Abstract
Reducing the time taken to run qPCR assays on today’s qPCR cyclers is rather straightforward and requires no specialised reagents or instruments. As the first article in a new series of short technical reports, I demonstrate that it is possible to reduce significantly both denaturation temperatures and cycling times, whilst retaining sensitivity and specificity of the original qPCR conditions.
Keywords: Real-time PCR, Taq polymerase, Denaturation, Annealing, Polymerisation
1. Introduction
Most users of the polymerase chain reaction (PCR) would describe it as a fairly fast technique, taking about 45 min to an hour to complete 40 cycles, depending on the particular protocol and instrument used. The first description of a PCR reaction carried out using thermostable DNA polymerase amplified targets ranging from 110 to 408 base pairs (bp) with one-minute denaturation, two-minute annealing and 30 s polymerisation steps [1]. Since then, there have been numerous and significant improvements to both instruments and reagents. These include the introduction of real-time (qPCR) and digital PCR (dPCR) technologies, new fluorophores, specialist master mixes and fast thermal cyclers, leading to an enormous expansion of practical uses, not least high throughput applications.
Despite these improvements, what has not changed a great deal are the protocols used to run mainstream qPCR reactions. A quick perusal of the peer-reviewed literature reveals that current standard methods, if they are published at all, include near universal denaturation temperatures and times of 95 °C and 15–30 s, respectively, annealing/polymerisation times of 15 s–1 min and optional separate polymerisation times of 45 s–2 min. These add up to around an hour for a typical 40-cycle reaction. In addition to these times, the speed of a PCR run is also dependent on ramp rates, i.e. how fast the PCR instrument can change from one temperature to another and especially how fast the cooling process works. Hence any attempt to reduce significantly PCR run times must look at reducing both cycle times and minimising the differences in temperatures for the various steps.
There have been several reports of PCR methods that significantly increase PCR reactions times significantly over those in mainstream use. However, whilst these can reduce PCR cycle times to a few minutes or even seconds, they use specialised equipment and procedures and generally compromise PCR efficiency [2], [3], [4], [5], [6]. Interestingly, a recent a series of publications by Carl Wittwer’s group has investigated the kinetics of PCR and its conclusions suggest that it is possible to use significantly modified protocols to achieve faster PCR results without compromising the sensitivity or specificity of the PCR assay. Although the extension rates of native Taq polymerases ranged from 10 to 45 nucleotides/second, some polymerases achieved up to 155 nucleotides/second [7]. Maximum extension rates were achieved by using optimised extension temperatures (Tm −5 °C) on somewhat G/C-rich templates (around 60%) without secondary structures [8] in the presence of minimal monovalent cations [9]. More recently, super-fast PCR reaction times were achieved by increasing primer and polymerase concentrations to around 20-fold above typical concentrations, increasing annealing/extension temperatures to around 75 °C and reducing denaturation temperatures to below 90 °C, so allowing amplification of short PCR amplicons less than 15 s [10].
Clearly, there is potential to reduce the time taken to complete standard qPCR reaction times using regular reagents and instruments. Therefore, if the speed of a PCR reaction is an important consideration, it is worth modifying legacy PCR procedures to incorporate these findings into a mainstream fast PCR protocol. This technical note describes such a modification, which reduces PCR reaction times on standard PCR instruments without compromising either its sensitivity or specificity.
2. Materials and methods
All pipetting was carried out using 0.1–3 μL Biohit mLine (Sartorius) manual pipettes for volumes up to 3 μL, 0.5–10 μL pipettes for volumes between 3 and 10 μL, 2–10 μL pipettes for volumes between 10 μL and 20 μL and 10–100 μL pipettes for volumes between 10 and 100 μL. qPCR reactions were carried out on the CFX Connect (Biorad) and Mic (Biomolecular Systems) qPCR cyclers.
2.1. cDNA synthesis
cDNA was prepared from 1 μg of RNA, which had been manually extracted from MCF 7 tissue culture cells using an RNeasy (Qiagen) RNA extraction kit with a DNase step according to the manufacturer’s protocol. Total RNA quality was assessed using a Bioanalyser (Agilent) and purity was assessed using the SPUD assay [11]. Reverse transcription was carried out using random primers and Superscript IV (ThermoFisher) in three separate 20 μL reactions at 23 °C for 10 min and 55 °C for 10 min, followed by an incubation at 80 °C for 10 min. The three samples were pooled, diluted with 540 μL of water, aliquoted and stored at −80 °C.
2.2. qPCR
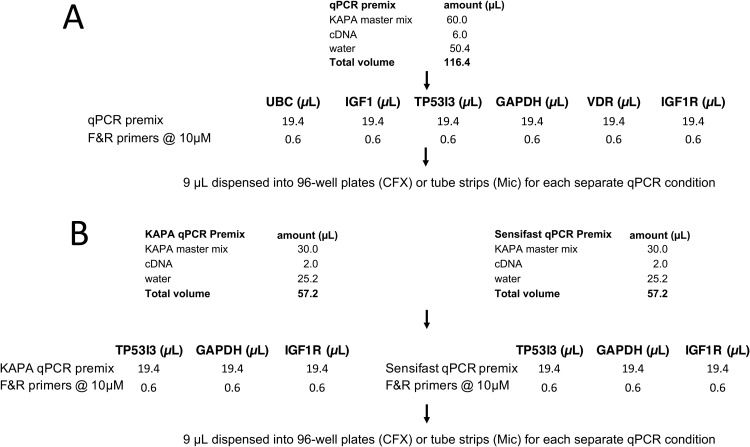
Six qPCR assays targeting Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), insulin-like growth factor 1 (IGF-1), insulin-like growth factor 1 receptor 1 (IGF-1R), tumour protein p53 inducible protein 3 (TP53IP3), Ubiquitin C (UBC) and Vitamin D receptor (VDR) were chosen at random from a selection of over 60 assays kept in the freezer and are described in Table 1. All primers were synthesised by Sigma-Aldrich and used at a final concentration of 300 nM. The effects of varying cycling times and temperatures were determined using KAPA (Sigma Aldrich), using the workflow shown in Fig. 1A. Three of the assays were chosen for comparison between SensiFAST (Bioline) and KAPA master mixes using the workflow shown in Fig. 1B.
Table 1.
qPCR assay features. All six assays were designed using Beacon Designer (Premier Biosoft).
| Targets | Primers | Size (bp) | Accession no | Start | End | CG | AT | %GC | |
|---|---|---|---|---|---|---|---|---|---|
| UBC2 | F: | AGGAAAAGTAGTCCCTTCTC | 177 | NM_021009 | 295 | 471 | 99 | 78 | 56% |
| R: | CGAAGATCTGCATTGTCAAG | ||||||||
| IGF1 | F: | CCTTTCAAGCCACCCATTGAC | 100 | NM_000376 | 1166 | 1265 | 62 | 38 | 62% |
| R: | AGCAGCGGGTACAAGATAAATATCC | ||||||||
| TP53I3 | F: | CTGCTGCCGGTTCTGGAC | 96 | NM_004881 | 1778 | 1873 | 51 | 45 | 53% |
| R: | CAGGACGATCTTGCCTATGTT | ||||||||
| GAPDH | F: | GCACAAGAGGAAGAGAGAGACC | 84 | NM_000875 | 1089 | 1172 | 52 | 32 | 62% |
| R: | AGGGGAGATTCAGTGTGGTG | ||||||||
| VDR1 | F: | ATCTGCATCGTCTCCCCAGAT | 104 | NM_001111283 | 6731 | 6834 | 44 | 60 | 42% |
| R: | AGCGGATGTACGTCTGCAGTG | ||||||||
| IGF1R | F: | CTCCTGTTTCTCTCCGCCG | 78 | NM_002046 | 1225 | 1302 | 47 | 31 | 60% |
| R: | ATAGTCGTTGCGGATGTCGAT |
Fig. 1.
qPCR workflow. All reaction components except for the primers were dispensed to individual microfuge tubes, to which target-specific forward and reverse primer mixes were added. Reaction mixtures were kept on ice until subjected to PCR. Sufficient qPCR premixes were prepared for each of the six targets to run six assays in duplicate. A. Workflow for initial investigation using KAPA master mix. B. Workflow for comparison between KAPA and SensiFast master mixes.
2.3. Data analysis
Data were analysed using instrument default settings and quantification cycles (Cqs) were calculated automatically. On the Mic the data were analysed using the in-built dynamic algorithm option, whilst the CFX data were analysed using the regression method option, both without manual intervention.
3. Results and discussion
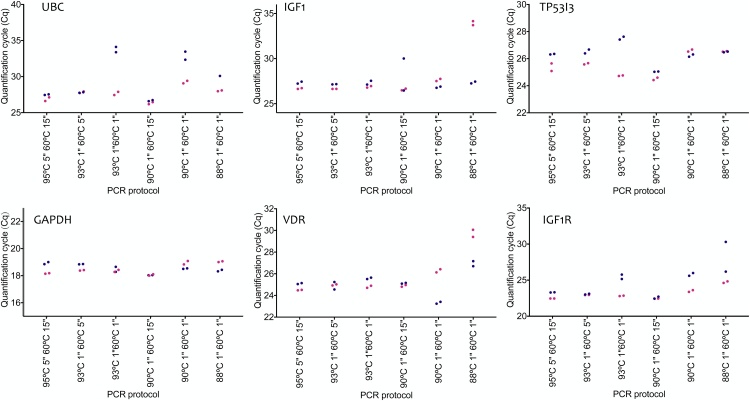
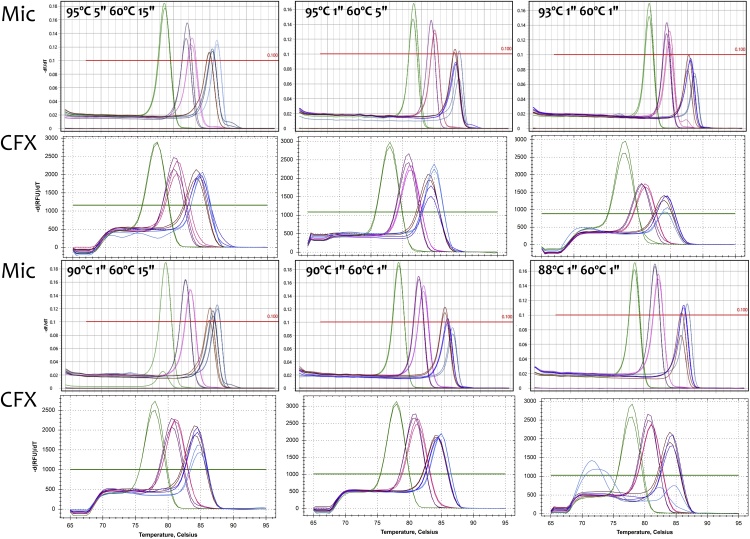
The quality of the RNA extracted from the MCF 7 cells was very high, with a RIN value of 10 and there was no inhibition as assessed by the SPUD assay. The Cqs recorded by the two qPCR instruments for the six assays are listed in Table 2 and plotted in Fig. 2. They show that successful PCR results can be achieved over a wide range of temperature and time conditions. Furthermore, the results are not necessarily platform-dependent, since these equivalent results were generated by two technically different instruments. Taking the baseline conditions of a 5 s at 95 °C denaturation and 15 s at 60 °C annealing/polymerisation step, all six assays assay generate approximately the same Cqs, regardless of which cycler was used. Melt curve analysis (Fig. 3) shows that the same amplicons are being generated at all temperatures and for all amplicons, with the exception of UBC on the CFX at the 88 °C denaturation temperature, where the major product appears to have a Tm of around 72 °C rather than 84 °C. Interestingly, the melt curve for UBC on the Mic cycler does not show that difference and records the same Tm as those obtained with the other conditions.
Table 2.
Quantification cycles recorded at each of the different qPCR reaction conditions. Assays were run in duplicate. A. Results from assays carried on the Mic cycler. N/A indicates that no Cq was obtained. B. Results from assays carried out in parallel on the CFX Connect cycler.
| (A) | ||||||
|---|---|---|---|---|---|---|
| 95 °C 5” 60 °C 15” | 93 °C 1” 60 °C 5” | 93 °C 1“60 °C 1” | 90 °C 1” 60 °C 15” | 90 °C 1” 60 °C 1” | 88 °C 1” 60 °C 1” | |
| UBC | 27.45 | 27.75 | 33.38 | 26.74 | 32.34 | 30.09 |
| 27.54 | 27.83 | 34.11 | 26.59 | 33.46 | N/A | |
| IGF1 | 27.23 | 27.14 | 27.12 | 30.01 | 26.76 | 27.25 |
| 27.44 | 27.17 | 27.54 | 26.46 | 26.89 | 27.45 | |
| TP53I3 | 26.31 | 26.67 | 27.62 | 25.05 | 26.31 | 26.52 |
| 26.35 | 26.39 | 27.41 | 25.03 | 26.14 | 26.47 | |
| GAPDH | 18.85 | 18.86 | 18.65 | 18.04 | 18.54 | 18.43 |
| 19.00 | 18.84 | 18.28 | 18.03 | 18.50 | 18.32 | |
| VDR | 25.14 | 25.24 | 25.51 | 25.16 | 23.24 | 26.70 |
| 25.05 | 24.55 | 25.64 | 25.09 | 23.41 | 27.16 | |
| IGF1R | 23.28 | 23.11 | 25.76 | 22.42 | 25.60 | 26.17 |
| 23.31 | 23.00 | 25.15 | 22.70 | 25.98 | 30.29 | |
| (B) | ||||||
|---|---|---|---|---|---|---|
| 95 °C 5” 60 °C 15” | 93 °C 1” 60 °C 5” | 93 °C 1“60 °C 1” | 90 °C 1” 60 °C 15” | 90 °C 1” 60 °C 1” | 8 °C 1” 60 °C 1” | |
| UBC | 26.59 | 27.93 | 27.86 | 26.15 | 29.39 | 27.95 |
| 27.10 | 27.70 | 27.42 | 26.43 | 29.04 | 28.06 | |
| IGF1 | 26.70 | 26.62 | 26.93 | 26.46 | 27.73 | 33.70 |
| 26.61 | 26.61 | 26.76 | 26.65 | 27.49 | 34.14 | |
| TP53I3 | 25.07 | 25.65 | 24.74 | 24.40 | 26.66 | 26.54 |
| 25.63 | 25.56 | 24.70 | 24.58 | 26.51 | 26.50 | |
| GAPDH | 18.13 | 18.40 | 18.41 | 17.99 | 19.07 | 18.99 |
| 18.17 | 18.36 | 18.26 | 18.09 | 18.82 | 19.05 | |
| VDR | 24.50 | 25.05 | 24.92 | 24.99 | 26.44 | 29.41 |
| 24.54 | 24.95 | 24.73 | 24.83 | 26.15 | 30.07 | |
| IGF1R | 22.53 | 23.03 | 22.93 | 22.52 | 23.69 | 24.69 |
| 22.53 | 23.01 | 22.87 | 22.52 | 23.45 | 24.91 |
Fig. 2.
Comparison of the Cqs recorded in Table 2 for individual targets. Blue circles mark Cqs obtained using the Mic cycler, pink circles those with the CFX Connect.
Fig. 3.
Melt curves obtained following the runs shown in Table 2 and Fig. 1.
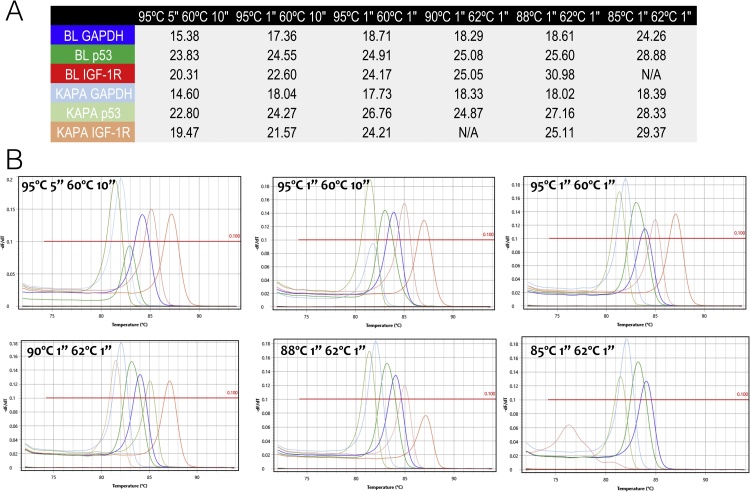
The comparison of two master mixes shown in Fig. 4 shows very similar results across a 10 °C denaturation temperature range. The melt curve profiles indicate amplification of the appropriate PCR amplicons, except for IGF-1R at the denaturation temperature of 85 °C, which does not amplify at all with the Bioline master mix and generates an incorrect melt curve with KAPA master mix.
Fig. 4.
Comparison of results obtained using Sensifast (BL) or KAPA master mixes. A. Quantification cycles recorded for each of the two master mixes at the different qPCR reaction conditions. B. Melt curves for these assays. The colours in Fig. 3A correspond to the colours of the melt curves.
These results indicate that it is perfectly feasible to obtain good PCR results with short denaturation and annealing/polymerisation times and at low denaturation temperatures. This has a direct effect on the overall time it takes to complete 40 cycles of a PCR assay, since the time taken to run the assay drops from 49 to 33 min for the CFX and from 51 to 31 min for the Mic. Clearly, these experimental conditions are appropriate for a PCR cycler not immediately associated with speed (CFX), and work well with both a 96 well and a rotary-based format.
Combining the published data on PCR kinetics [7], [8], [9] with the results presented above allows a summary of the conditions most likely to generate robust and reproducible data in the shortest time possible without using specialised equipment:
-
•
PCR amplicon sizes of around 100 bp are optimal, although the use of longer amplicons, such as UBC at 177, is possible.
-
•
Since substitution of TTP with dUTP lowers nucleotide incorporation rate, even if its concentration is increased, master mixers that use dUTP should not be used if speed is the main consideration.
-
•
If possible, amplicons with higher GC contents (around 60%) should be chosen since these templates have the highest extension rates.
-
•
Two-step PCR is faster than three-step PCR. Extension rates are maximal at 5 °C below the Tm of the primers. Given that the optimal temperature of extension is between 70 °C and 75 °C, the fastest two-step PCR would occur with primer Tms at 65 °C or above.
Speed demons can look forward to significant reductions in the time taken to complete PCR reactions. Already there is a new commercial PCR cycler, NexgenPCR (Molecular Biosystems) that can amplify a 100 bp target in just under two minutes (http://www.nextgenpcr.com/wp-content/uploads/2016/05/MBS-application-note-2-minute-pcr.pdf). A combination of such innovative cycler design, smaller volumes and optimised assay conditions will revolutionise the time taken to complete PCR reactions. Meanwhile, I have demonstrated above that it is fairly easy to reduce significantly the time taken to run current qPCR reactions with standard thermal cyclers and reagents.
Conflict of interest
There is no conflict of interest.
Acknowledgement
All experiments were carried out and analysed by the author. Thank you to Dr Sara Kirvell for preparing the mRNA.
Handled by Jim Huggett
References
- 1.Saiki R.K., Gelfand D.H., Stoffel S. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 2.Kopp M.U., Mello A.J., Manz A. Chemical amplification: continuous-flow PCR on a chip. Science. 1998;280:1046–1048. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- 3.Hühmer A.F., Landers J.P. Noncontact infrared-mediated thermocycling for effective polymerase chain reaction amplification of DNA in nanoliter volumes. Anal. Chem. 2000;72:5507–5512. doi: 10.1021/ac000423j. [DOI] [PubMed] [Google Scholar]
- 4.Zhang C., Xing D. Miniaturized PCR chips for nucleic acid amplification and analysis: latest advances and future trends. Nucleic Acids Res. 2007;35:4223–4237. doi: 10.1093/nar/gkm389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler E.K., Hara C.A., Frank J. Under-three minute PCR: probing the limits of fast amplification. Analyst. 2011;136:3707–3712. doi: 10.1039/c1an15365j. [DOI] [PubMed] [Google Scholar]
- 6.Fuchiwaki Y., Nagai H., Saito M., Tamiya E. Ultra-rapid flow-through polymerase chain reaction microfluidics using vapor pressure. Biosens. Bioelectron. 2011;27:88–94. doi: 10.1016/j.bios.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 7.Montgomery J.L., Rejali N., Wittwer C.T. Stopped-flow DNA polymerase assay by continuous monitoring of dNTP incorporation by fluorescence. Anal. Biochem. 2013;441:133–139. doi: 10.1016/j.ab.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Montgomery J.L., Rejali N., Wittwer C.T. The influence of nucleotide sequence and temperature on the activity of thermostable DNA polymerases. J. Mol. Diagn. 2014;16:305–313. doi: 10.1016/j.jmoldx.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Montgomery J.L., Wittwer C.T. Influence of PCR reagents on DNA polymerase extension rates measured on real-time PCR instruments. Clin. Chem. 2014;60:334–340. doi: 10.1373/clinchem.2013.212829. [DOI] [PubMed] [Google Scholar]
- 10.Farrar J.S., Wittwer C.T. Extreme PCR: Efficient and Specific DNA Amplification in 15–60 Seconds. Clin. Chem. 2015;61:145–153. doi: 10.1373/clinchem.2014.228304. [DOI] [PubMed] [Google Scholar]
- 11.Nolan T., Hands R.E., Ogunkolade B.W., Bustin S.A. SPUD: a qPCR assay for the detection of inhibitors in nucleic acid preparations. Anal. Biochem. 2006;351:308–310. doi: 10.1016/j.ab.2006.01.051. [DOI] [PubMed] [Google Scholar]