Abstract
Theory predicts that horizontal gene transfer (HGT) expands the selective conditions under which genes spread in bacterial populations. Whereas vertically inherited genes can only spread by positively selected clonal expansion, mobile genetic elements can drive fixation of genes by infectious HGT. We tested this using populations of Pseudomonas fluorescens and the conjugative mercury resistance (HgR) plasmid pQBR57. HGT expanded the selective conditions allowing the spread of HgR: Chromosomal HgR only increased in frequency under positive selection, whereas plasmid-encoded HgR reached fixation with or without positive selection. Tracking plasmid dynamics over time revealed that the mode of HgR inheritance varied across mercury environments. Under mercury selection, the spread of HgR was driven primarily by clonal expansion while in the absence of mercury HgR dynamics were dominated by infectious transfer. Thus, HGT is most likely to drive the spread of resistance genes in environments where resistance is useless.
Microbial populations reproduce clonally by vertical descent but can also exchange genes by horizontal gene transfer (HGT). HGT is an important process in bacterial evolution, accelerating adaptation by allowing the spread of ecologically and clinically relevant traits between lineages (Frost et al., 2005; Thomas and Nielsen, 2005). Therefore, the balance of vertical versus horizontal inheritance is expected to have important effects on bacterial evolution and thus function (Smith et al., 1993; Cordero and Polz, 2014; Shapiro, 2016). Comparative genomics has revealed that bacterial species undergo dramatic shifts in the balance of vertical versus horizontal inheritance over time (Cordero and Polz, 2014). These shifts may be due to changes in selection on inherited traits, as theory predicts that vertical inheritance is favoured by strong positive selection, increasing clonality via genome-wide selective sweeps (clonal expansion), whereas horizontally inherited genes can spread even in the absence of positive selection (Tazzyman and Bonhoeffer, 2014), maintaining population genomic diversity. However, experimental data addressing this issue are lacking.
To test how selection alters the balance of vertical versus horizontal transmission of bacterial genes, we quantified the dynamics of mercury resistance (HgR) in populations of the soil bacterium Pseudomonas fluorescens SBW25 (Rainey and Bailey, 1996) with the HgR operon mer encoded either chromosomally or carried on an HgR plasmid pQBR57 (Lilley and Bailey, 1997). We established 36 replicate populations of SBW25, 18 with HgR encoded on their chromosome (non-horizontally transferable), 18 with HgR encoded on pQBR57 (horizontally transferable). Each population was mixed 50:50 with a mercury-sensitive differentially marked SBW25 strain and then propagated by serial transfer every 24 h for 8 days. Populations were grown in one of three mercury environments, 0, 20 and 40 μM HgCl2 (six replicates per treatment); this represents a selective gradient wherein plasmid-encoded HgR is under, respectively, strong negative selection, weak positive selection and strong positive selection, due to the balance between the cost of plasmid carriage and the benefits of HgR (Supplementary Figure S1; Hall et al., 2015). Because pQBR57 is maintained at low copy number (Hall et al., 2015), the chromosomal and plasmid-encoded HgR genes provide equivalent levels of resistance (Supplementary Figure S2). Every 2 days we determined the proportion of HgR cells within each population by plate counts. Furthermore, as our donors and recipients were differentially marked we were able to track the frequency of pQBR57 in both donor and recipient populations (for full methods see Supplementary Information).
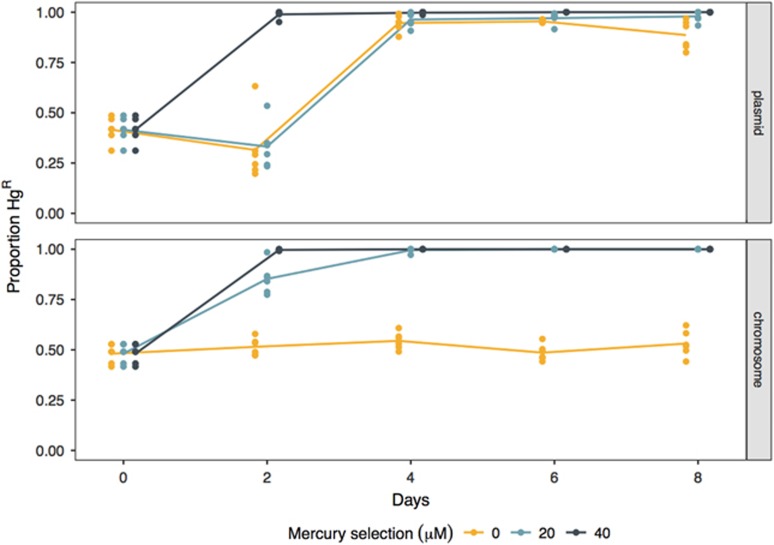
The end point proportion of mercury-resistant (HgR) cells in the population was significantly affected by the horizontal transmissibility of HgR (Figure 1; main effect of mobility at 0 μM HgCl2: F1,10=74.34, P<0.001). Where HgR was encoded on the chromosome, positive selection was required to drive the spread of resistance: HgR rapidly became fixed within the population in the 20 and 40 μM HgCl2 environments, whereas in the 0 μM HgCl2 environment chromosomal HgR remained at ~50% prevalence. In contrast, when HgR was encoded on the conjugative plasmid pQBR57, and thus horizontally transferable, HgR reached high frequencies across all mercury environments (0, 20 and 40 μM HgCl2). Thus, the opportunity for horizontal transfer expanded the selective conditions allowing the fixation of HgR such that this occurred both with and without positive selection for resistance.
Figure 1.
Horizontal transmission had a significant impact on the proportion of HgR. The proportion of chromosome- and plasmid-encoded HgR was determined over time across the three mercury treatments (0, 20 and 40 μM HgCl2). Points represent replicate populations and are slightly offset by treatment on the x axis to prevent over plotting. Lines represent means (n=6).
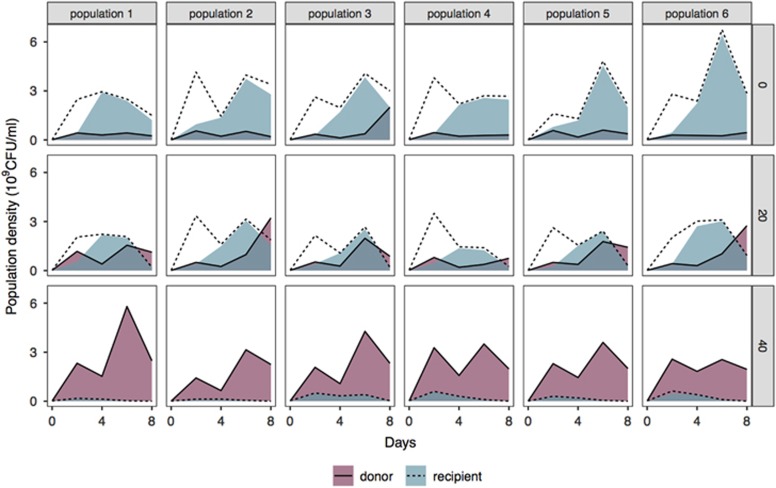
Tracking plasmid dynamics over time revealed that the strength of positive selection determined the balance of horizontal versus vertical inheritance of plasmid-encoded HgR in bacterial populations. HGT played a significantly greater role as the strength of selection decreased (Figure 2; Main effect of mercury: F1,16=392.72, P<0.001). Under strong positive selection (40 μM HgCl2), HgR swept through the population by clonal expansion of the original HgR donor population. This was presumably due to the high toxicity of the environment strongly selecting against plasmid-free recipients, limiting the opportunity for HGT via plasmid conjugation as a consequence. The contribution of vertical inheritance to the spread of HgR reduced with weakening positive selection. Under weak positive selection (20 μM HgCl2), HgR spread through the population by a mixture of vertical clonal expansion of donor cells and horizontal transmission of the plasmid into the recipient subpopulation. Under negative selection (0 μM HgCl2) HgR spread by conjugative plasmid transfer into available plasmid-free recipient cells. Therefore, while strong positive selection favoured vertical inheritance, the contribution of horizontal transfer to the spread of resistance genes increased as positive selection weakened.
Figure 2.
Selection determines the balance of horizontal versus vertical inheritance of plasmid-encoded HgR. Plasmid transfer in each of six replicate populations was tracked over time across the three mercury treatments (0, 20 and 40 μM HgCl2). Dotted lines indicate densities of recipient populations; solid lines indicate densities of donor populations. For each population, shaded regions represent plasmid prevalence within donor (purple) and recipient (blue) subpopulations.
Our data are consistent with theory that HGT can overcome selective barriers to drive the spread of resistance genes in the absence of positive selection, whereas resistance genes spread through vertical transmission only under positive selection. Thus, whereas positive selection for resistance would purge genomic diversity via genome-wide sweeps of resistance (Wiedenbeck and Cohan, 2011), negative selection against resistance coupled with infectious HGT of resistance genes can spread resistance genes into diverse genomic backgrounds. Consequently, the sharing of resistance genes between lineages is most likely to occur in environments without positive selection, and therefore where resistance genes have little use.
Acknowledgments
We thank D Guymer for technical support, and V Friman and J Pitchford for valuable comments. This work was supported by an ERC Starting Grant from the European Research Council awarded to MAB (StG-2012-311490-COEVOCON), a Leverhulme Prize from the Leverhulme Trust awarded to MAB (PLP-2014-242), the University of York and a NERC studentship to CS supervised by MAB and AJW.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Cordero OX, Polz MF. (2014). Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Micro 12: 263–273. [DOI] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. (2005). Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3: 722–732. [DOI] [PubMed] [Google Scholar]
- Hall JPJ, Harrison E, Lilley AK, Paterson S, Spiers AJ, Brockhurst MA. (2015). Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects. Environ Microbiol 17: 5008–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley AK, Bailey MJ. (1997). The acquisition of indigenous plasmids by a genetically marked pseudomonad population colonizing the sugar beet phytosphere is related to local environmental conditions. Appl Environ Microbiol 63: 1577–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey PB, Bailey MJ. (1996). Physical and genetic map of the Pseudomonas fluorescens SBW25 chromosome. Mol Microbiol 19: 521–533. [DOI] [PubMed] [Google Scholar]
- Shapiro BJ. (2016) How clonal are bacteria over time? Curr Opin Microbiol 31: 116–123. [DOI] [PubMed] [Google Scholar]
- Smith JM, Smith NH, ORourke M, Spratt BG. (1993). How clonal are bacteria? Proc Natl Acad Sci USA 90: 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazzyman SJ, Bonhoeffer S. (2014). Plasmids and evolutionary rescue by drug resistance. Evolution 68: 2066–2078. [DOI] [PubMed] [Google Scholar]
- Thomas CM, Nielsen KM. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3: 711–721. [DOI] [PubMed] [Google Scholar]
- Wiedenbeck J, Cohan FM. (2011). Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev 35: 957–976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.