Summary
Hyperpolarization-activated cyclic nucleotide-gated (HCN) channels underlie the control of rhythmic activity in cardiac and neuronal pacemaker cells. In HCN, the polarity of voltage dependence is uniquely reversed. Intracellular cAMP levels tune the voltage response, enabling sympathetic nerve stimulation to increase the heart rate. We present cryo-electron microscopy structures of the human HCN channel in the absence and presence of cAMP at 3.5 Å resolution. HCN channels contain a K+ channel selectivity filter-forming sequence from which the amino acids create a unique structure that explains Na+ and K+ permeability. The voltage sensor adopts a depolarized conformation and the pore is closed. An S4 helix of unprecedented length extends into the cytoplasm, contacts the C-linker and twists the inner helical gate shut. cAMP binding rotates cytoplasmic domains to favor opening of the inner helical gate. These structures advance understanding of ion selectivity, reversed polarity gating and cAMP regulation in HCN channels.
eTOC
Cryo-EM structures of the human HCN1 hyperpolarization-activated ion channel provides mechanistic insights on voltage-dependent gating with reversed polarity.
Introduction
Rhythmic cycles, such as spontaneous and repetitive firing patterns in excitable cells, are essential timing mechanisms that regulate various biological processes. Not unexpectedly, the pacemaker current (also known as funny current If or hyperpolarization-activated current Ih) was first described in the heart (Brown et al., 1979), the most reliable rhythmic organ of the body. Later also characterized in nerves (Mayer and Westbrook, 1983), this current was found to be mediated by hyperpolarization-activated cyclic nucleotide-gated (HCN) ion channels (Gauss et al., 1998; Ludwig et al., 1998; Santoro et al., 1998). HCN channels contribute to cardiac and neuronal pacemaker activity. In sinoatrial node cells, these channels account for the initial phase of the action potential and hence regulate the firing rate. HCN channels are modulated by intracellular cyclic nucleotides (DiFrancesco and Tortora, 1991), including cyclic adenosine monophosphate (cAMP). The activity of channels is augmented in the presence of cAMP and such facilitation is crucial to accelerating the heart rate under sympathetic stimulation (Brown et al., 1979; DiFrancesco and Tortora, 1991). In the nervous system, widely expressed HCN channels are involved in neuronal excitability and network activity (Benarroch, 2013). Given their multiple roles, dysfunction of HCN channels has been implicated in a spectrum of diseases (DiFrancesco and DiFrancesco, 2015; Verkerk and Wilders, 2015), for instance arrhythmias, epilepsies, and neuropathic pain, making HCN channels attractive new targets for therapeutic exploration (Postea and Biel, 2011; Tibbs et al., 2016). One HCN channel blocker, ivabradine, was recently approved for clinical use to treat chronic heart failure (Tibbs et al., 2016).
HCN channels belong to the superfamily of six-transmembrane segment channels, and are related to cyclic nucleotide-gated (CNG) channels and voltage-dependent K+ (Kv) channels Kv10 to Kv12. In addition to their physiological significance, intriguing biophysical properties of HCN channels motivate us to observe their molecular architecture. First, the HCN channel, despite having a selectivity filter sequence that contains all of the essential amino acids required for K+ selectivity, allows both K+ and Na+ to pass through its pore, yielding a reversal potential of −20 to −30 mV under physiological conditions (Gauss et al., 1998; Ludwig et al., 1998; Santoro et al., 1998). Thus, when the HCN channel opens cations flow into the cell and cause depolarization, in contrast to K+ channels whose opening generally repolarizes the cell. Second, the polarity of voltage-dependent gating is reversed in HCN channels compared to most other voltage-dependent ion channels: depolarization causes closure and hyperpolarization causes opening. Finally, the open probability of HCN can be increased by cAMP, but unlike CNG channels, cyclic nucleotides are not a prerequisite for channel opening. To understand the molecular basis of ion selectivity, reversed polarity gating and cAMP modulation, we determined structures of the human HCN channel in the ligand-free and cAMP-bound states using single- particle cryo-electron microscopy (cryo-EM).
Results and Discussion
HCN1 channel architecture
We determined structures of the human HCN1 channel expressed without its C-terminus to improve its stability in detergent (Figure S1). The truncated channel, HCN1EM, is activated by membrane hyperpolarization, is cation permeable like the full-length channel, and is sensitive to cAMP (Figure S2A–D).
Reconstructions of HCN1EM in the ligand-free and cAMP-bound states were carried out to an overall resolution of 3.5 Å (Figures S3 and S4; Table S1). Nearly complete atomic models for the structured core of the protein were built (Figures S1 and S5). Density for N-terminal residues 1 to 93 was not visible because these residues are most likely unstructured and disordered (nearly 30% are glycines). The refined models correlate well with the density maps and have good stereochemistry (Table S1). While significant conformational changes were observed in the cytoplasmic domains, the ligand-free and cAMP-bound structures are similar in the transmembrane domains (root mean square deviation (RMSD) of Cα = 0.43 Å). We will describe the ligand-free structure first and then discuss conformational changes induced by cAMP binding.
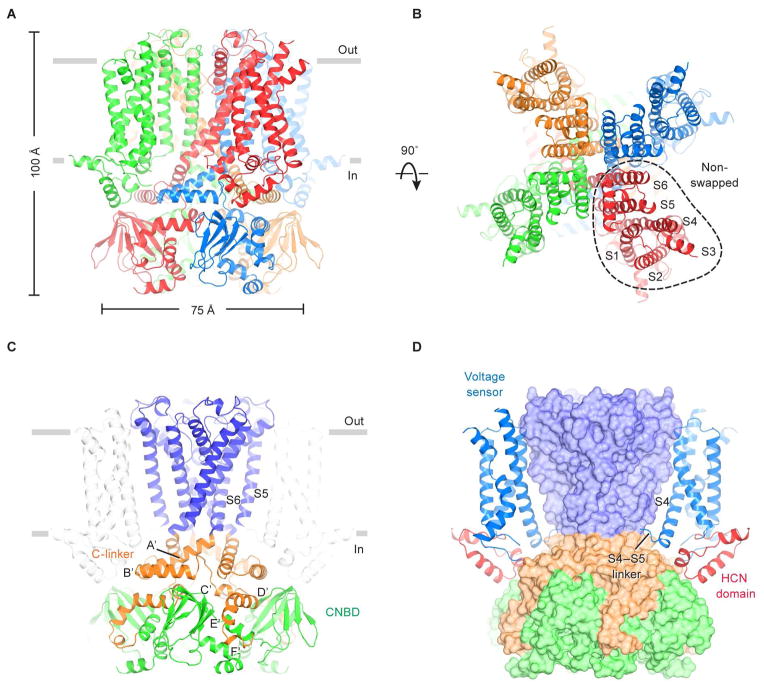
The HCN channel is ~100 Å in length perpendicular to the membrane and ~75 Å in width when viewed from within the plane of the membrane (Figure 1A and 1B). Four identical subunits form a four-fold symmetric pore (helices S5 through S6), which is surrounded by membrane-embedded voltage sensors (helices S1 through S4), as in Kv channels. The voltage sensors in HCN1 are “non-swapped”, which means each voltage sensor contacts the pore through amino acids from the same polypeptide chain (i.e. same subunit), as depicted (Figure 1B). In contrast, based on known molecular structures and a structure-based analysis of Kv channel amino acid sequences (Whicher and MacKinnon, 2016), in Kv1–9 and voltage-dependent Na+ and Ca2+ channels the voltage sensors interact with an adjacent subunit (i.e. they are domain-swapped). The domain-swapped voltage sensors permit a long α-helical S4–S5 linker, which likely functions as a mechanical lever through which voltage-sensor conformational changes impart force to open or close the pore’s gate (Long et al., 2005, 2007). In HCN1 the S4–S5 linker is much shorter and not α-helical (Figure 1D). In the absence of an α-helical lever, non-swapped voltage sensors must apparently work in a different manner to regulate the gate.
Figure 1. Architecture of the human HCN1 channel.
(A and B) Structure of the channel tetramer in the ligand-free state, viewed parallel to the membrane (A) or from the extracellular side (B). Each subunit is shown in a different color. Gray bars represent approximate boundaries of the membrane bilayer.
(C and D) Domain organization of the HCN channel. In (C), the pore domain (helices S5 and S6) is colored blue, the C-linker disk (A′- and B′-helices) and four following helices (C′- to F′-helices) are colored orange, and the CNBD is colored green. In (D), the voltage sensors and the HCN domains are shown as ribbons, whereas the pore and C-terminal domains are in surface representation. The voltage sensor from the subunit nearest to the viewer is removed for clarity.
See also Figures S1, S2, S3 and S5.
The pore-lining S6 inner helices form a right-handed, tightly packed bundle within the membrane’s inner leaflet (Figure 1C). At the level of the intracellular membrane-water interface the S6 helices make a sharp bend and give rise to a helix-turn-helix ‘C-linker’, which forms an α-helical disk just below the membrane. C-terminal to the C-linker, the polypeptide chain then gives rise to five additional short α-helices (C′- to F′- and A-helices) and a β-jelly roll that is characteristic of cyclic nucleotide binding domains (CNBDs) (Figures 1C and S1) (Lolicato et al., 2011; Zagotta et al., 2003). Four CNBDs – one from each subunit – are docked onto the cytoplasmic face of the C-linker disk (Figure 1C). As we describe later, cAMP binding to the CNBD induces conformational changes to affect gating.
The HCN channel exhibits an additional unique feature that may also contribute to this channel’s very unusual gating. The 45 amino acids preceding S1 form a 3 α-helical domain that is wedged between the voltage sensor and the cytoplasmic domains (Figure 1D). As far as we can tell this domain is unique to HCN channels and therefore we call it the HCN domain. It contacts the S4 helix (near the short S4–S5 linker) from the same subunit, and the C-linker and CNBD from an adjacent subunit. In the context of the tetramer, the HCN domains appear like pincers that constrict the channel at the level of the C-linker disk.
The selectivity filter
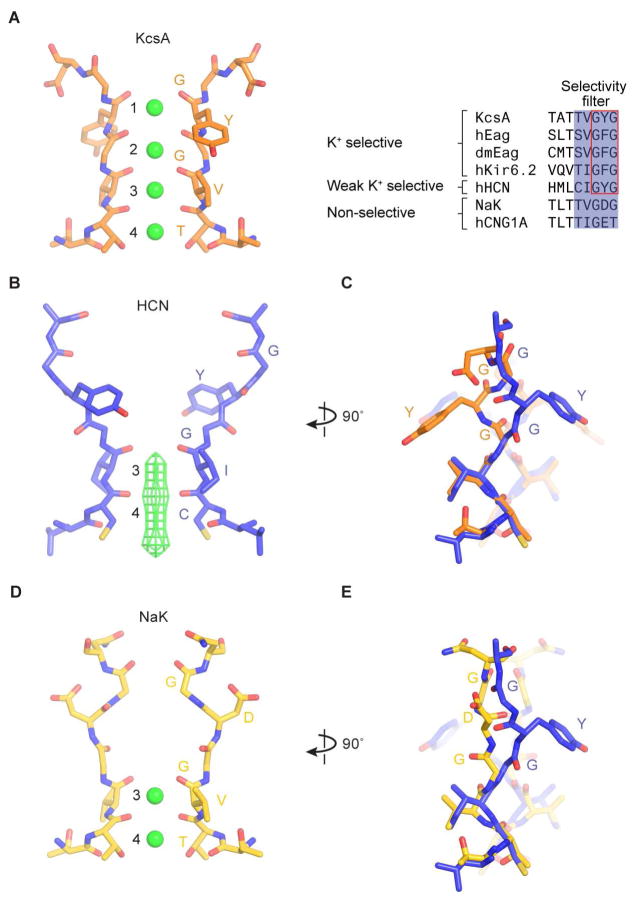
HCN channels are non-selective monovalent cation channels, permitting K+ over Na+ by a ratio ~ 4:1 (Gauss et al., 1998; Ludwig et al., 1998; Santoro et al., 1998), not very different from NaK and hCNG1A channels, which permit K+ over Na+ by a ratio of 1:1 (Derebe et al., 2011; Kaupp et al., 1989). In comparison, K+ channels select K+ over Na+ by a ratio greater than 1000:1 (Neyton and Miller, 1988; Yellen, 1984). This difference in selectivity is especially notable when one considers that HCN channels actually contain the essential amino acids that are required to form the K+ selectivity filter. In the K+ channel, protein atoms from the sequence (T/S)XG(Y/F)G directly coordinate K+ ions (Figure 2A), and furthermore, the first residue (T/S) is mutable to C (the residue present in HCN) without alteration of selectivity (Zhou and MacKinnon, 2004). Therefore we ask, why is this close relative of the K+ channel non-selective? This deviation from K+ selectivity is key to its function as a membrane-depolarizing channel.
Figure 2. Selectivity filter of the HCN1 channel.
(A) Structure of the KcsA filter, a K+-selective filter (PDB ID: 1K4C). Discrete K+ binding sites (1 to 4) are labeled. K+ ions within the filter are represented as green spheres. A sequence alignment of the filter regions from selected channels is shown on the right.
(B) Structure of the HCN1 filter, a weak K+-selective filter. The density of K+ ions is represented as green mesh. The map is sharpened with a b-factor of −120 Å2, and contoured at 5.5 σ.
(C) Comparison of the HCN1 and KcsA filters. The superposition of the HCN (blue) and KcsA filters (orange) is based on Cα atoms of residues 357 to 359 of the HCN channel. K+ ions in the KcsA channel are removed for clarity.
(D) Structure of the NaK filter, a non-selective filter (PDB ID: 2AHZ).
(E) Comparison of the HCN1 and NaK filters.
See also Figure S6.
Potassium selectivity comes from the precise geometry of the selectivity filter, in which K+ ions are coordinated by protein oxygen atoms (Figure 2A). Four ion binding sites, 1 through 4, exist within the filter (Figure 2A) (Zhou et al., 2001). When selectivity filters from distantly related K+ channels are compared, deviations in structure are so small that estimates are limited by measurement error, which in the best case is less than 0.2 Å, underscoring the importance of the K+ filter’s specific geometry (Nishida et al., 2007). In contrast, the selectivity filter in HCN is strikingly different than in the K+ channel (Figures 2B and S6A), with an RMSD of 2.9 Å for Cα atoms. In HCN, the Tyr side chain in the GYG sequence is reoriented almost 180 degrees compared to the Tyr in the K+ selectivity filter (Figure 2C). The main chain carbonyl oxygen atoms, which ordinarily would form sites 1 and 2, are no longer directed toward the ion pathway in HCN (Figure 2B and 2C). Consequently, HCN preserves only K+ binding sites 3 and 4. The outer half of the selectivity filter in HCN is dilated and sites 1 and 2 are absent.
The conformational difference between HCN and the K+ selectivity filter is likely due to changes in surrounding amino acids that interact with the filter. For example, the Tyr side chain in the GYG sequence in many K+ channels interacts with specific amino acids on the pore helix (i.e. Trp68 and Thr72 in the KcsA K+ channel) that form hydrogen bonds with the Tyr hydroxyl (Figure S6B) (Doyle et al., 1998; Shealy et al., 2003; Zhou et al., 2001). In HCN, the Trp and Thr on the pore helix are replaced by Lys and His (Figures S6B), respectively, thus offering a possible explanation for reorientation of the filter Tyr.
The observation that only sites 3 and 4 are maintained in HCN explains its non-selectivity. The multi ion nature of the K+ selectivity filter (four sites allow 2 K+ ions to bind simultaneously) is essential to K+ selectivity for the following reason. A filter in which only a single ion can bind, even if the binding is thermodynamically favorable for K+ over Na+, will not exhibit strong kinetic selectivity because an entering Na+ ion, which will only reside very briefly, will rapidly exit to either side (i.e. it can permeate). Multi K+ ion occupancy will increase the probability that a Na+ will exit from the same side it entered (because there is a K+ ion blocking the other side) and thus permit kinetic selectivity. This behavior has been demonstrated through elegant experiments in the NaK channel (Derebe et al., 2011; Lockless, 2015; Shi et al., 2006), which also has only sites 3 and 4 in its selectivity filter (Figure 2D). The structure of the NaK filter is clearly different than the filter in HCN (Figure 2D and 2E), but the two channels have in common only two monovalent cation binding sites.
Voltage sensor control of the pore
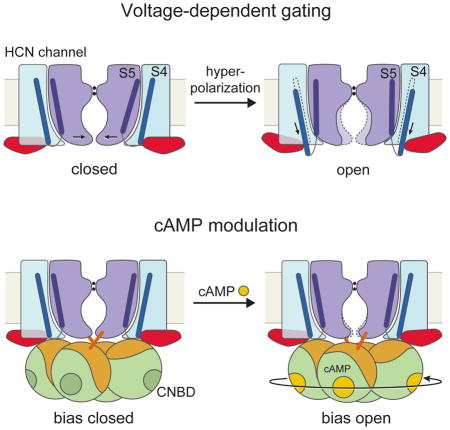
The other intriguing question is HCN’s reversed polarity of voltage-dependence, the structural basis of which has never been observed. Most voltage-dependent ion channels open when the membrane is depolarized. HCN channels respond to voltage in the opposite manner, closing with depolarization and opening with hyperpolarization (Gauss et al., 1998; Ludwig et al., 1998; Santoro et al., 1998). But puzzlingly, in most obvious respects the amino acid sequence of HCN channels would seem to be similar to other voltage-dependent K+ channels. What can the structures tell us about voltage dependence with reversed polarity?
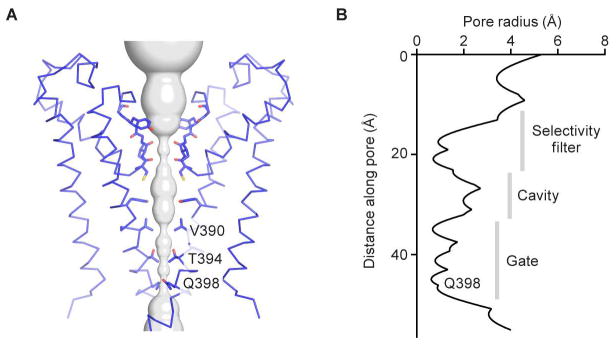
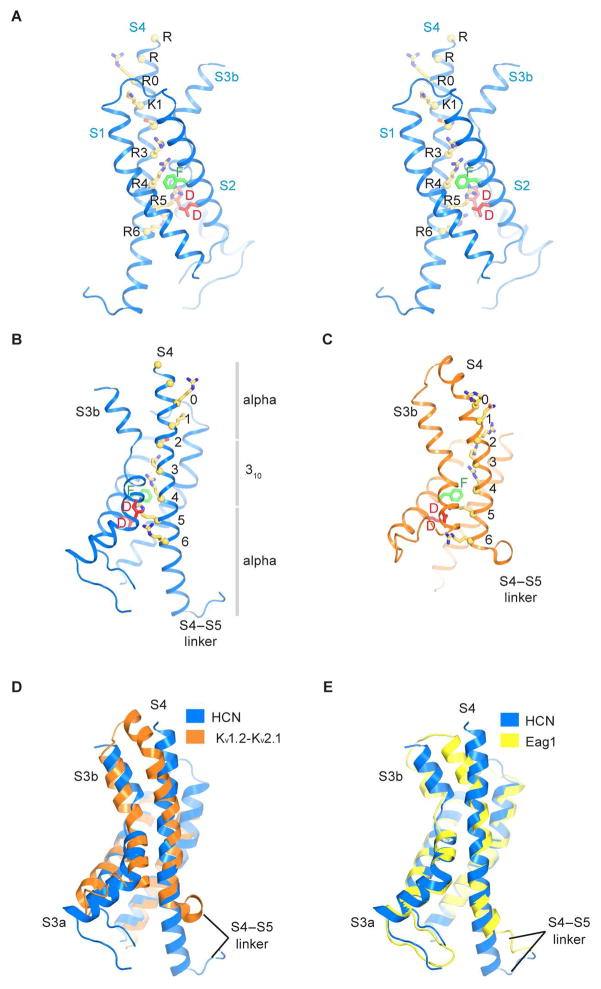
The gate of HCN1 is closed by a tightly packed inner helical bundle that constricts the pore to a radius of about 1 Å at amino acid positions Val390, Thr394 and Gln398 (Figure 3A and 3B). In fact the inner helices are so tightly packed that the volume of the central cavity in the HCN channel is much smaller than in the KcsA channel (Doyle et al., 1998). Concomitantly, the voltage sensors are in a depolarized conformation (Figure 4A and 4B). This conclusion is based on a direct comparison of the HCN1 voltage sensor with that of the Kv1.2–2.1 paddle chimera (Figure 4C–D), whose voltage sensor is known to be in a depolarized conformation (Long et al., 2007; Tao et al., 2010). S4 helix positions labeled R0 through R4 are located on the extracellular side of the gating charge transfer center (Figure 4B, green and red residues; Figure S6C), as in the depolarized Kv1.2–2.1 voltage sensor. A depolarized voltage sensor conformation together with a closed pore corresponds to the expected physiological state of an HCN channel at 0 mV (Figure S2). How do these voltage sensor and pore conformations fit together into a coherent mechanistic scheme and how might the voltage sensor allow the pore to open when the membrane is hyperpolarized?
Figure 3. Pore of the HCN1 channel.
(A) Ion pore with only two subunits shown, viewed from within the membrane. The minimal radial distance from the center axis to the protein surface is colored in gray. Selected residues facing the pore are in stick representation and constricting residues are labeled.
(B) Radius of the pore. The van der Waals radius is plotted against the distance along the pore axis.
Figure 4. Voltage sensor of the HCN1 channel.
(A) Stereo view of the voltage sensor. Yellow spheres denote Cα positions of Arg252, Arg255, Arg258 (R0), Lys261 (K1), Ser264, Arg267 (R3), Arg270 (R4), Arg273 (R5) and Arg276 (R6). Resides that form the gating charge transfer center are labeled (Phe186, green; Asp189 and Asp225, red).
(B and C) Comparison of the HCN1 and Kv1.2–2.1 voltage sensors (PDB ID: 2R9R). The gray bars in (B) show the extents of α- versus 310-helical transitions in the S4 helix of the HCN1 channel.
(D) Superposition of the HCN1 and Kv1.2–2.1 voltage sensors using Cα atoms of S1 and S2 helices. The S1–S2 loop of Kv1.2-Kv2.1 is not shown for clarity.
(E) Superposition of the HCN1 and Eag1 voltage sensors (PDB ID: 5K7L), using Cα atoms of S1 to S3 helices.
See also Figure S6.
The unique feature of the HCN1 voltage sensor, which we believe accounts for its reversed polarity, is the extraordinary length of its S4 helix (Figure 4A). The S4 helix is the structural element that contains positive charged amino acids for sensing the membrane electric field (Aggarwal and MacKinnon, 1996; Seoh et al., 1996). Compared with other voltage-dependent channels, the S4 helix of HCN1 contains two additional helical turns on the cytoplasmic side (Figure 4D and 4E). Consequently, it extends further than the S1–S3 helices, beyond the membrane surface into the cytoplasm. The extension of S4 in HCN1 brings the S4–S5 linker into contact with the C-linker of a neighboring subunit (Figure 5A). Force exerted by S4 in this position onto the C-linker has the correct orientation to twist the C-linker disk in a direction that will wrap the right-handed helical bundle formed by the S6 helices, which form the pore’s gate, closed. This contact can be made in the depolarized conformation of the voltage sensor only because the S4 is so long. We propose that this is one of the features of the HCN channel that allows it to stabilize a closed pore when the voltage sensor is depolarized. In keeping with this hypothesis, in the Eag1 (Kv10.1) channel, which is related to HCN but opens with depolarization, the S4, because it is shorter (Figure 4E), does not extend into the cytoplasm and does not contact the C-linker in its depolarized conformation (Whicher and MacKinnon, 2016). In fact, we suggest that in Eag1 when the membrane is hyperpolarized, its S4–S5 linker will move into the cytoplasm to make contacts with the C-linker that are similar to those observed here in HCN1 in the depolarized conformation.
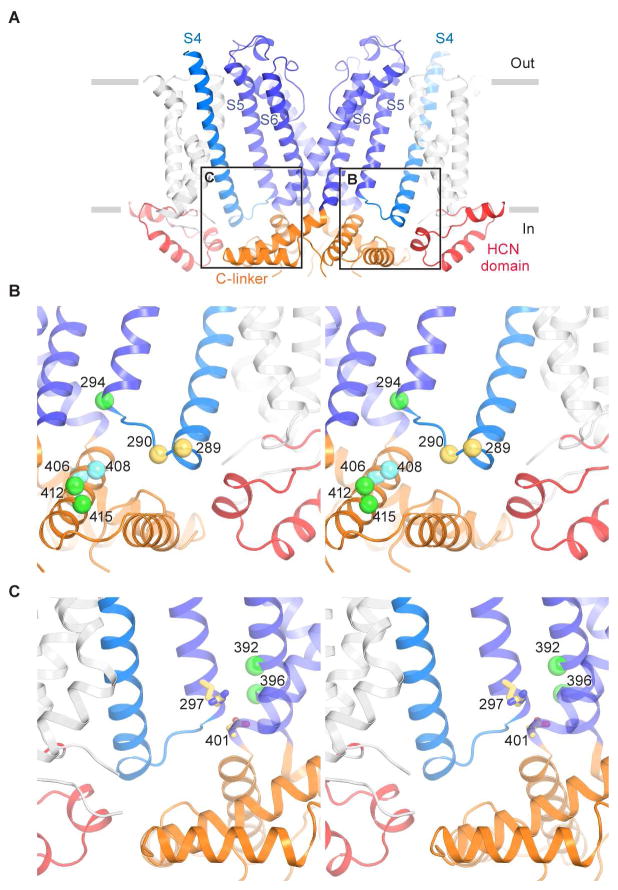
Figure 5. Coupling between the voltage sensor and pore.
(A) Interactions between the voltage sensor, pore and cytoplasmic domains. S1 to S5 helices from the subunits nearest and farthest to the viewer are removed for clarity. The boxes indicate approximate regions of the views shown in (B) and (C).
(B) Stereo view of interactions between the S4–S5 linker, S5 helix and C-linker. Residues 289 and 290 that may interact with the HCN domain are shown as yellow spheres. Residues on the Clinker that “lock-open” the channel when cross-linked with residue 294 are represented as green spheres. Residues on the C-linker that “lock-close” the channel when cross-linked with residue 294 are represented as cyan spheres.
(C) Stereo view of interactions between S4, S5 and S6 helices. The salt bridge forming residues, Arg297 and Asp401, are shown as yellow sticks. Residues that “lock-open” the channel after Cd2+ crosslinking are represented as green spheres.
See also Figure S6.
We observe another aspect of HCN’s architecture that may also support pore closure upon depolarization. Helices S6, S5 and S4 are in close contact with each other almost in a plane (Figures 5A and S6D). S5 and S6 are so well-packed that amino acid side chains interlock like the elements of a zipper (Figure S6D). S4, while not as near, directly contacts S5 for almost its entire length. In addition, in the cytoplasm the HCN domain appears to buttress this plane of 3 helices and force the intracellular end of S4 in towards the pore axis (Figure 5A–C), while it also contacts the outer edge of the C-linker disk. These features of the structure raise the possibility that lateral contacts from the voltage sensors and HCN domains compress the pore so that it is held closed.
The structural features described above lead us to hypothesize that the closed pore is stabilized by the voltage sensors and HCN domains in the depolarized conformation. If we add to this proposal a conjecture that the S6 helices (which form the gate) are in a higher energy conformation when closed (i.e. they would open if unbiased by the voltage sensor and S5) then a plausible model for HCN voltage-dependent gating is as follows: S4 helix displacement, driven by hyperpolarization, would release the constraints on the C-linker and S6 imposed by S4, allowing the pore to open.
This hypothesis is supported by numerous functional studies in which mutational perturbations and cross-links have been used to probe the activity of HCN channels. Mutations on the S4–S5 linker (Tyr289 and Asp290) cause HCN to open with less negative hyperpolarizing voltages (Chen et al., 2001a). This fits with the idea that the S4–S5 linker serves to hold the pore closed and that these mutations interfere with this function (Figure 5B). Similarly, mutations involving Arg297 and Asp401 also favor channel opening (Decher et al., 2004; Nava et al., 2014). These amino acids form a salt bridge between S5 and S6 helices (Figure 5C), appearing therefore to stabilize the closed channel. A series of lock-open and lock-closed Cd2+-mediated cross links by Yellen and colleagues also lend support to the proposed model of gating (Kwan et al., 2012; Rothberg et al., 2003). Cd2+ reaction with Cys residues at positions 392 and 396 on the S6 helix (from the same but not adjacent subunit) “lock-open” the pore (Figure 5C, green spheres). These positions lie on the face of S6 contacting S5, buried at the interface in the closed conformation. If upon hyperpolarization S5 is pulled by S4 away from S6 and Cd2+ then reacts it would presumably destabilize the closed channel by preventing the S5 helix from again adopting its packed-against-S6 closed channel conformation. Cross bridges between positions 294 at the bottom of S5 and positions 412 and 415 on the C-linker also lock the channel open (Figure 5B, green spheres). This is also consistent with the idea that S5 is pulled out and down by S4. In this case, rotation of the C-linker disk associated with gate opening (due to unwrapping of the right-handed bundle of S6 helices) would bring the C-linker residues in close contact with the down-displaced bottom of S5. Concordantly, cross links between the same position 294 and 406 and 408, which occur just C-terminal to the bend where S6 gives rise to the C-linker (Figure 5B, green and cyan spheres), lock HCN in a closed conformation.
It has been shown that a truncation mutation devoid of the C-linker and cyclic nucleotide binding domains still retains hyperpolarization-activated gating (Wainger et al., 2001). It thus appears in this mutant the forces on S6 imposed by S4 and S5 are alone sufficient to gate the channel.
Cyclic nucleotide modulation
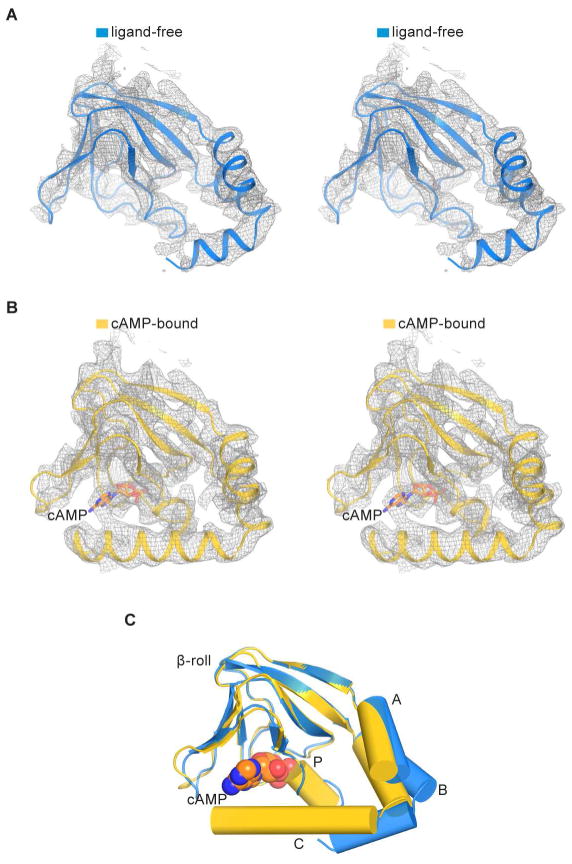
In addition to membrane voltage, the gating of HCN channels is also influenced by cyclic nucleotides. Intracellular cAMP binds to CNBDs, accelerates activation kinetics and shifts the activation curve to more positive voltages (DiFrancesco and Tortora, 1991; Gauss et al., 1998; Ludwig et al., 1998; Santoro et al., 1998). To understand the mechanism of cAMP modulation, we determined the structure of HCN1 in the presence of cAMP. The quality of the density map is sufficient to identify the ligand and associated protein conformational changes (Figures 6 and S4F). The overall structure of the cAMP-bound CNBD is similar to that described in crystal structures of isolated domains (Figure S7), and cAMP binds at a location consistent with previous studies (Figure 6B) (Chen et al., 2001b; Lolicato et al., 2011; Zagotta et al., 2003). Local superposition of the ligand-free and cAMP-bound CNBDs (using the β-jelly rolls) shows the conformational changes near the binding pocket that are induced by cAMP (Figure 6C). Specifically, binding of cAMP stabilizes the P-helix and causes the A-, B-, and C-helices to approach the β-jelly roll, as has been suggested based on both spectroscopic and crystallographic studies of isolated domains (Goldschen-Ohm et al., 2016; Puljung et al., 2014; Saponaro et al., 2014). In addition, helices D and E, not observed in the ligand-free cryo-EM structure, become ordered in the cAMP-bound state (compare Figures 7A and 1C). These helices may play a role in the binding and action of TRIP8b, a protein that regulates HCN trafficking and gating (DeBerg et al., 2015; Santoro et al., 2009; Saponaro et al., 2014; Zolles et al., 2009).
Figure 6. Cyclic AMP-induced conformational changes in the CNBD.
(A) Stereo view of CNBD density in the absence of cAMP. The density map (gray mesh) is sharpened with a b-factor of −90 Å2 and contoured at 6.5 σ.
(B) Stereo view of CNBD density in the presence of cAMP. The density map (gray mesh) is sharpened with a b-factor of −120 Å2 and contoured at 6.5 σ. The cAMP is represented as sticks with carbon atoms in orange, oxygen in red and nitrogen in blue.
(C) Superposition of CNBDs in the ligand-free (blue) and cAMP-bound states (yellow) based on Cα atoms of the β-jelly roll. A-, B-, C- and P-helices are shown in cylinders and cAMP is shown in spheres.
See also Figures S4 and S7.
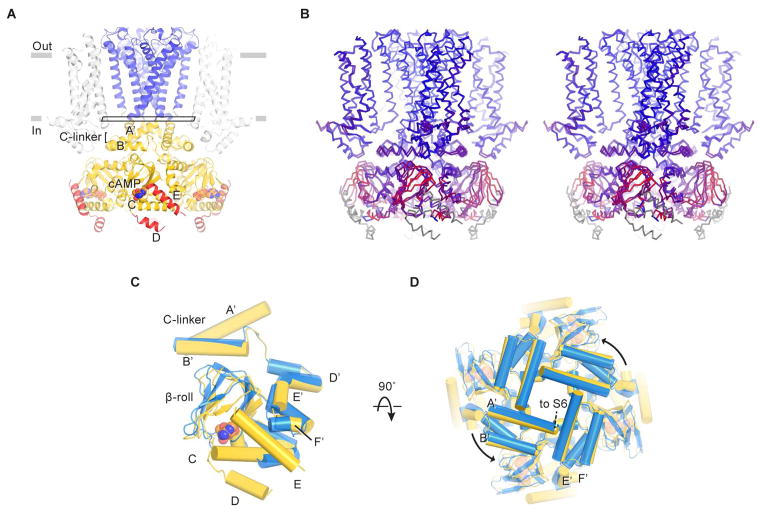
Figure 7. Cyclic AMP modulation of the HCN1 channel.
(A) Structure of the channel in the cAMP-bound state. D- and E-helices are colored red. The rectangle near the gate indicates the plane of the “slice” used for the view in (D).
(B) Stereo view of the superposition of ligand-free and cAMP-bound structures based on Cα atoms of the pore helices. The channel is colored on a red-to-blue spectrum according to the displacement of Cα atoms between the two structures. Blue color represents minimal displacements and red color represents displacements up to 4.5 Å. Gray color indicates residues only present in one structure and thus are not aligned.
(C and D) Superposition of C-linkers and CNBDs in the ligand-free (blue) and cAMP-bound states (yellow). Only single subunits are shown in (C) for clarity. (D) is viewed from the extracellular side.
See also Figures S4 and S7.
Prior studies using isolated domains (without transmembrane regions), although valuable, provide limited structural information on how cAMP binding modulates the ion channel pore. Here, global superposition (using the pore helices) of two complete channel structures shows how the local conformational changes within the CNBDs propagate to the pore via the C-linkers (Figure 7B–D). As can be seen in the stereo image (Figure 7B), the displacement of S6 is small, consistent with the fact that cAMP binding alone in the absence of a hyperpolarizing membrane voltage is insufficient to open the channel (Gauss et al., 1998; Ludwig et al., 1998; Santoro et al., 1998). It is noteworthy, however, that the C-linker rotation and S6 helix displacement is in the direction of pore opening (i.e. the right-handed bundle formed by S6 in the closed conformation is being unwrapped) (Figure 7C and 7D). These ligand-induced conformation changes are consistent with the known functional effects of cAMP to favor voltage-dependent opening (Gauss et al., 1998; Ludwig et al., 1998; Santoro et al., 1998).
Conclusion
Cryo-EM structures have addressed three essential properties of HCN channels. First, we show that these channels, despite having the essential amino acids to form a K+ selectivity filter, permit Na+ permeability because the filter adopts a non-canonical conformation that contains two instead of four cation binding sites. The filter structure thus explains Na+ permeability, which is a fundamental property that enables HCN channels to depolarize a cell’s membrane potential.
Second, several unique structural features have led us to posit a plausible hypothesis to explain voltage dependent gating with reversed polarity. These features include (i) an unusually long S4 helix that contacts a C-linker in the cytoplasm to stabilize a closed gate in the setting of a depolarized voltage sensor, (ii) a packing arrangement of S4, S5 and S6 helices that enable the depolarized voltage sensor to stabilize a closed pore, and (iii) an HCN domain that also stabilizes the closed pore in the setting of a depolarized voltage sensor. These structural features support a mechanism in which voltage-driven downward displacement of the S4 helix upon membrane hyperpolarization will disrupt these stabilizing interactions, allowing the S6 helices to spontaneously open.
Third, we observe that cAMP binding induces local conformational changes that are propagated to the channel. These initiate a rotation of the gate-forming inner helices toward opening. This would explain why cAMP favors HCN channel opening.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for reagents may be directed to the lead contact Roderick MacKinnon (mackinn@rockefeller.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Chinese hamster ovary (CHO)-K1 cells were cultured in DMEM/F-12 medium (Gibco) supplemented with 10% fetal bovine serum at 37 °C. Sf9 cells were cultured in Sf-900 II SFM medium (Gibco) at 28 °C. HEK293S GnTI− cells were cultured in Freestyle 293 medium supplemented with 2% fetal bovine serum at 37 °C.
METHOD DETAILS
Construct design
A DNA segment encoding the human HCN1 channel was synthesized and cloned into a pEG BacMam vector (Goehring et al., 2014), where HCN1 is placed after an N-terminal green fluorescent protein (GFP) tag and a PreScission protease cleavage site. To improve the biochemical stability and expression level of HCN1, we truncated a segment near its C-terminus (amino acid residues 636–865), which is only present in HCN1 but not HCN2–4. This construct, denoted HCN1EM (Figure S1), was used in all following experiments.
Electrophysiological recording
CHO-K1 cells were cultured on coverslips placed in a 6-well plate. Cells in each well were transiently transfected with 2 μg plasmid DNA of HCN1EM using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. After 36–60 h, the coverslips were transferred to a recording chamber containing the external solution (55 mM KCl, 100 mM NaCl, 1 mM CaCl2, 10 mM glucose, and 10 mM HEPES pH 7.4; 310 mOsm). Borosilicate micropipettes (OD 1.5mm, ID 0.86 mm, Sutter) were pulled and fire polished. Pipettes were then filled with the internal solution (85 mM KCl, 70 mM KF, 5 mM EGTA, and 10 mM HEPES pH 7.2; 297 mOsm). Where indicated, the internal solution also contained 500 μM cAMP (as a sodium salt). Pipettes had a resistance of 2–5 MΩ. Whole-cell recordings were obtained at room temperature (−23 °C) using an Axopatch 200B amplifier, a Digidata 1550 digitizer and pCLAMP software (Molecular Devices). The recordings were low-pass filtered at 1 kHz and sampled at 20 kHz. The cells were initially held at −30 mV. To obtain the activation curves, cells were hyperpolarized from −30 to −130 mV in 10 mV decrements, followed by a test pulse at −130 mV (Figure S2A and S2B). To measure the reversal potential, cells were first hyperpolarized to −130 mV, followed by a series of test pulses from 30 to −130 mV in 20 mV decrements (Figure S2C and S2D). The permeability ratio of K+ over Na+ (PK/PNa) was calculated using the Goldman Hodgkin Katz equation. Nonlinear regressions were performed in Prism (GraphPad).
Protein Expression and purification
HCN1EM was expressed in HEK293S GnTI− cells using BacMam method (Goehring et al., 2014). Briefly, a bacmid carrying HCN1EM was generated by transforming E. coli DH10Bac cells with the HCN1EM construct according to the manufacturer’s instructions (Bac-to-Bac; Invitrogen). Baculoviruses were produced by transfecting Spodoptera frugiperda Sf9 cells with the bacmid using Cellfectin II (Invitrogen). Baculoviruses after two rounds of amplification were used for cell transduction. Suspension cultures of HEK293S GnTI− cells were grown at 37 °C to a density of ~3 × 106 cells/ml and baculoviruses were added (10–12% v/v) to initiate the transduction. After 12 h, 10 mM sodium butyrate was supplemented and the temperature was shifted to 30 °C. Cells were harvested at 60 h post-transduction.
HCN1EM protein was purified at 4 °C. The cell pellet from 1 L culture was resuspended in 5 ml of 30 % glycerol for 10 min. The cell suspension was then mixed with 200 ml hypotonic lysis buffer (20 mM KCl, 0.5 mM MgCl2, 2 mM DTT, 0.1 mg/ml DNase, and 10 mM Tris pH 8) for 25 min, and the lysate was spun at 39800 × g for 35 min to sediment crude membranes. The membrane pellet was mechanically homogenized and solubilized in extraction buffer (10 mM lauryl maltose neopentyl glycol, 2 mM cholesteryl hemisuccinate, 300 mM KCl, 2 mM DTT, and 20 mM Tris pH 8) for 1.5 h. Solubilized membranes were clarified by centrifugation at 39800 × g for 35 min. The supernatant was applied to the GFP nanobody-coupled Sepharose resin (Kirchhofer et al., 2010), which was subsequently washed with 10 column volumes of wash buffer (0.05 % digitonin, 300 mM KCl, 2 mM DTT, and 20 mM Tris pH 8). The washed resin was incubated overnight with PreScission protease at a target protein to protease ratio of 40:1 (w/w) to cleave off GFP and release the protein from the resin. The protein was eluted with wash buffer, concentrated by Amicon Ultra centrifugal filter (MWCO 100 kDa), and then injected to a Superose 6 column (GE Healthcare) equilibrated with SEC buffer (0.05 % digitonin, 150 mM KCl, 2 mM DTT, and 20 mM Tris pH 8) (Figure S2E and S2F). Peak fractions were pooled and concentrated to 4–5 mg/ml. All buffers contain protease inhibitors (2 μg/ml leupepetin, 1 μg/ml pepstatin, 50 μg/ml benzamidine, 10 μg/ml aprotinin, and 1 mM AEBSF). To purify the channel in the cAMP-bound state, 0.1 % digitonin and 300 mM KCl were used in the wash and SEC buffer, and all buffers were supplemented with 5 μM cAMP. In addition, 5 mM cAMP was spiked to the protein sample prior to EM grid preparation.
EM data acquisition
3.5 μl of purified HCN channel samples at 4–5 mg/ml were applied to glow-discharged Quantifoil R1.2/1.3 400 mesh Au grids. After a 15-s incubation, the grids were blotted for 1 s and plunged into liquid ethane, using a Vitrobot Mark IV (FEI) operated at 22 °C and 100% humidity. The grids were then loaded onto a Titan Krios transmission electron microscope (FEI) operated at 300 keV. Micrographs were recorded on a K2 Summit direct electron detector (Gatan) in super-resolution counting mode using SerialEM (Mastronarde, 2005). Images have a calibrated physical pixel size of 1.3 Å (a super-resolution pixel size of 0.65 Å) and a nominal defocus range of −1.5 to −3.3 μm. For the ligand-free dataset, a dose rate of 8.5 electrons per pixel per second was used. The exposure time for each image was 10 s fractionated over 40 frames, resulting in a total cumulative dose of ~50 electrons per Å2 (1.26 electrons per Å2 per frame). For the cAMP dataset, a dose rate of 10 electrons per pixel per second was used. The exposure time for each image was 15 s fractionated over 50 frames, resulting in a total cumulative dose of ~89 electrons per Å2 (1.78 electrons per Å2 per frame). Acquisition parameters are summarized in Table S1.
Image processing
Super-resolution image frames were gain-normalized and binned 2 by 2 to a pixel size of 1.3 Å with Fourier cropping (Grant and Grigorieff, 2015a). The frames were then aligned using UnBlur (Grant and Grigorieff, 2015b) to correct beam-induced motions. Gctf (Zhang, 2016) was used to estimate the defocus parameters on the summed image of motion-corrected frames generated by UnBlur. The summed image of motion-corrected frames after the exposure filter (also by UnBlur) was subjected to subsequent processing in RELION (Scheres, 2012). About 3,000 particles were manually picked and processed by reference-free 2D classification in RELION to generate 7 classes for templated-based particle picking using Gautomatch (written by Kai Zhang). Auto-picked particles were visually examined to remove false positives and further cleaned up by multiple rounds of 2D classification. An initial 3D model with C4 symmetry was generated from representative 2D class averages by EMAN2 (Tang et al., 2007). For the ligand-free dataset, a 3D classification into four classes was carried out without applying symmetry in RELION. The major class (53 % of the input; 55,870 particles) contained the most structural details and was refined to 3.6 Å resolution. These particles were subjected to particle-based motion correction and radiation-damage weighing (particle polishing) in RELION (Scheres, 2014). The ‘shiny’ particles were used for the final reconstruction, resulting in a map of overall 3.5 Å resolution. For the cAMP dataset, particles from two sessions of data collection were first processed separately. Particles of session 1 were subjected to two rounds of 3D classification into ten classes without applying symmetry. The classes with the most structural details were combined (36 % of the input; 103,356 particles) and refined to 4.2 Å. The particle polishing improved the resolution to 3.9 Å. The 103,356 particles were subjected to another 3D classification into nine classes without alignment. Two major resulting classes were combined (94,222 particles) and refined to 3.8 Å resolution. Particles of session 2 were subjected to a 3D classification into six classes without applying symmetry. Three major classes were combined (61 % of the input; 110,341 particles) and refined to 3.9 Å resolution. The particle polishing improved the resolution to 3.6 Å. At this stage, particles from session 1 and 2 were merged and subjected to another 3D classification into eight classes without alignment. Two major resulting classes were combined (125,339 particles) and refined to 3.5 Å resolution. All refinements were performed with C4 symmetry imposed and followed the gold-standard Fourier shell correlation (FSC) procedure (Scheres and Chen, 2012). The FSC curves were calculated with soft masks that exclude the detergent micelles and the effects of masking were corrected by the post-processing procedure in RELION (Chen et al., 2013). The reported resolutions were based on the FSC = 0.143 criterion on corrected FSC curves (Figures S3D, S4D and Table S1). Local resolutions of unfiltered density maps were estimated by Blocres (Cardone et al., 2013) with a kernel size of 16 or by ResMap (Kucukelbir et al., 2014). In both datasets, ResMap estimated higher resolutions and most regions fell between 3.0–3.5 Å. Only the estimations from Blocres were shown (Figures S3F and S4F).
Model building
A homology model for one subunit of the transmembrane domains was generated by SWISS- MODEL server (Biasini et al., 2014) using the Eag1 channel as a reference (Whicher and MacKinnon, 2016). This homology model and the crystal structure of mouse HCN1 cytoplasmic domains (Lolicato et al., 2011) (PDB ID: 3U0Z) were docked into the summed density map of the cAMP dataset using UCSF chimera (Pettersen et al., 2004). A tetramer model of the channel was obtained by a symmetry operation of the monomer. The model was first refined with phenix.real_space_refine (Afonine et al., 2013). Subsequent model building and real space refinement were done in COOT (Emsley et al., 2010). The resulting model was used as an initial model for the ligand-free structure, which was then refined and rebuilt following the same procedure. Both tetramer models were further refined against their respective summed maps in reciprocal space using REFMAC v5.8 with a script developed for cryo-EM model refinements (Brown et al., 2015; Murshudov et al., 2011) (Table S1). Four-fold symmetry was imposed during refinement using strong non-crystallographic symmetry restraints. Secondary structure restraints generated by ProSMART were also applied (Nicholls et al., 2014). A refinement weight of 0.001 or 0.0005 was used for the ligand-free dataset or cAMP dataset, respectively. For cross-validation (Brown et al., 2015), the refined structures were randomly displaced up to 0.5 Å. The displaced models were then refined against one of the two half maps produced by RELION. The FSChalf1 curves were calculated between the refined models and the half maps used during refinement. The FSChalf2 curves were calculated between the refined models and the other half maps not used during the refinement. Small differences between FSChalf1 and FSChalf2 suggest that the models were not over fitted (Figures S3E and S4E). The final ligand-free structure includes residues 94–200, 203–242, and 252–586, whereas the cAMP-bound structure includes additional residues 587–608 and the E-helices (Figure S1). Side chains were not modeled for residues with poor density. In the cAMP-bound structure, we could not accurately register the E-helix, which was thus built as a separate, poly-alanine chain for each subunit (chain E, F, G, and H). The quality of the final models was evaluated by MolProbity (Chen et al., 2010) and EMRinger (Barad et al., 2015) (Table S1). The pore radius was calculated by HOLE (Smart et al., 1996). Figures were prepared using PyMOL (Schrodinger LLC, 2015).
QUANTIFICATION AND STATISTICAL ANALYSIS
See METHODS DETAILS for details on cryo-EM analysis
DATA AND SOFTWARE AVAILABILITY
Data Resources
Cryo-EM density maps of HCN1 and HCN1 in the cAMP-bound state have been deposited in the electron microscopy data bank under accession code EMD-8511 and EMD-8512, respectively. Atomic coordinates of HCN1 and HCN1 in the cAMP-bound state have been deposited in the protein data bank under accession code 5U6O and 5U6P, respectively.
Supplementary Material
The full-length human HCN1 and core regions of human HCN2–4 (hHCN1–4) and sea urchin HCN1 (SpHCN1) are shown. Secondary structures are represented by ribbons (α-helices), arrows (β-strands), and lines (loops). Residues of the human HCN1EM (hHCN1EM) construct not modeled in the cryo-EM structures are denoted by dash lines. Relevant residues are labeled and numbered based on the human HCN1 (hHCN1) sequence. For brevity, a portion of the hHCN1 sequence at the C-terminus is removed (represented by//). A precise registration of the E-helix is not feasible for the current study, which is indicated by an asterisk. Amino acid sequences are from UniProtKB using the following entries: hHCN1, O60741; hHCN2, Q9UL51; hHCN3, Q9P1Z3; hHCH4, Q9Y3Q4; and SpHCN1, O76977.
(A) Voltage-dependent activation of HCN1EM. A representative electrophysiological recording from the CHO-K1 cell expressing HCN1EM is shown. Tail currents used to generate activation curves are indicated by an arrow.
(B) Activation curves of HCN1EM in the absence or presence of internal cAMP. Fraction of the maximum current (n = 6–11; means ± SEM) is plotted against the hyperpolarization voltage and fitted with a single Boltzmann function to obtain the midpoint voltage (V1/2).
(C and D) Ion selectivity of HCN1EM. A representative recording is shown in (C). Tail currents used to estimate the reversal potential are indicated by an arrow. In (D), fraction of the normalized current (n = 6; means ± SEM) is plotted against the holding voltage to show the reversal potential of HCN1EM. The current for each holding voltage is normalized to the current at −130 mV. The calculated PK/PNa is about 3.
(E) Representative size-exclusion chromatography (SEC) trace of HCN1EM. Peak fractions were used for cryo-EM analysis.
(F) SDS-PAGE gel of peak fractions stained with Coomassie blue. The predicted molecular weight based on protein sequence for a single HCN1EM subunit is 75 kDa.
(A) Representative raw micrograph of the channel in the ligand-free state.
(B) Selected 2D class averages.
(C) Euler angle distribution of particles for the final 3D reconstruction. The radius of the gray sphere is proportional to the number of particles assigned to that specific orientation.
(D) Fourier shell correlation (FSC) curves between two half maps before (blue) and after (black) the post-processing in RELION.
(E) FSC curves for cross-validation: model versus summed map (black), model versus half map 1 (FSChalf1, red), model versus half map 2 (FSChalf2, blue). The model was refined against halfmap 1 but not half map 2. Small differences between the FSChalf1 and FSChalf2 curves suggestlittle effect of over-fitting.
(F) Local resolution of density map estimated by Blocres. The map shown is low-pass filtered to 3.50 Å and sharpened with a b-factor of −90 Å2.
(A) Representative raw micrograph of the channel in the cAMP-bound state.
(B) Selected 2D class averages.
(C) Euler angle distribution of particles for the final 3D reconstruction. The radius of the gray sphere is proportional to the number of particles assigned to that specific orientation.
(D) Fourier shell correlation (FSC) curves between two half maps before (blue) and after (black) the post-processing in RELION.
(E) FSC curves for cross-validation: model versus summed map (black), model versus half map 1 (FSChalf1, red), model versus half map 2 (FSChalf2, blue). The model was refined against half map 1 but not half map 2. Small differences between the FSChalf1 and FSChalf2 curves suggest little effect of over-fitting.
(F) Local resolution of density map estimated by Blocres. The map shown is low-pass filtered to 3.51 Å and sharpened with a b-factor of −120 Å2.
The atomic model of each domain is in different colors (HCN domain, red; S1–S4, blue; S5–S6, purple; C-linker, orange; and CNBD, green). For the S4–S5 linker, residues from S4 and S5 helices are also included to emphasize the continuity of the density at this region. The map is low-pass filtered to 3.50 Å, sharpened with a b-factor of −90 Å2, and contoured at 5.5–6 σ.
(A) Stereo view of the selectivity filter in the HCN1 channel. Only two subunits of the channel are shown. The density map (gray mesh) is sharpened with a b-factor of −120 Å2 and contoured at 5.5 σ.
(B) Cross section of KcsA and HCN1 channels viewed from the extracellular side, showing the packing of residues (thin sticks) surrounding the selectivity filter (thick sticks). Residues W68 and Y78 in the KcsA channel correspond to residues K351 and Y361 in the HCN1 channel.
(C) Stereo view of the gating charge transfer center in the HCN1 channel. The density map (gray mesh) is sharpened with a b-factor of −90 Å2 and contoured at 5.5 σ.
(D) Stereo view of S4–S6 helices in the HCN1 channel. The density map (gray mesh) is sharpened with a b-factor of −90 Å2 and contoured at 5.5 σ.
(A and C) Superposition of the cytoplasmic domains (single subunit) from the cAMP-bound cryo-EM structure (yellow) and from crystal structures of isolated domains (blue and green) (mHCN2, PDB ID: 1Q43; mHCN1, PDB ID: 3U0Z). The CNBDs align well, whereas the Clinker regions have larger displacements. D- and E-helices of the CNBD are absent in the crystal structures.
(B and D) Superposition of the cytoplasmic tetramers based on Cα atoms of β-jelly rolls, in the same view as Figure 7D. The C-terminal fragments of mHCN forms a tetramer in the crystal, which creates a larger vestibule than the corresponding region of hHCN1. This is perhaps due to lack of constraints from the transmembrane domains in crystal structures of isolated domains.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM/F-12 medium | Gibco | 11330-032 |
| Sf-900 II SFM medium | Gibco | 10902-088 |
| Freestyle 293 medium | Gibco | 12338-018 |
| Lipofectamine 3000 reagent | Invitrogen | L3000015 |
| Cellfectin II reagent | Invitrogen | 10362100 |
| lauryl maltose neopentyl glycol | Anatrace | NG310 |
| Cholesteryl hemisuccinate | Anatrace | CH210 |
| Digitonin | Calbiochem | 300410 |
| 3′,5′-cyclic AMP sodium salt | Sigma-Aldrich | A6885 |
| Critical Commercial Assays | ||
| CNBR-activated Sepharose beads | GE Healthcare | 17-0430-01 |
| Superose 6, 10/300 GL | GE Healthcare | 17-5172-01 |
| Deposited Data | ||
| Human HCN1EM density map | This study | EMDB: EMD-8511 |
| Human HCN1EM with cAMP density map | This study | EMDB: EMD-8512 |
| Human HCN1EM atomic model | This study | PDB: 5U6O |
| Human HCN1EM with cAMP atomic model | This study | PDB: 5U6P |
| Experimental Models: Cell Lines | ||
| CHO-K1 | ATCC | CRL-9618 |
| Sf9 | ATCC | CRL-1711 |
| HEK293S GnTI- | ATCC | CRL-3022 |
| Recombinant DNA | ||
| Human HCN1EM | This study | N/A |
| Software and Algorithms | ||
| Seriel EM | Mastronarde, 2005 | http://bio3d.colorado.edu/SerialEM |
| Unblur | Grant and Grigorieff, 2015b | http://grigoriefflab.janelia.org/unblur |
| Gctf | Zhang, 2016 | http://www.mrc-lmb.cam.ac.uk/kzhang/ |
| RELION | Scheres, 2012 | http://www2.mrc-lmb.cam.ac.uk/relion |
| Gautomatch | Kai Zhang | http://www.mrc-lmb.cam.ac.uk/kzhang/ |
| EMAN2 | Tang et al., 2007 | http://blake.bcm.edu/emanwiki/EMAN2 |
| Blocres | Cardone et al., 2013 | https://lsbr.niams.nih.gov/bsoft/programs/blocres.html |
| ResMap | Kucukelbir et al., 2014 | http://resmap.sourceforge.net/ |
| SWISS-MODEL | Biasini et al., 2014 | https://swissmodel.expasy.org/ |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera |
| PHENIX | Afonine et al., 2013 | https://www.phenix-online.org |
| COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| REFMAC | Brown et al., 2015; Murshudov et al., 2011 | https://www2.mrc-lmb.cam.ac.uk/groups/murshudov/content/em_fitting/em_fitting.html |
| ProSMART | Nicholls et al., 2014 | https://www2.mrc-lmb.cam.ac.uk/groups/murshudov/content/prosmart/prosmart.html |
| MolProbity | Chen et al., 2010 | http://molprobity.biochem.duke.edu |
| EMRinger | Barad et al., 2015 | http://fraserlab.com/2015/02/18/EMringer/ |
| HOLE | Smart et al., 1996 | http://www.smartsci.uk/hole |
| PyMOL | Schrodinger LLC, 2015 | http://www.pymol.org |
| Other | ||
| R1.2/1.3 400 mesh Au holey carbon grids | Quantifoil | 1210627 |
Highlights.
Structures of the human HCN1 hyperpolarization-activated channel at 3.5 Å resolution
Channel filter adopts a particular conformation, allowing both K+ and Na+ to permeate
Long S4 helix and unusual coupling to the pore mediate reversed polarity gating
Cyclic AMP induces a rotation of cytoplasmic domains to favor channel opening
Acknowledgments
We thank M. Ebrahim at the Evelyn Gruss Lipper Cryo-EM Resource Center at Rockefeller University for assistance in data collection, members of the MacKinnon laboratory and Gary Yellen for helpful discussions and Jue Chen for comments on the manuscript. This work was supported in part by National Institutes of Health grant GM43949. C-H.L. is supported by the Jane Coffin Childs Memorial Fund fellowship (#61-1632). R.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions
C-H.L. performed the experiments. C-H.L. and R.M. designed the experiments, analyzed the results, and prepared the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afonine PV, Headd JJ, Terwilliger TC, Adams PD. New tool: phenix.real_space_refine. Comput Crystallogr Newsl. 2013;4:43–44. [Google Scholar]
- Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- Barad BA, Echols N, Wang RYR, Cheng Y, DiMaio F, Adams PD, Fraser JS. EMRinger: side chain–directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch EE. HCN channels: function and clinical implications. Neurology. 2013;80:304–310. doi: 10.1212/WNL.0b013e31827dec42. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Long F, Nicholls RA, Toots J, Emsley P, Murshudov G. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HF, Dfirancesco D, Noble SJ. How does adrenaline accelerate the heart? Nature. 1979;280:235–236. doi: 10.1038/280235a0. [DOI] [PubMed] [Google Scholar]
- Cardone G, Heymann JB, Steven AC. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J Struct Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Mitcheson JS, Tristani-Firouzi M, Lin M, Sanguinetti MC. The S4–S5 linker couples voltage sensing and activation of pacemaker channels. Proc Natl Acad Sci U S A. 2001a;98:11277–11282. doi: 10.1073/pnas.201250598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang J, Siegelbaum SA. Properties of hyperpolarization-activated pacemaker current defined by coassembly of HCN1 and HCN2 subunits and basal modulation by cyclic nucleotide. J Gen Physiol. 2001b;117:491–504. doi: 10.1085/jgp.117.5.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SHW, Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerg HA, Bankston JR, Rosenbaum JC, Brzovic PS, Zagotta WN, Stoll S. Structural Mechanism for the Regulation of HCN Ion Channels by the Accessory Protein TRIP8b. Structure. 2015;23:734–744. doi: 10.1016/j.str.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decher N, Chen J, Sanguinetti MC. Voltage-dependent Gating of Hyperpolarization-activated, Cyclic Nucleotide-gated Pacemaker Channels: MOLECULAR COUPLING BETWEEN THE S4–S5 AND C-LINKERS. J Biol Chem. 2004;279:13859–13865. doi: 10.1074/jbc.M313704200. [DOI] [PubMed] [Google Scholar]
- Derebe MG, Sauer DB, Zeng W, Alam A, Shi N, Jiang Y. Tuning the ion selectivity of tetrameric cation channels by changing the number of ion binding sites. Proc Natl Acad Sci. 2011;108:598–602. doi: 10.1073/pnas.1013636108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFrancesco D, Tortora P. Direct activation of cardiac pacemaker channels by intracellular cyclic AMP. Nature. 1991;351:145–147. doi: 10.1038/351145a0. [DOI] [PubMed] [Google Scholar]
- DiFrancesco JC, DiFrancesco D. Dysfunctional HCN ion channels in neurological diseases. Front Cell Neurosci. 2015;6:174. doi: 10.3389/fncel.2015.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Seifert R, Kaupp UB. Molecular identification of a hyperpolarization-activated channel in sea urchin sperm. Nature. 1998;393:583–587. doi: 10.1038/31248. [DOI] [PubMed] [Google Scholar]
- Goehring A, Lee CH, Wang KH, Michel JC, Claxton DP, Baconguis I, Althoff T, Fischer S, Garcia KC, Gouaux E. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschen-Ohm MP, Klenchin VA, White DS, Cowgill JB, Cui Q, Goldsmith RH, Chanda B. Structure and dynamics underlying elementary ligand binding events in human pacemaking channels. Elife. 2016;5:e20797. doi: 10.7554/eLife.20797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Automatic estimation and correction of anisotropic magnification distortion in electron microscopes. J Struct Biol. 2015a;192:204–208. doi: 10.1016/j.jsb.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. Elife. 2015b;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp UB, Niidome T, Tanabe T, Terada S, Bönigk W, Stühmer W, Cook NJ, Kangawa K, Matsuo H, Hirose T, et al. Primary structure and functional expression from complementary DNA of the rod photoreceptor cyclic GMP-gated channel. Nature. 1989;342:762–766. doi: 10.1038/342762a0. [DOI] [PubMed] [Google Scholar]
- Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan DCH, Prole DL, Yellen G. Structural changes during HCN channel gating defined by high affinity metal bridges. J Gen Physiol. 2012;140:279–291. doi: 10.1085/jgp.201210838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless SW. Determinants of cation transport selectivity: Equilibrium binding and transport kinetics. J Gen Physiol. 2015;146:3–13. doi: 10.1085/jgp.201511371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolicato M, Nardini M, Gazzarrini S, Möller S, Bertinetti D, Herberg FW, Bolognesi M, Martin H, Fasolini M, Bertrand JA, et al. Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels. J Biol Chem. 2011;286:44811–44820. doi: 10.1074/jbc.M111.297606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- Ludwig A, Zong X, Jeglitsch M, Hofmann F, Biel M. A family of hyperpolarization-activated mammalian cation channels. Nature. 1998;393:587–591. doi: 10.1038/31255. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL. A voltage-clamp analysis of inward (anomalous) rectification in mouse spinal sensory ganglion neurones. J Physiol. 1983;340:19–45. doi: 10.1113/jphysiol.1983.sp014747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC 5 for the refinement of macromolecular crystal structures. Acta Crystallogr Sect D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava C, Dalle C, Rastetter A, Striano P, de Kovel CGF, Nabbout R, Cancès C, Ville D, Brilstra EH, Gobbi G, et al. De novo mutations in HCN1 cause early infantile epileptic encephalopathy. Nat Genet. 2014;46:640–645. doi: 10.1038/ng.2952. [DOI] [PubMed] [Google Scholar]
- Neyton J, Miller C. Potassium blocks barium permeation through a calcium-activated potassium channel. J Gen Physiol. 1988;92:549–567. doi: 10.1085/jgp.92.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RA, Fischer M, McNicholas S, Murshudov GN. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr Sect D Biol Crystallogr. 2014;70:2487–2499. doi: 10.1107/S1399004714016241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida M, Cadene M, Chait BT, MacKinnon R. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 2007;26:4005–4015. doi: 10.1038/sj.emboj.7601828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Postea O, Biel M. Exploring HCN channels as novel drug targets. Nat Rev Drug Discov. 2011;10:903–914. doi: 10.1038/nrd3576. [DOI] [PubMed] [Google Scholar]
- Puljung MC, DeBerg HA, Zagotta WN, Stoll S. Double electron-electron resonance reveals cAMP-induced conformational change in HCN channels. Proc Natl Acad Sci U S A. 2014;111:9816–9821. doi: 10.1073/pnas.1405371111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg BS, Shin KS, Yellen G. Movements near the Gate of a Hyperpolarization-activated Cation Channel. J Gen Physiol. 2003;122:501–510. doi: 10.1085/jgp.200308928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro B, Liu DT, Yao H, Bartsch D, Kandel ER, Siegelbaum SA, Tibbs GR. Identification of a Gene Encoding a Hyperpolarization-Activated Pacemaker Channel of Brain. Cell. 1998;93:717–729. doi: 10.1016/s0092-8674(00)81434-8. [DOI] [PubMed] [Google Scholar]
- Santoro B, Piskorowski RA, Pian P, Hu L, Liu H, Siegelbaum SA, Blatch GL, Lassle M, Bonifacino JS, Traub LM, et al. TRIP8b splice variants form a family of auxiliary subunits that regulate gating and trafficking of HCN channels in the brain. Neuron. 2009;62:802–813. doi: 10.1016/j.neuron.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saponaro A, Pauleta SR, Cantini F, Matzapetakis M, Hammann C, Donadoni C, Hu L, Thiel G, Banci L, Santoro B, et al. Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function. Proc Natl Acad Sci. 2014;111:14577–14582. doi: 10.1073/pnas.1410389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.8. 2015. [Google Scholar]
- Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- Shealy RT, Murphy AD, Ramarathnam R, Jakobsson E, Subramaniam S. Sequence-function analysis of the K+-selective family of ion channels using a comprehensive alignment and the KcsA channel structure. Biophys J. 2003;84:2929–2942. doi: 10.1016/S0006-3495(03)70020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi N, Ye S, Alam A, Chen L, Jiang Y. Atomic structure of a Na+− and K+− conducting channel. Nature. 2006;440:570–574. doi: 10.1038/nature04508. [DOI] [PubMed] [Google Scholar]
- Smart OS, Neduvelil JG, Wang X, Wallace BAA, Sansom MSP. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A Gating Charge Transfer Center in Voltage Sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbs GR, Posson DJ, Goldstein PA. Voltage-Gated Ion Channels in the PNS: Novel Therapies for Neuropathic Pain? Trends Pharmacol Sci. 2016;37:522–542. doi: 10.1016/j.tips.2016.05.002. [DOI] [PubMed] [Google Scholar]
- Verkerk A, Wilders R. Pacemaker Activity of the Human Sinoatrial Node: An Update on the Effects of Mutations in HCN4 on the Hyperpolarization-Activated Current. Int J Mol Sci. 2015;16:3071–3094. doi: 10.3390/ijms16023071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger BJ, DeGennaro M, Santoro B, Siegelbaum SA, Tibbs GR. Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature. 2001;411:805–810. doi: 10.1038/35081088. [DOI] [PubMed] [Google Scholar]
- Whicher JR, MacKinnon R. Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science. 2016;353:664–669. doi: 10.1126/science.aaf8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellen G. Relief of Na+ block of Ca2+-activated K+ channels by external cations. J Gen Physiol. 1984;84:187–199. doi: 10.1085/jgp.84.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta WN, Olivier NB, Black KD, Young EC, Olson R, Gouaux E. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature. 2003;425:200–205. doi: 10.1038/nature01922. [DOI] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, MacKinnon R. A Mutant KcsA K+ Channel with Altered Conduction Properties and Selectivity Filter Ion Distribution. J Mol Biol. 2004;338:839–846. doi: 10.1016/j.jmb.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- Zolles G, Wenzel D, Bildl W, Schulte U, Hofmann A, Müller CS, Thumfart JO, Vlachos A, Deller T, Pfeifer A, et al. Association with the auxiliary subunit PEX5R/Trip8b controls responsiveness of HCN channels to cAMP and adrenergic stimulation. Neuron. 2009;62:814–825. doi: 10.1016/j.neuron.2009.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The full-length human HCN1 and core regions of human HCN2–4 (hHCN1–4) and sea urchin HCN1 (SpHCN1) are shown. Secondary structures are represented by ribbons (α-helices), arrows (β-strands), and lines (loops). Residues of the human HCN1EM (hHCN1EM) construct not modeled in the cryo-EM structures are denoted by dash lines. Relevant residues are labeled and numbered based on the human HCN1 (hHCN1) sequence. For brevity, a portion of the hHCN1 sequence at the C-terminus is removed (represented by//). A precise registration of the E-helix is not feasible for the current study, which is indicated by an asterisk. Amino acid sequences are from UniProtKB using the following entries: hHCN1, O60741; hHCN2, Q9UL51; hHCN3, Q9P1Z3; hHCH4, Q9Y3Q4; and SpHCN1, O76977.
(A) Voltage-dependent activation of HCN1EM. A representative electrophysiological recording from the CHO-K1 cell expressing HCN1EM is shown. Tail currents used to generate activation curves are indicated by an arrow.
(B) Activation curves of HCN1EM in the absence or presence of internal cAMP. Fraction of the maximum current (n = 6–11; means ± SEM) is plotted against the hyperpolarization voltage and fitted with a single Boltzmann function to obtain the midpoint voltage (V1/2).
(C and D) Ion selectivity of HCN1EM. A representative recording is shown in (C). Tail currents used to estimate the reversal potential are indicated by an arrow. In (D), fraction of the normalized current (n = 6; means ± SEM) is plotted against the holding voltage to show the reversal potential of HCN1EM. The current for each holding voltage is normalized to the current at −130 mV. The calculated PK/PNa is about 3.
(E) Representative size-exclusion chromatography (SEC) trace of HCN1EM. Peak fractions were used for cryo-EM analysis.
(F) SDS-PAGE gel of peak fractions stained with Coomassie blue. The predicted molecular weight based on protein sequence for a single HCN1EM subunit is 75 kDa.
(A) Representative raw micrograph of the channel in the ligand-free state.
(B) Selected 2D class averages.
(C) Euler angle distribution of particles for the final 3D reconstruction. The radius of the gray sphere is proportional to the number of particles assigned to that specific orientation.
(D) Fourier shell correlation (FSC) curves between two half maps before (blue) and after (black) the post-processing in RELION.
(E) FSC curves for cross-validation: model versus summed map (black), model versus half map 1 (FSChalf1, red), model versus half map 2 (FSChalf2, blue). The model was refined against halfmap 1 but not half map 2. Small differences between the FSChalf1 and FSChalf2 curves suggestlittle effect of over-fitting.
(F) Local resolution of density map estimated by Blocres. The map shown is low-pass filtered to 3.50 Å and sharpened with a b-factor of −90 Å2.
(A) Representative raw micrograph of the channel in the cAMP-bound state.
(B) Selected 2D class averages.
(C) Euler angle distribution of particles for the final 3D reconstruction. The radius of the gray sphere is proportional to the number of particles assigned to that specific orientation.
(D) Fourier shell correlation (FSC) curves between two half maps before (blue) and after (black) the post-processing in RELION.
(E) FSC curves for cross-validation: model versus summed map (black), model versus half map 1 (FSChalf1, red), model versus half map 2 (FSChalf2, blue). The model was refined against half map 1 but not half map 2. Small differences between the FSChalf1 and FSChalf2 curves suggest little effect of over-fitting.
(F) Local resolution of density map estimated by Blocres. The map shown is low-pass filtered to 3.51 Å and sharpened with a b-factor of −120 Å2.
The atomic model of each domain is in different colors (HCN domain, red; S1–S4, blue; S5–S6, purple; C-linker, orange; and CNBD, green). For the S4–S5 linker, residues from S4 and S5 helices are also included to emphasize the continuity of the density at this region. The map is low-pass filtered to 3.50 Å, sharpened with a b-factor of −90 Å2, and contoured at 5.5–6 σ.
(A) Stereo view of the selectivity filter in the HCN1 channel. Only two subunits of the channel are shown. The density map (gray mesh) is sharpened with a b-factor of −120 Å2 and contoured at 5.5 σ.
(B) Cross section of KcsA and HCN1 channels viewed from the extracellular side, showing the packing of residues (thin sticks) surrounding the selectivity filter (thick sticks). Residues W68 and Y78 in the KcsA channel correspond to residues K351 and Y361 in the HCN1 channel.
(C) Stereo view of the gating charge transfer center in the HCN1 channel. The density map (gray mesh) is sharpened with a b-factor of −90 Å2 and contoured at 5.5 σ.
(D) Stereo view of S4–S6 helices in the HCN1 channel. The density map (gray mesh) is sharpened with a b-factor of −90 Å2 and contoured at 5.5 σ.
(A and C) Superposition of the cytoplasmic domains (single subunit) from the cAMP-bound cryo-EM structure (yellow) and from crystal structures of isolated domains (blue and green) (mHCN2, PDB ID: 1Q43; mHCN1, PDB ID: 3U0Z). The CNBDs align well, whereas the Clinker regions have larger displacements. D- and E-helices of the CNBD are absent in the crystal structures.
(B and D) Superposition of the cytoplasmic tetramers based on Cα atoms of β-jelly rolls, in the same view as Figure 7D. The C-terminal fragments of mHCN forms a tetramer in the crystal, which creates a larger vestibule than the corresponding region of hHCN1. This is perhaps due to lack of constraints from the transmembrane domains in crystal structures of isolated domains.