Abstract
ATR is a key regulator of cell cycle checkpoints and homologous recombination (HR). Paradoxically, ATR inhibits CDKs during checkpoint responses, but CDK activity is required for efficient HR. Here, we show that ATR promotes HR after CDK-driven DNA end resection. ATR stimulates the BRCA1-PALB2 interaction after DNA damage, and promotes PALB2 localization to DNA damage sites. ATR enhances BRCA1-PALB2 binding at least in part by inhibiting CDKs. The optimal interaction of BRCA1 and PALB2 requires phosphorylation of PALB2 at S59, an ATR site, and hypo-phosphorylation of S64, a CDK site. The PALB2-S59A/S64E mutant is defective for localization to DNA damage sites and HR, whereas the PALB2-S59E/S64A mutant partially bypasses ATR for its localization. Thus, HR is a biphasic process requiring both high-CDK and low-CDK periods. As exemplified by the regulation of PALB2 by ATR, ATR promotes HR by orchestrating a “CDK-to-ATR switch” post resection, directly coupling the checkpoint with HR.
eTOC Blurb
Activation of ATR inhibits CDKs, but paradoxically it also promotes homologous recombination (HR), a CDK-dependent repair process. Buisson et al. show that after the CDK-driven DNA end resection, ATR promotes the HR function of PALB2 by phosphorylating PALB2 and suppressing CDK-mediated PALB2 phosphorylation, directly coupling checkpoint-mediated CDK inhibition to HR.

Introduction
Cell cycle checkpoints are pathways that control the order and timing of cell cycle progression, and they are particularly important when the genome is facing DNA damage and replication stress (Elledge, 1996). The ATR (ataxia telangiectasia mutated and rad3-related) kinase is a master regulator of DNA damage signaling (Cimprich and Cortez, 2008; Marechal and Zou, 2013). The activation of ATR in S-phase cells suppresses firing of replication origins and restrains DNA synthesis, whereas the activation of ATR in G2 cells promotes the G2/M cell-cycle arrest. The ability of ATR to promote checkpoint responses is at least in part attributed to its effects on CDKs. Through Chk1-mediated degradation or inhibition of CDC25 phosphatases (Busino et al., 2003; Jin et al., 2003; Mailand et al., 2000; Peng et al., 1997; Sanchez et al., 1997), the ATR-Chk1 pathway suppresses activities of CDK2 and CDK1, thereby hindering origin firing and mitotic entry. Checkpoint responses are thought to provide time for DNA repair (Elledge, 1996), but whether they directly participate in DNA repair is not known.
In addition to checkpoint responses, ATR also regulates DNA repair. ATR is implicated in homologous recombination (HR) (Adamson et al., 2012; Wang et al., 2004), a pathway that repairs DNA double-stranded breaks (DSBs). The function of ATR in HR has not been clearly defined (see discussion). Paradoxically, although ATR is known to inhibit CDKs, CDK activity is required for efficient HR. CDKs promote resection of DNA ends at DSBs (Huertas et al., 2008; Huertas and Jackson, 2009), which gives rise to the single-stranded DNA (ssDNA) necessary for HR. These seemingly contradictory findings raise the question as to whether ATR-mediated checkpoint responses are compatible with HR and, if so, how the functions of ATR in checkpoint responses and HR are coordinated.
We previously showed that ATM and ATR kinases are consecutively activated by DSBs, and that ATR activation is driven by resection, raising the possibility that ATR has a post-resection function in HR (Shiotani and Zou, 2009). Here, we investigate the function of ATR in HR. During HR, BRCA1 recruits the PALB2-BRCA2 complex to DSBs (Orthwein et al., 2015; Sy et al., 2009; Zhang et al., 2009), promoting RAD51 filament formation (Buisson et al., 2010; Jensen et al., 2010; Liu et al., 2010; Thorslund et al., 2010). We find that ATR enhances the BRCA1-PALB2 interaction after DNA damage, and that ATR is required for efficient PALB2 accumulation at DSBs. Unexpectedly, the optimal interaction between BRCA1 and PALB2 not only requires the phosphorylation of PALB2 at Serine 59 (S59), an ATR site, but also the hypo-phosphorylation of Serine 64 (S64), a CDK site, suggesting that ATR regulates HR at least in part through controlling the S59–S64 “phosphorylation switch”. Our results suggest that HR occurs in two consecutive phases: high CDK activity drives resection and ATR activation in the first, and ATR suppresses CDKs and phosphorylates HR substrates in the second (see Fig. S4G). This CDK-to-ATR switch is necessary for optimal BRCA1-PALB2 interaction and possibly other HR events. Thus, ATR promotes HR by phosphorylating HR substrates and by inhibiting CDKs, directly coupling the checkpoint to HR.
Results
ATR promotes PALB2 accumulation at DNA damage sites
To assess the function of ATR in HR, we first tested the effects of a panel of ATR inhibitors on ionizing radiation (IR)-induced RAD51 focus formation. U2OS and HeLa cells were treated with three different ATR inhibitors (ATRi: VE-821; ATRi#2: AZ20; ATRi#3: EPT46464) (Foote et al., 2013; Reaper et al., 2011; Toledo et al., 2011), irradiated with IR, and RAD51 foci were analyzed in 2–4 hours (Fig. 1A–B, S1A–B). All three ATR inhibitors drastically reduced RAD51 foci, confirming that ATR promotes RAD51 accumulation at DSBs (Fokas et al., 2012; Prevo et al., 2012). To test if ATR has a post-resection function in HR, we sought to inhibit ATR after resection (Fig. S1C). RPA foci were readily detected 1 hour after IR (Fig. S1D), showing resection of DSBs (Gruz-Garcia et al., 2014). When ATR inhibitors were added 1 hour after IR, RAD51 foci were still reduced (Fig. 1C, S1E–G). Notably, ATR inhibitors reduced RAD51 foci in RPA foci-positive cells (Fig. 1C, S1G), highlighting a post-resection role of ATR in HR.
Fig. 1. ATR promotes PALB2 localization to DSBs after resection.
A. BRCA1 and RAD51 localization in U2OS cells 2 h after IR (4 Gy). Cells were pre-treated with DMSO or ATRi for 30 min before IR. B. Quantification of cells displaying >5 RAD51 foci in A. Error bar: S.D. (n=3). C. RPA and RAD51 localization in HeLa cells 4 h after IR (10 Gy). Cells were pre-treated with DMSO of ATRi 30 min before IR. D. BRCA1 and PALB2 localization in U2OS cells 2 h after IR (4 Gy). Cells were pre-treated with DMSO or ATRi 30 min before IR. E. Quantification of cells displaying >10 BRCA1 foci in D. Error bar: S.D. (n=3). F. The fractions of BRCA1 foci-positive cells displaying >10 PALB2 foci were quantified after DMSO or ATRi treatment. Error bar: S.D. (n=3). G. Localization of YFP-PALB2WT to laser-induced DNA damage sites was analyzed by time-lapse after indicated treatment. The fluorescence intensity of YFP-PALB2WT at DNA damage sites was quantified over time. For each cell analyzed, the signal in an unirradiated nuclear area in the same cell was determined as background. Error bar: S.E. (n > 50).
To pinpoint the execution point of ATR in HR, we analyzed the effects of ATRi on BRCA1 and PALB2. During the IR response, both BRCA1 and PALB2 are recruited to DSBs, and BRCA1 promotes formation of PALB2 foci. ATRi did not affect BRCA1 foci (Fig. 1A, DE), but it diminished PALB2 foci (Fig. 1D). While BRCA1 foci were readily detected in ATRi-treated cells, the PALB2 in BRCA1 foci was reduced (Fig. 1F). To test the effects of ATR inhibition on PALB2 recruitment more rigorously, we analyzed the recruitment of YFP-tagged wild-type PALB2 (PALB2WT) to sites of laser-induced DNA damage in living cells. Two different ATR inhibitors reduced the localization of PALB2 to DNA damage sites in time-lapse experiments (Fig. 1G, S1H). These results suggest that ATR acts between BRCA1 and PALB2 to promote the accumulation of PALB2 at DSBs.
ATR promotes the BRCA1-PALB2 interaction after DNA damage
Given that BRCA1 interacts with PALB2 and promotes PALB2 recruitment to DNA damage sites, we next tested if ATR inhibition affects the interaction between BRCA1 and PALB2. HeLa cells were treated with IR, and Flag-tagged PALB2WT was immunoprecipitated at different time points after irradiation (Fig. 2A). While an interaction between BRCA1 and PALB2 was detected in undamaged cells (Buisson and Masson, 2012; Sy et al., 2009; Zhang et al., 2009), this interaction was enhanced from 2 to 4 hours after IR, coinciding with the colocalization of BRCA1 and PALB2 in nuclear foci (Fig. 1D, 2A, S2A). To test if the enhancement of BRCA1-PALB2 interaction is dependent on the ATR-Chk1 pathway, we treated cells with two different ATR inhibitors and two Chk1 inhibitors (Chk1i: MK-8766; Chk1i#2: UNC-01) (Fig. 2B). All of these ATR and Chk1 inhibitors suppressed the enhancement of BRCA1-PALB2 binding after IR. In contrast, ATRi and Chk1i did not affect the interactions of PALB2 with BRCA2 and RAD51. ATRi did not alter the cell cycle when it suppressed the IR-induced BRCA1-PALB2 interaction (Fig. S2B). The BRCA1-PALB2 interaction is inhibited by KEAP1-mediated PALB2 ubiquitination in G1 (Orthwein et al., 2015). A PALB2 mutant unable to bind KEAP1 (PALB2-ΔETGE) still interacted with BRCA1 in an IR-induced and ATR-regulated manner (Fig. 2C, S2C) (Ma et al., 2012), showing that ATR and KEAP1 regulate the BRCA1-PALB2 interaction interdependently. These results suggest that ATR promotes the BRCA1-PALB2 interaction during S and G2 phases when the KEAP1 inhibitory mechanism is inactive and HR is active.
Fig. 2. ATR promotes the BRCA1-PALB2 interaction through CDK inhibition and an additional mechanism.
A. Immunoprecipitation of PALB2 from HeLa cells after IR (10 Gy) at the indicated time points. Co-immunoprecipitated BRCA1 was analyzed by Western blot. B. HeLa cells were treated with ATRi or Chk1i prior to IR (10 Gy). PALB2 was immunoprecipitated 2 h after IR, and levels of co-immunoprecipitated BRCA1 and BRCA2 were analyzed by Western blot. C. HeLa cells were treated with ATR inhibitor prior to IR (4 Gy). PALB2-ΔETGE was immunoprecipitated 2 h after IR, and levels of co-immunoprecipitated BRCA1 were analyzed by Western blot. D. HeLa cells were irradiated (10 Gy) and levels of the indicated proteins were analyzed by Western blot at the shown time points. E. HeLa cells were treated with ATRi prior to IR (4 Gy). Endogenous PALB2 was immunoprecipitated and levels of pS/TP were analyzed by Western blot. F. HeLa cells were treated with Wee1 inhibitor prior to IR (10 Gy). PALB2 was immunoprecipitated 2 h after IR, and levels of co-immunoprecipitated BRCA1 were analyzed by Western blot. G-I. U2OS cells were treated with DMSO or ATRi 1 h before IR, and then with DMSO, Roscovitine, or PHA-793887 1 h after IR. Fractions of BRCA1 foci-positive cells displaying >10 PALB2 foci (H) or >5 RAD51 foci (I) were quantified. Error bar: S.D. (n=2 in H, n=3 in I)
CDK inhibition is required to promote the BRCA1-PALB2 interaction
Activation of the ATR-Chk1 pathway inhibits CDK activity. The involvement of this pathway in the BRCA1-PALB2 interaction raised the possibility that CDK inhibition may promote BRCA1-PALB2 binding. To test this idea, we first analyzed if ATRi prevents the inhibition of CDKs during the IR response. In DMSO-treated control cells, CDC25A was gradually degraded after IR as Chk1 became phosphorylated (Fig. 2D). Concomitantly, the inhibitory phosphorylation of CDK2 at Y15 gradually accumulated. Moreover, CDK-mediated phosphorylation of BRCA2 at S3291 declined after IR (Esashi et al., 2005). These results suggest that CDK activity is gradually inhibited during the IR response when the ATR-Chk1 pathway is activated. Importantly, in ATRi-treated cells, Chk1 was not phosphorylated and CDC25A did not decline (Fig. 2D). Furthermore, CDK2 pY15 remained low, and BRCA2 pS3291 stayed high. These results confirm that ATR promotes CDK inhibition during the IR response.
Next, we tested if ATR affects the CDK phosphorylation of PALB2 during the IR response. Using anti-pS/TP antibodies, we detected phosphorylation events on immunoprecipitated endogenous PALB2 (Fig. 2E, S2D–E). The pS/TP signals of PALB2 were inhibited by roscovitine, an inhibitor of CDKs, and enhanced by Wee1 inhibitor (Wee1i), an inducer of CDK1/2 activities (Hughes et al., 2013), confirming that they reflect CDK phosphorylation events (Fig. S2D–E). The CDK phosphorylation of PALB2 was suppressed by IR in an ATR-dependent manner (Fig. 2E). To assess the relevance of CDK inhibition to the enhanced BRCA1-PALB2 interaction after DNA damage, we sought to elevate CDK activity after IR. In cells treated with Wee1i, the IR-induced enhancement of BRCA1-PALB2 interaction was diminished (Fig. 2F), suggesting that CDK inhibition is necessary to promote the BRCA1-PALB2 interaction during the IR response.
Post-resection CDK inhibition is not sufficient to promote PALB2 recruitment
Since CDK activity is required for efficient DNA end resection, we asked if the inhibition of CDKs during the IR response occurs after resection. Consistent with the positive role of CDKs in resection, treatment of cells with CDK inhibitors roscovitine and PHA-793887 prior to IR reduced the phosphorylation of RPA32 and Chk1 (Fig. S2F–G). Furthermore, inhibition of CDKs prior to IR reduced RAD51 foci (Fig. 2G). As indicated by the robust Chk1 phosphorylation 1 hour after IR (Fig. S2F), substantial resection had occurred by this time. Adding ATRi 1 hour after IR did not reduce RAD51 foci as much as pretreating cells with ATRi prior to IR (Fig. 2G). Furthermore, adding CDK inhibitors 1 hour after IR did not affect PALB2 foci (Fig. 2H). Thus, while CDK activity is important for resection and ATR activation, the requirement of CDKs for PALB2 recruitment and HR is alleviated after resection.
The requirement of ATR activity and ATR-mediated CDK inhibition for the DNA damage-induced BRCA1-PALB2 interaction raised the possibility that the sole function of ATR in PALB2 recruitment is to inhibit CDKs. However, PALB2 and RAD51 foci did not form efficiently in cells treated with both ATR and CDK inhibitors after resection (Fig. 2H–I), showing that CDK inhibition cannot bypass the role of ATR in PALB2 recruitment. Therefore, in addition to CDK inhibition, ATR must have another role in promoting PALB2 recruitment and HR.
Phosphorylation of PALB2 during the IR response
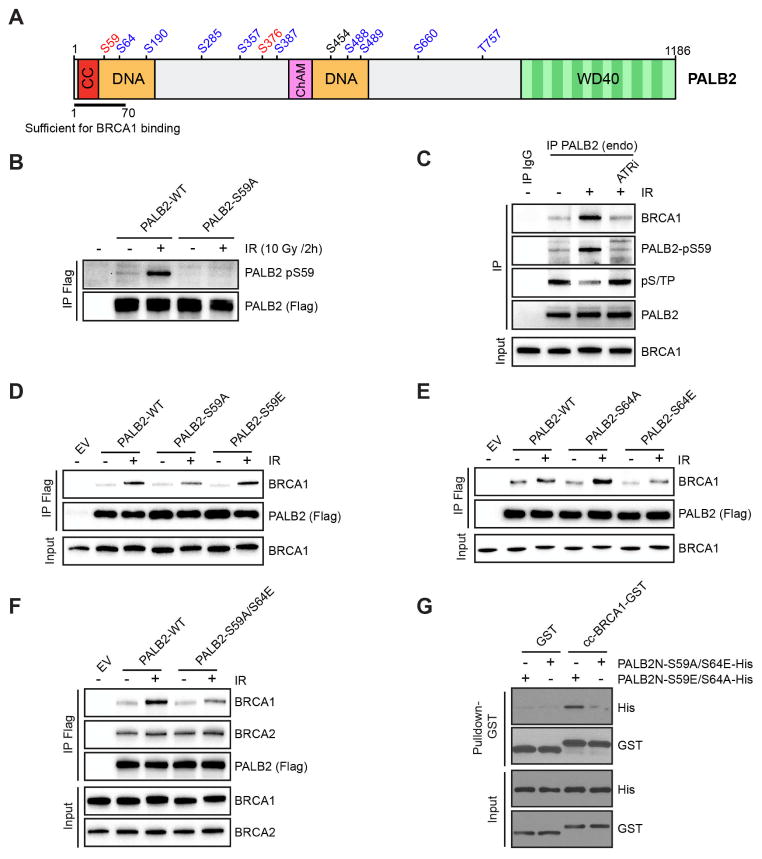
To understand how PALB2 recruitment is regulated by ATR and CDKs, we used tandem-mass spectrometry to identify the phosphorylation sites on endogenous PALB2 before and after IR. We identified a total of 12 phosphorylation sites on PALB2 (Fig. 3A), a subset of which was shown previously (Ahlskog et al., 2016; Guo et al., 2015; Matsuoka et al., 2007). The N-terminal coiled-coil domain of PALB2 interacts with BRCA1 directly (Fig. 3A) (Sy et al., 2009; Zhang et al., 2009). A truncated PALB2 mutant lacking exon 4 is functional for RAD51 recruitment (Xia et al., 2007), suggesting that the first 70 amino acids at the N terminus are sufficient to target PALB2 to sites of DNA damage. Among the sites that we identified, S59 and S64 are located within this small N-terminal region of PALB2 (Fig. 3A). S59 and S64 reside in SQ and SP motifs, respectively, suggesting that they are substrates of ATM/ATR and CDKs.
Fig. 3. PALB2 phosphorylation at S59 and S64 affects BRCA1 binding.
A. A schematic representation of PALB2 and the 12 phosphorylation sites identified by mass spectrometry. S/TQ motifs are shown in red and S/TP motifs are shown in blue. CC : Coil –coiled domain; DNA: DNA binding domain; ChAM: Chromatin-Association Motif; WD40: WD40-repeat-containing domain. B. Flag-tagged PALB2WT or PALB2S59A were transiently expressed in HeLa cells. Flag immunoprecipitation was performed 2 h after IR (10 Gy), and levels of pS59 were analyzed by Western blot. C. HeLa cells were treated with ATRi prior to IR (4 Gy). At 4 h after irradiation, endogenous PALB2 was immunoprecipitated, and levels of pS59, pS/TP, and co-immunoprecipitated BRCA1 were analyzed by Western blot. D-F. Indicated Flag-tagged PALB2 variants were transiently expressed in HeLa cells. Flag immunoprecipitation was performed 2 h after IR, and levels of co-immunoprecipitated BRCA1 or BRCA2 were analyzed by Western blot. G. Purified GST-tagged BRCA1 coiled-coil domain (BRCA1-cc) was incubated with purified His-tagged PALB2N fragments (PALB2N) containing the indicated mutations. The direct interaction between BRCA1-cc and PALB2N was analyzed by GST pulldown.
Next, we generated phospho-specific antibodies against PALB2 pS59 to further characterize this phosphorylation event. The pS59 antibody recognized Flag-PALB2WT but not Flag-PALB2S59A after IR (Fig. 3B). Endogenous PALB2 was also phosphorylated at S59 in an IR-induced manner (Fig. 3C). ATRi reduced the phosphorylation of PALB2 at S59 (Fig. 3C), confirming that it is an ATR-mediated event. Notably, the ATR-mediated phosphorylation of PALB2 at S59 and the CDK-mediated phosphorylation of PALB2 (pS/TP signals) were inversely regulated during the IR response. ATR-mediated S59 phosphorylation was induced by IR, but CDK phosphorylation was reduced (Fig. 3C). Furthermore, while S59 phosphorylation was inhibited by ATRi, CDK phosphorylation was enhanced (Fig. 3C). Our mass spectrometry analysis of endogenous PALB2 revealed that the phosphorylation of S64 was reduced after IR (Fig. S3A–B). Moreover, S64 phosphorylation was reduced by roscovitine and enhanced by Wee1i (Fig. S3C), confirming that it is a CDK-mediated event. Thus, through suppressing CDKs and phosphorylating its own substrates, ATR drives a phosphorylation switch at S59 and S64 on PALB2 during the IR response.
The CDK phosphorylation of PALB2 is inhibited by IR in an ATR-dependent manner (Fig. 3C). Although it is unlikely that the phosphorylation of PALB2 by ATR at S59 affects the ability of ATR to inhibit CDKs, S59 phosphorylation may be influenced by the nearby CDK site S64. To test this possibility, we expressed Flag-tagged PALB2WT, PALB2S64A and PALB2S64E in cells, isolated the proteins before or after IR, and analyzed them using anti-pS59 antibody. S59 phosphorylation was similarly induced by IR in PALB2WT, PALB2S64A, and PALB2S64E (Fig S3D). These results show that S59 phosphorylation occurs independently of the phosphorylation status of S64, suggesting that these two phosphorylation sites of PALB2 are regulated by ATR independently of each other.
Phosphorylation of PALB2 at S59 and S64 influences BRCA1 binding
To understand how the phosphorylation status of PALB2 at S59 and S64 effects the BRCA1-PALB2 interaction, we analyzed a set of PALB2 mutants including PALB2S59A, PALB2S59E, PALB2S64A, and PALB2S64E. In IR-treated cells, the interactions of PALB2S59A and PALB2S64E with BRCA1 were reduced compared to that of PALB2WT, whereas PALB2S64A interacted with BRCA1 more efficiently (Fig. 3D–E, S3E). These results suggest that the phosphorylation of S59 favors the BRCA1-PALB2 interaction, while the phosphorylation of S64 is unfavorable. Consistent with this idea, the PALB2S59A/S64E double mutant interacted with BRCA1 less efficiently than PALB2WT and PALB2S64E (Fig. 3F, S3E). PALB2S59A/S64E was only defective for the interaction with BRCA1 but not BRCA2, showing that S59A and S64E mutations specifically affect the ability of PALB2 to bind BRCA1.
The proximity of S59 and S64 to the coiled-coil domain of PALB2 raised the possibility that the phosphorylation status of these sites may affect the direct interaction with BRCA1. To test this idea, we generated an N-terminal PALB2 fragment (PALB2N) containing the coiled-coil domain as well as S59 and S64 for expression in E. coli. The mutant derivatives of this PALB2 fragment, PALB2NS59E/S64A and PALB2NS59A/S64E, were purified to homogeneity (Fig. S3F). In a direct binding assay, PALB2NS59E/S64A bound to the coiled-coil domain of BRCA1 more efficiently than PALB2NS59A/S64E (Fig. 3G). This result suggests that the phosphorylation status of S59 and S64 indeed affects the direct interaction between the coiled-coil domains of PALB2 and BRCA1.
Phosphorylation of PALB2 at S59 and S64 regulates its localization and function
To test whether the phosphorylation status of S59 and S64 affects PALB2 localization and function in cells, we analyzed cells expressing PALB2WT, PALB2S59A/S64E and PALB2S59E/S64A. In cells microirradiated with laser, YFP-tagged PALB2S59E/S64A was recruited to sites of DNA damage more rapidly and efficiently than PALB2WT (Fig. 4A). In contrast, the recruitment of PALB2S59A/S64E was delayed and attenuated compared to PALB2WT (Fig. 4A). These results lend further support to the notion that the combination of S59 phosphorylation and S64 hypo-phosphorylation is necessary for the optimal accumulation of PALB2 at DSBs.
Fig. 4. The S59/S64 phosphorylation switch on PALB2 regulates its localization and HR function.
A. Localization of YFP-PALB2 variants to laser-induced DNA damage sites was analyzed by time-lapse. The fluorescence intensity of YFP-PALB2 at DNA damage sites was quantified as in Fig. 1G. Error bar: S.E. (n > 50). B. Indicated Flag-tagged PALB2 variants were transiently expressed in HeLa cells. Cells were treated DMSO or ATRi, irradiated with IR, and Flag immunoprecipitation was performed 2 h after IR. Levels of co-immunoprecipitated BRCA1 were analyzed by Western blot. C. Representative images of GFP-PALB2WT or GFP-PALB2S59E/S64A foci in DMSO or ATRi-treated cells 2 h after IR. D. The intensity of PALB2 staining in BRCA1 foci was determined in the indicated cell populations. Cells expressing GFP-tagged PALB2WT, PALB2S59E/S64A, or PALB2S59A/S64E were treated with DMSO or ATRi as shown. ***, P< 0.001 and ****, P< 0.0001. E. Localization of YFP-PALB2 variants to laser-induced DNA damage sites in the presence of ATRi (1 μM VE-821). Error bar: S.E. (n > 50). F-G. Flag-tagged PALB2 variants were stably integrated in U2OS cells under the control of a Tet-on promoter. Cells were treated with PALB2 siRNA to deplete endogenous PALB2, and with Doxycycline to induce the expression of the PALB2 variants. Gene-targeting efficiency were quantified (mClover positive cells) after the indicated treatments. *, P<0.05; **, P< 0.01 and ****, P< 0.0001.
If the role of ATR in PALB2 recruitment were to promote S59 phosphorylation and suppress S64 phosphorylation, one would expect that this function of ATR could be bypassed by S59E and S64A mutations. Indeed, ATRi reduced IR-induced interaction of BRCA1 with PALB2S64A, but not PALB2S59E/S64A (Fig. 4B). PALB2S59E also partially bypassed the requirement of ATR for IR-induced BRCA1 binding, although the bypass was less complete compared to that of PALB2S59E/S64A (Fig. S4A). To further test if PALB2S59E/S64A bypasses ATR, we compared the recruitment of PALB2WT, PALB2S59E/S64A, and PALB2S59A/S64E to IR-induced BRCA1 foci in the absence or presence of ATRi (Fig. 4C–D). ATRi significantly diminished the PALB2WT signals in BRCA1 foci (Fig. 4C, 4D lanes 1–2), but only modestly reduced the PALB2S59E/S64A signals (Fig. 4C, 4D lanes 3–4), suggesting that PALB2S59E/S64A partially bypasses ATR for localization to sites of DNA damage. Compared to PALB2WT and PALB2S59E/S64A, PALB2S59A/S64E displayed a defect in localization to BRCA1 foci even in the absence of ATRi (Fig. 4D lanes 1, 3, 5). Nonetheless, ATRi further reduced the PALB2S59A/S64E signals in BRCA1 foci (Fig. 4D lanes 5–6), suggesting that ATR has additional roles in PALB2 recruitment independent of the phosphorylation states of S59 and S64.
In addition to analyzing PALB2 foci, we used time-lapses to compare the recruitment of YFP-tagged PALB2WT, PALB2S59E/S64A, and PALB2S59A/S64A to laser-induced DNA damage (Fig. 4E). Even in the presence of ATRi, PALB2S59E/S64A accumulated at sites of DNA damage more efficiently than PALB2WT (Fig. 4E). In contrast, PALB2S59A/S64E was recruited less efficiently than PALB2WT. Notably, the recruitment of all three PALB2 proteins was reduced by ATRi (Fig. 4A, 4E), suggesting that ATR has additional functions in PALB2 recruitment independent of the phosphorylation switch at S59 and S64. Nonetheless, PALB2S59E/S64A indeed bypasses, although partially, the function of ATR in PALB2 recruitment.
Finally, to determinate if the phosphorylation of PALB2 at S59 and S64 regulates its HR function, we generated stable cell lines that inducibly express PALB2WT, PALB2S59A/S64E, or PALB2S59E/S64A at levels slightly higher than endogenous PALB2 (Fig. S4B). All the PALB2 variants were expressed at the same levels and localized to the nucleus efficiently (Fig. S4B–C). To measure the HR function of the PALB2 variants, we preformed Cas9/mClover assays after depletion of endogenous PALB2 (Fig. S4D) (Pinder et al., 2015). The Cas9/mClover assay measures the HR-dependent insertion of a mClover-containing cassette into Cas9-generated DSBs in the LMNA gene, which results in green lamin A/C (Fig. S4D–E). Using the Cas9/mClover assay, we confirmed that ATR is required for efficient HR (Fig. S4F). PALB2WT efficiently restored HR in cells lacking endogenous PALB2 (Fig. 4F). PALB2S59A/S64E, which is compromised for BRCA1 binding, was defective for HR restoration. In contrast, PALB2S59E/S64A, which binds BRCA1 more efficiently than PALB2WT, elevated HR to a higher level than PALB2WT did (Fig. 4G). Even after ATRi treatments, HR efficiency was still higher in cells expressing PALB2S59E/S64A than in cells expressing PALB2WT (Fig. 4G), supporting the idea that S59E and S64A mutations of PALB2 partially bypass ATR. Together, these results show that ATR promotes HR in part by driving the S59–S64 phosphorylation switch on PALB2, highlighting the importance of the CDK-to-ATR switch in HR regulation.
Discussion
The relationship of checkpoint and DNA repair
In the context of DNA damage and replication stress responses, the term “checkpoint” is often used to describe pathways that arrest or slow the cell cycle when genomic stability is challenged (Elledge, 1996). The DNA damage-signaling pathways mediated by ATM and ATR are both critical for checkpoint responses. Checkpoints are important for preventing cells carrying DNA damage from going through the cell cycle, thereby reducing the chance of passing incorrect genetic information to daughter cells. In addition, it was proposed that checkpoint-mediated cell cycle arrest provides time for DNA repair, allowing cells to fix or overcome genomic problems before entering the next cell-cycle phase. The inhibition of CDKs by checkpoints is important for the functions of these pathways in G1, S, and G2 phases. Although the role of checkpoints in cell-cycle control has been long appreciated, whether they function directly in DNA repair is not known. In this study, we find evidence that ATR-mediated CDK inhibition directly contributes to regulation of PALB2, a key HR protein. This finding reveals a surprising link between the checkpoint response and DNA repair. While our current results are limited to the ATR pathway and HR, it is tempting to speculate that checkpoints function broadly in various DNA repair processes. It is conceivable that the inhibition of CDKs by checkpoints coordinates DNA repair and cell cycle progression, giving cells the best chance to resolve DNA lesions or other genomic problems before they damage the cells irreversibly.
CDK and two critical phases of HR
HR primarily occurs in S and G2 phases of the cell cycle, a period when CDK2 and CDK1 are active. Nonetheless, how CDK activity influences HR is not completely understood. On one hand, CDK activity is clearly required for DNA end resection, one of the initial events in HR (Ferretti et al., 2013; Huertas et al., 2008; Huertas and Jackson, 2009; Wang et al., 2013). On the other hand, some CDK-mediated phosphorylation events are inhibitory to proteins involved in HR (Davies and Pellegrini, 2007; Esashi et al., 2005; Luo et al., 2015; Yata and Esashi, 2009). The results from this study suggest that CDKs play both positive and negative roles in HR. During the initial phase of HR, CDK activity promotes DNA end resection and activation of the ATR-Chk1 pathway (Fig. S4G). As the ATR pathway is gradually activated, it functions to inhibit CDKs, driving HR into the second phase. During the second phase of HR, CDK-mediated phosphorylation of HR proteins gradually declines, allowing these proteins to interact with their functional partners more efficiently. The opposite effects of CDK activity in these two consecutive phases of HR may be important for keeping specific molecular events in order, preventing formation of inappropriate intermediates before DNA ends are properly processed. In addition, the requirement of both high- and low-CDK phases for HR may prevent the initiation of HR in G1, when CDK1/2 activities are low. Importantly, the model of two-phase HR elucidates how HR is intertwined with checkpoint responses, explaining how this CDK-driven DNA repair pathway is compatible with checkpoint-mediated CDK inhibition.
The roles of ATR in HR
Our results not only shed light on the process of HR but also on the role of ATR in this pathway. The inhibition of CDKs by ATR is required for efficient BRCA1-PALB2 interaction and possibly other HR events. A recent study suggested that CDKs are recruited to DSBs to stimulate resection (Chen et al., 2016), which may create a dependency on ATR to prevent excessive CDK phosphorylation of HR proteins after resection. The dephosphorylation of various HR proteins at CDK sites might take place at different rates, depending on activities of their respective phosphatases. Regardless of the dephosphorylation rates of various HR proteins, the inhibition of CDKs by ATR likely prevents CDK-mediated phosphorylation and maintains a pool of HR proteins that is hypo-phosphorylated at CDK sites. The presence of a pool of CDK-hypo-phosphorylated HR proteins at DNA damage sites may be critical for the second phase of HR. In addition to CDK inhibition, ATR also promotes HR by phosphorylating its substrates in the HR pathway. Furthermore, some of the functions of ATR in HR may be mediated by its effector kinase Chk1. In the context of PALB2 regulation, the phosphorylation of PALB2 by ATR plays a positive role in PALB2 recruitment.
While the phosphorylation status of S59 and S64 of PALB2 is important for optimal PALB2 recruitment and may explain at least part of ATR’s function in HR, the regulation of PALB2 is probably more complex. Notably, the recruitment of PALB2S59E/S64A still requires ATR (Fig. 4G). ATR may have additional roles in PALB2 regulation. A recent study suggested that the phosphorylation of PALB2 at S157 and S376, two putative ATM/ATR sites, is dispensable for HR but required for the recovery from the DNA damage response (Guo et al., 2015). Another study proposed that the phosphorylation of PALB2 by ATM/ATR at S59, S157, and S376 promotes HR by enhancing the interaction between PALB2 and RAD51 (Ahlskog et al., 2016). However, we did not observe a reduction in the PALB2-RAD51 or PALB2-BRCA2 interaction after inhibition of ATR and Chk1 (Fig. 2B). Although ATR has an execution point between BRCA1 and PALB2, it may have additional functions downstream of PALB2. The phosphorylation of XRCC3 by ATM/ATR and the phosphorylation of RAD51 by Chk1 have been suggested to promote RAD51 recruitment in specific contexts (Somyajit et al., 2013; Sorensen et al., 2005). Our study suggests that efficient HR not only requires ATR-mediated phosphorylation of ATR/Chk1 substrates, but also ATR-dependent CDK inhibition. Future studies investigating different types of ATR-regulated phosphorylation events in HR proteins will further elucidate the role of ATR in this critical DNA repair pathway.
Supplementary Material
Highlights.
ATR acts after DNA end resection to promote HR.
The CDK-to-ATR switch after resection is important for efficient HR.
ATR promotes the BRCA1-PALB2 interaction and PALB2 localization to DSBs.
ATR regulates PALB2 by driving a phosphorylation switch on S59 and S64.
Acknowledgments
We thank the members of Zou and Dyson labs for helpful discussions. R.B. is supported by a Marsha Rivkin Scholar Award and a Susan G. Komen Fellowship. L.Z. is the James and Patricia Poitras Endowed Chair in Cancer Research, and supported by a Jim & Ann Orr Massachusetts General Hospital Research Scholar Award. J.Y.M. is a FRQS Chercheur National and FRQS Chair. This work is supported by grants from the NIH (GM076388 and CA197779 to L.Z., CA138804 and CA188096 to B.X.), Federal Share of Program Income (to L.Z.), and CIHR (to J.Y.M.).
Footnotes
Author contributions
R.B., C.K.H. and L.Z. designed the study. R.B., N.J., A.R., C.K.H., J.K., T.K.F., E.J.-L.H. performed the experiments. W.H., B.X., J.-Y.M., and L.Z. supervised the experiments. G.D. provided critical reagents. R.B. and L.Z. wrote the manuscript with help from other authors.
Methods
Methods and materials are described in Supplemental Information.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlskog JK, Larsen BD, Achanta K, Sorensen CS. ATM/ATR-mediated phosphorylation of PALB2 promotes RAD51 function. EMBO Rep. 2016;17:671–681. doi: 10.15252/embr.201541455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, Stasiak A, Xia B, Masson JY. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson R, Masson JY. PALB2 self-interaction controls homologous recombination. Nucleic Acids Res. 2012;40:10312–10323. doi: 10.1093/nar/gks807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- Chen X, Niu H, Yu Y, Wang J, Zhu S, Zhou J, Papusha A, Cui D, Pan X, Kwon Y, et al. Enrichment of Cdk1-cyclins at DNA double-strand breaks stimulates Fun30 phosphorylation and DNA end resection. Nucleic Acids Res. 2016;44:2742–2753. doi: 10.1093/nar/gkv1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- Ferretti LP, Lafranchi L, Sartori AA. Controlling DNA-end resection: a new task for CDKs. Front Genet. 2013;4:99. doi: 10.3389/fgene.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokas E, Prevo R, Pollard JR, Reaper PM, Charlton PA, Cornelissen B, Vallis KA, Hammond EM, Olcina MM, Gillies McKenna W, et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis. 2012;3:e441. doi: 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote KM, Blades K, Cronin A, Fillery S, Guichard SS, Hassall L, Hickson I, Jacq X, Jewsbury PJ, McGuire TM, et al. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-y l}-1H-indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J Med Chem. 2013;56:2125–2138. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- Guo Y, Feng W, Sy SM, Huen MS. ATM-dependent Phosphorylation of the Fanconi Anemia Protein PALB2 Promotes the DNA Damage Response. J Biol Chem. 2015;290:27545–27556. doi: 10.1074/jbc.M115.672626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes BT, Sidorova J, Swanger J, Monnat RJ, Jr, Clurman BE. Essential role for Cdk2 inhibitory phosphorylation during replication stress revealed by a human Cdk2 knockin mutation. Proc Natl Acad Sci U S A. 2013;110:8954–8959. doi: 10.1073/pnas.1302927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, Harper JW. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Deng M, Li Y, Wu C, Xu Z, Yuan J, Lou Z. CDK-mediated RNF4 phosphorylation regulates homologous recombination in S-phase. Nucleic Acids Res. 2015;43:5465–5475. doi: 10.1093/nar/gkv434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Cai H, Wu T, Sobhian B, Huo Y, Alcivar A, Mehta M, Cheung KL, Ganesan S, Kong AN, et al. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol Cell Biol. 2012;32:1506–1517. doi: 10.1128/MCB.06271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- Pinder J, Salsman J, Dellaire G. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015;43:9379–9392. doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevo R, Fokas E, Reaper PM, Charlton PA, Pollard JR, McKenna WG, Muschel RJ, Brunner TB. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther. 2012;13:1072–1081. doi: 10.4161/cbt.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, Golec JM, Pollard JR. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somyajit K, Basavaraju S, Scully R, Nagaraju G. ATM- and ATR-mediated phosphorylation of XRCC3 regulates DNA double-strand break-induced checkpoint activation and repair. Mol Cell Biol. 2013;33:1830–1844. doi: 10.1128/MCB.01521-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shi LZ, Wong CC, Han X, Hwang PY, Truong LN, Zhu Q, Shao Z, Chen DJ, Berns MW, et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013;9:e1003277. doi: 10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004;64:7139–7143. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- Yata K, Esashi F. Dual role of CDKs in DNA repair: to be, or not to be. DNA Repair (Amst) 2009;8:6–18. doi: 10.1016/j.dnarep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.