal cellular function and what goes wrong in diseases such as cancer, paving the way for novel therapies.
Acknowledgments
We apologize to those whose papers have not been cited here because of space limitations.
Research in the Hammarskjold and Rekosh laboratories is funded by core support from the Thaler Center for AIDS and Human Retrovirus Research and by National Institutes of Health grants GM110009 and CA206275.
The authors declare no competing financial interests.
References
- Änkö M.L. 2014. Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin. Cell Dev. Biol. 32:11–21. 10.1016/j.semcdb.2014.03.011 [DOI] [PubMed] [Google Scholar]
- Botti V., McNicoll F., Steiner M.C., Richter F.M., Solovyeva A., Wegener M., Schwich O.D., Poser I., Zarnack K., Wittig I., et al. 2017. Cellular differentiation state modulates the mRNA export activity of SR proteins. J. Cell Biol. 10.1083/jcb.201610051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres J.F., Misteli T., Screaton G.R., Spector D.L., and Krainer A.R.. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138:225–238. 10.1083/jcb.138.2.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S., Hafner M., and Tuschl T.. 2014. A census of human RNA-binding proteins. Nat. Rev. Genet. 15:829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow E.J., Flynn R.A., Chavez S.L., Bayless N.L., Wossidlo M., Wesche D.J., Martin L., Ware C.B., Blish C.A., Chang H.Y., et al. 2015. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 522:221–225. 10.1038/nature14308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Bor Y.C., Misawa Y., Xue Y., Rekosh D., and Hammarskjöld M.L.. 2006. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 443:234–237. 10.1038/nature05107 [DOI] [PubMed] [Google Scholar]
- Li Y., Bor Y.C., Fitzgerald M.P., Lee K.S., Rekosh D., and Hammarskjold M.L.. 2016. An NXF1 mRNA with a retained intron is expressed in hippocampal and neocortical neurons and is translated into a protein that functions as an Nxf1 cofactor. Mol. Biol. Cell. 27:3903–3912. 10.1091/mbc.E16-07-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney M.W. 1993. P19 embryonal carcinoma cells. Int. J. Dev. Biol. 37:135–140. [PubMed] [Google Scholar]
- Sanford J.R., Gray N.K., Beckmann K., and Cáceres J.F.. 2004. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18:755–768. 10.1101/gad.286404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz J.E., Bor Y.C., Misawa Y., Rekosh D., and Hammarskjold M.L.. 2007. The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J. Biol. Chem. 282:19844–19853. 10.1074/jbc.M701660200 [DOI] [PubMed] [Google Scholar]
- Wang B., Rekosh D., and Hammarskjold M.L.. 2015. Evolutionary conservation of a molecular machinery for export and expression of mRNAs with retained introns. RNA. 21:426–437. 10.1261/rna.048520.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Wang Q., and Huang Y.. 2007. Fragile X mental retardation protein FMRP and the RNA export factor NXF2 associate with and destabilize Nxf1 mRNA in neuronal cells. Proc. Natl. Acad. Sci. USA. 104:10057–10062. 10.1073/pnas.0700169104 [DOI] [PMC free article] [PubMed] [Google Scholar]