1. Introduction
For more than two decades, the apolipoprotein E (APOE) gene has been consistently identified as the primary risk gene for late onset Alzheimer’s disease (LOAD), accounting for approximately 50% of the genetic risk for AD [1–3]. Despite the strength of the APOEε4 risk variant in predicting Alzheimer’s disease (AD), population studies of APOE allele frequency among AD patients indicate that 36–50% of patients do not carry the ε4 variant [4], and that other, significant genetic contributions to disease risk and pathological progression remain unidentified or uncharacterized for their role in AD [5]. Possession of the ε4 variant of the APOE gene does not provide sufficient sensitivity, selectivity, or power to be used as a predictive tool for AD diagnosis[6] and much of the last decade of genetics research in AD has focused on identifying other genetic markers related to disease risk and age of onset in the hopes of identifying those more likely to experience future cognitive decline.
Several studies implicate APOE’s neighbor on chromosome 19, the translocase of outer mitochondrial membrane 40 homolog (TOMM40) gene, in risk for AD [4–7]. The variant (rs10524523, “523”) in intron 6 of the TOMM40 gene is a variable length poly-T sequence with lengths classified as short (14–20 repeats; i.e. ‘S’), long (21–29 repeats, i.e., ‘L’) or very long (>29 repeats, i.e., ‘VL’). The number of T residues in the homopolymer, “N,” is 35 and the specific variation described by rs10524523 is a 19-base pair deletion, making the homopolymer T16 (N=16 T residues) the variant allele [11]. One of the earliest studies of this variant concluded that the longer length poly-T allele increases risk for AD and decreases age of onset [8]; this finding was replicated in an independent study a few years later [12]. Linkage disequilibrium between TOMM40 and APOE genes ensures that the L poly-T repeat in the TOMM40 gene is almost always (with rare exceptions) inherited with the APOEε4 allele. However, the ε3 allele is most commonly with either a VL or an S poly-T variant, again, with rare exceptions [8,13]. In individuals homozygous for the ε3 variant, the VL variant was found to be associated with a higher risk and earlier age of onset for AD, whereas S variant carriers had a later age of onset [8]. A review of the APOE-TOMM40 phylogenetic field suggests that the discovery of the polymorphism in TOMM40 may improve AD-risk prediction [14].
Recent results suggest the protein encoded by the TOMM40 gene may impact the development of AD via mitochondrial function [15,16]. The protein that TOMM40 encodes, TOM40, is a mitochondrial import channel protein that facilitates the transport of amyloid-β protein precursor (APP) and amyloid-β (Aβ) transport to the mitochondria [17,18]. The TOM40 protein acts as a chaperone, expediting the movement of preproteins through the channel and assembling them post-translationally in the mitochondrion [19]. Because APOEε4, Aβ, and APP have been found to influence function and motility of mitochondria, it has been postulated that APOE and TOMM40 genes might mediate disease risk, and lower age of onset, through mitochondrial dysfunction [8,17,20,21]. Mitochondrial dysfunction is a well-documented factor in the pathogenesis of several age-related diseases, including Parkinson’s disease, Amyotrophic lateral sclerosis, Huntington’s disease, and AD [22–25].
In homozygous APOEε3/ε3 subjects, a phylogenetic experiment suggested that possession of the VL poly-T repeat was associated with increased disease risk and earlier age of onset [8]. A separate study revealed similar findings; possession of the VL poly-T variant in subjects homozygous for ε3 was linked to developing AD at a higher rate when they were ≥ 79 years old [12]. The authors conclude that in the absence of ε4, longer poly-T variants increase the likelihood of developing AD, where the ε3 allele may be linked to either an S or VL poly-T repeat [8].
In order to pinpoint the biologically relevant endophenotypes that relate to the TOMM40 gene, it is crucial to investigate healthy individuals using sensitive metrics to assess the very earliest manifestations of pathophysiological changes in the brain, before the onset of clinical symptoms. While hippocampal volume is a hallmark brain imaging phenotype in AD, substantial work has shown that the first brain changes in AD begin in entorhinal cortex (ERC) [26]. Additionally, subregional analysis of the medial temporal lobe (MTL), especially in ERC, can be more sensitive to possible preclinical morphology differences in both nondemented APOEε4 subjects [27] and MCI patients [28] than volumetric measures. Our group [29–31], and others [32,33] have used high-resolution MRI combined with a cortical unfolding technique which improves visibility of the MTL in order to investigate sub-regional changes in this area, even in non-demented, cognitively intact subjects who carry the at-risk ε4 variant [27].
The aim of the current study was to use high-resolution imaging combined with sub-regional data analysis techniques in non-demented, older subjects in order to investigate the impact of rs10524523 poly-T alleles on the MTL in vivo, which is the site of the very earliest structural changes in AD [34]. We focused analyses on subjects who did not carry the ε4 risk variant of APOE, in order to investigate the contribution of the poly-T variant length in the absence of other known genetic risk attributable to the APOE gene.
2. Methods
2.1 Participants
The study was conducted with the approval of the University of California, Los Angeles Institutional Review Board; all subjects signed informed consent forms before participation. Participants were drawn from a larger study of predictors of cognitive decline by the UCLA Longevity Center [35,36]. Briefly, volunteers from the local community were recruited though local advertisements. Subjects were screened over the phone by research staff of the Longevity Center. However, subjects meeting criteria for AD or any other dementia were excluded from the study [37]. Subjects were also excluded for any history of substance abuse, head trauma or other major systemic disease affecting brain function, a history of neurological or psychiatric disorders, as well as hypertension or cardiovascular disease.
During their visit to the Longevity Center, subjects underwent neuropsychological testing and a clinical interview, in addition to a physical and medical examination, laboratory screening including blood tests to rule out medical conditions that could affect cognitive performance. The current study was conducted on a subset of 65 of these participants (see Table 1) who had successfully completed genotyping for both APOE and TOMM40, as well as cognitive and imaging procedures.
Table 1. Demographic and Clinical Characteristics of Study Participants.
Subjects were divided into ε4 carriers and non-carriers to begin with, then noncarriers were subdivided according to the combined length of both TOMM40 variants. There were no significant differences across the three TOMM40 variant length groups, or between APOE ε4 carrier and non-carriers groups, according to the following characteristics listed in the table.
| Cohort | APOE ε4 carriers | APOE ε4 non-carriers | ||
|---|---|---|---|---|
|
| ||||
| S/S Group | S/L* Group | L*/L* Group | ||
|
| ||||
| Summed TOMM40 poly-T Length < 35 | 35 <Summed TOMM40 poly-T Length < 65 | Summed TOMM40 poly-T Length > 65 | 35 < TOMM40 poly-T Length < 67 | |
|
|
||||
| n=10 | n=18 | n=13 | n=24 | |
| Mini-Mental State Examination score | 28.8±0.8 | 29.1±1.3 | 28.5±1.4 | 28.7±1.0 |
| Age, y | 66.1±11.0 | 62.27±7.7 | 62.2±9.0 | 64.4±9.9 |
| Educational achievement, y | 15.6±3.2 | 16.2±1.5 | 16.7±2.6 | 16.7±3.1 |
| Female sex, No (%) | 6 (60) | 15 (83.3) | 9 (69.2) | 13 (54.2) |
| Family history of dementia, No (%) | 6 (60) | 10 (55.5) | 11 (84.6) | 13 (54.2) |
| Hamilton Depression Scale score | 1.2±1.9 | 2.4±3.2 | 1.8±3.4 | 2.5±3.5 |
| Ethnicity (No. African-American (%), Caucasian (%), Asian (%), Latino (%)) | 3 (30), 0 (0), 7 (70), 0 (0) | 1 (6), 0 (0), 17 (94), 0 (0) | 0 (0), 1 (8), 11 (85), 1 (8) | 1 (4), 1 (4), 20 (83), 2(8) |
2.2 Neuropsychological Testing
Neuropsychiatric test scores were divided into the following domains of cognitive function: Processing Speed (Trailmaking test, part A; Stroop test, word reading speed; Weschler adult intelligence scale-III Digit Symbol), Memory Encoding (Buschke-Fuld selective reminding test, consistent long-term retrieval; Weschler memory scale-II, Logical memory I and Verbal Paired Associates I), Delayed Memory (Weschler memory scale: Logical Memory II and Verbal Paired Associates II; Rey Osterrieth Complex Figure, delayed recall; Buschke-Fuld selective reminding test, delayed recall), and Executive Functioning (Trailmaking test part B; Verbal Fluency FAS and animal naming tests; Stroop test, interference). Studies using these domains have been reported elsewhere [39–42]. We converted the raw scores to Z scores (Z=(raw score-mean)/standard deviation) and created a domain Z score by averaging the Z scores belonging to the cognitive tests in each domain.
2.3 DNA sampling and Genotyping
DNA samples were aliquoted on 96-well plates for determination of both APOE and TOMM40 genotypes. Genotyping for the APOE gene was done by the UCLA Center for Neurobehavioral Genetics (P.I.; D. Geshwind, M.D., Ph.D.) using standard methods [43]. Genotyping for TOMM40 using the rs10524523 (‘523’) allele was completed at Polymorphic DNA Technologies (Alameda, CA, USA; http://www.polymorphicdna.com). TOMM40 polymorphisms were analyzed using polymerase chain reaction (PCR) and bidirectional direct Sanger sequencing of the DNA templates on an Applied Biosystems 3730xl DNA Analyzer (Applied Biosystems Inc., Carlsbad, CA) followed by sequence data analysis. This polymorphism, ‘523,’ is a homopolymer length polymorphism (poly-T) located in an intronic region of TOMM40. The poly-T lengths for each chromosome were converted into the S, L and VL standard labeling [8].
2.4 MRI Acquisition
MRI scans were performed on a Siemens 3T Allegra head-only scanner. Two scans were acquired; 1) Sagittal T1-weighted magnetization prepared rapid acquisition gradient echo (MPRAGE) volumetric scans were acquired to serve as a guide in sulcal visualization during segmentation procedures in the same way an atlas is used as a visual reference (repetition time: 2,300ms, echo time: 2.93ms, slice thickness: 1 mm, 160 slices, in-plane voxel size 1.3×1.3mm, field of view (FOV) 256mm); 2) High-resolution oblique coronal T2-weighted fast-spin echo sequence scan (repetition time: 5,200 ms; echo time: 105 ms; slice thickness: 3mm; skip 0, 19 slices; in-plane voxel size: 0.39×0.39 mm. FOV: 200 mm).
2.5 Whole Brain Structural Imaging
In order to calculate intracranial volume (ICV) estimates to normalize subregional hippocampal thickness values, we used FreeSurfer [44] on whole brain T1-weighted scans. This software suite uses tissue contrast to determine the boundary between gray matter (GM), white matter (WM), and the pial surfaces of the brain in order to calculate the difference between vertices plotted as a mesh surface for each of the layers across the entire cortex. After the automated portion of the FreeSurfer pipeline is complete, each subject’s scan is visually checked for accuracy. Minimal manual edits were completed by a single individual when necessary (TMH). ICV values from FreeSurfer were used to normalize hippocampal complex thickness as detailed below.
2.6 High-resolution Hippocampal Structural Imaging
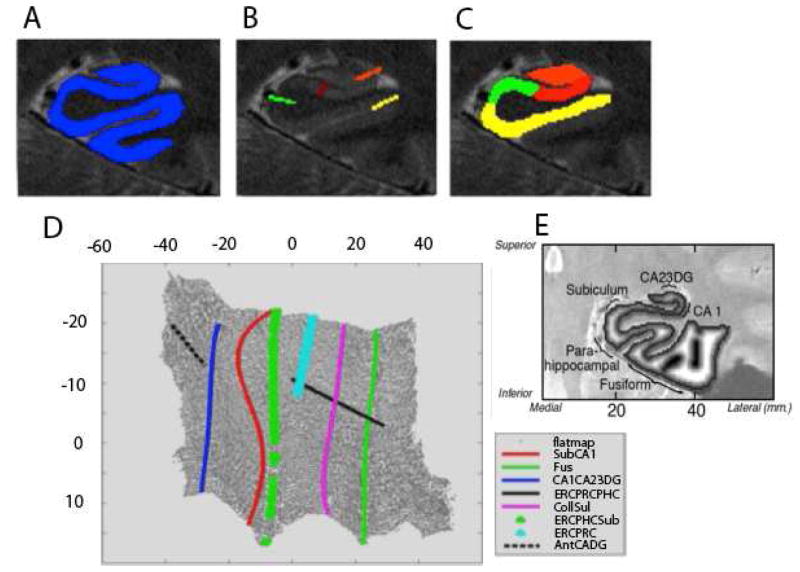
Cortical unfolding is used to improve visualization of the convoluted MTL cortex by flatting the entire 3-D volume into a 2-D flat map [27,29,31]. We use a technique that maximizes resolution in-plane (0.39 × 0.39 mm), where there is significant variability in subregional structure, and increase signal to noise ratio by using thicker slices in the long axis where there is less variability in structure. We acquired T2-images perpendicular to the long axis of the hippocampus to minimize the variability in slice-to-slice changes across the images. Thus, we maximize in-plane resolution and recover signal by increasing thickness in the invariant longitudinal axes, creating maximally resolved anisotropic voxels, while minimizing variability from slice-to slice[29–31,45]. We begin by manually defining WM and cerebral spinal fluid (CSF) on the in-plane oblique coronal images. In order to maximize segmentation, these original images are interpolated by a factor of 7. Then, up to 18 continuous layers of GM are grown out from the boundary of WM, using a region-expansion algorithm to cover all pixels of GM between WM and CSF space (Figure 1a). Boundary demarcations divided the following subregions encompassed by GM: cornu ammonis (CA) fields 1, 2, and 3, the dentate gyrus (DG), subiculum (Sub), ERC, perirhinal cortex (PRC), parahippocampal cortex (PHC), and the fusiform gyrus (Fus) (Figure 1B–C). Because of limits in resolution in CA fields 2, 3 and DG we treat these regions as a single entity (CA23DG). This strip of gray matter is used as the input for the unfolding procedure, an iterative algorithm based on multidimensional scaling (http://ccn.ucla.edu/wiki/index.php/Unfolding). Boundary delineations were projected to their corresponding coordinates in flat map space (Figure 1D).
Figure 1. High-Resolution Hippocampal Image Processing and Thickness Calculations.
A) The goal of high-resolution hippocampal image processing is to isolate the strip of gray matter in the medial temporal lobe that encompasses the subregions of the hippocampus proper and surrounding neocortex, shown in A in blue. This is done by manually segmenting cerebrospinal fluid and white matter and growing sequential layers of gray matter from the edge of white matter until the layer reach the CSF boundary. B) The boundaries between MTL subregions are marked according to anatomical landmarks. Demarcations shown here include CA23DG | CA1 (orange), CA1 | subiculum (red), parahippocampal gyrus | subiculum (light green), and fusiform gyrus (yellow). C) Each subregion is considered separately for cortical thickness calculations. D) Demarcations are projected from in-plane space to the corresponding location in 2-dimensional flat map space and then extended for form complete and smooth boundaries between subregions. E). Cortical thickness is visualized in in-plane space as a grayscale map of thickness values between maximum (white) and minimum (black) values.
In order to calculate thickness, we computed the distance for each voxel in in-plane space to the nearest non-gray matter voxel, we took the maximum distance value in 2D voxels of the corresponding 3D voxels across all layers and multiplied by two and calculated the mean thickness in subregions by averaging thickness of all 2D voxels (Figure 1E). Cortical thickness within subregions was averaged over both hemispheres.
We corrected for differences in head size across subjects by normalizing hippocampal thickness values to ICV estimates. The following formula was used to normalize thickness values: ICV-corrected thickness = [(thickness in mm/ICV in mm3) * 106]. Multiplying by 106 results in values at the same order of magnitude as original thickness estimates.
2.7 Statistical Analyses
Statistical models were used to investigate the effect of TOMM40 genetic variant lengths on subregional MTL thickness in the absence of the APOEε4 variant. As has been done previously in the literature to condense the largest number of potential genotype combinations into subgroups, the L and VL alleles were pooled into an L* group; participants with the S/S genotype were compared to those carrying only one S allele (pooled S/L and S/VL; hereinafter S/L*) and also compared to participants carrying no S alleles (pooled L/L, L/VL, and VL/VL; hereinafter L*/L*; [11]). In order to assess whether varying TOMM40 poly-T lengths were associated with thinner hippocampal cortex in individual subregions in the absence of APOEε4, we computed a multivariate analysis of covariance (MANCOVA) with thicknesses of all subregions as the dependent variables, categorical groups of additive poly-T lengths as the predictors (S/S, S/L* and L*/L*) and age, education, sex, and MMSE as covariates. In order to assess whether varying TOMM40 poly-T lengths were associated with cognitive performance in the absence of APOEε4, we computed a MANCOVA with Z-scaled cognitive performance scores as dependent variables, poly-T lengths as predictors (S/S, S/L* and L*/L*) and age, education, sex, and MMSE as covariates.
3. Results
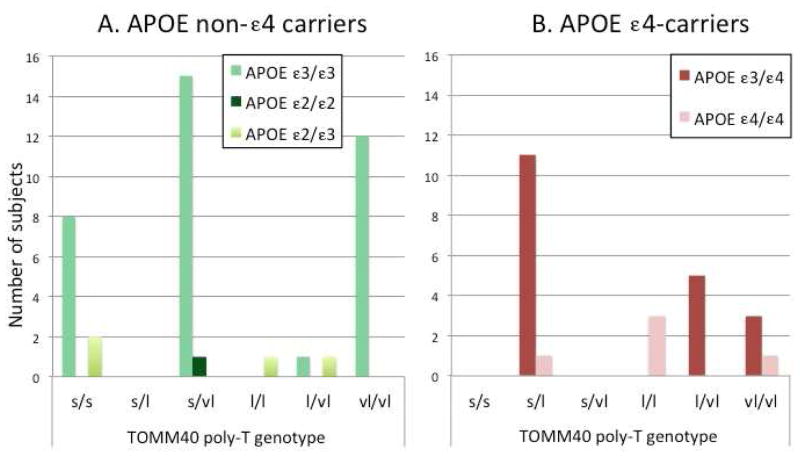
Of the 65 subjects enrolled, 24 subjects carried at least one copy of the ε4 variant for the APOE gene and 41 were non-ε4 carriers. Among the APOEε4 non-carriers, the APOE genotype was as follows: 1 APOE ε2/ε2, 3 APOEε2/ε3 and 37 APOE ε3/ε3 subjects. There were no differences in clinical and demographic variables across the groups (Table 1), however, we also included age, education, sex and MMSE score as covariates in the multivariate analysis. Ethnicity is also reported in Table 1, however, the number of subjects enrolled in each category was too small to study the effect of ethnicity on TOMM40 poly-T lengths and hippocampal thickness separately. Figure 2 shows the breakdown of TOMM40 variant lengths among subjects by APOE genotype.
FIGURE 2. Distribution of TOMM40 variant lengths by APOE group.
Subjects were categorized first by whether they were APOE ε4 carriers or non-carriers, then grouped according to the combined length of both TOMM40 poly-T length variants. The distribution of non-ε4 carrier subjects is shown in panel A while ε4 carriers are shown in panel B. Similar to previous reports [7,46], the majority of poly-T lengths in the ε3/ε3 cohort were either S (<21) or VL (>30). In agreement with previous reports that the ε4 variant is typically bound by linkage disequilibrium to L TOMM40 variants [8], the majority of ε4 carriers (n=20) possessed as least one L TOMM40 variant, although four subjects were homozygous for VL (Figure 2B). Non-ε4 carriers showed a distribution (Figure 2A) where the majority of subjects possessed two short S copies of the TOMM40 variant (S/S; n=10), two copies of the very long VL variant (n=12), or a heterozygous combination of S/VL (n=16). As demonstrated in this Figure, there were no non-APOE e4 subjects. However, for consistency’s sake, we chose to continue the nomenclature for S/L and S/VL carriers to be pooled into an S/L* group much like participants carrying no S alleles (L/L, L/VL, and VL/VL) were pooled into an L*/L* group.
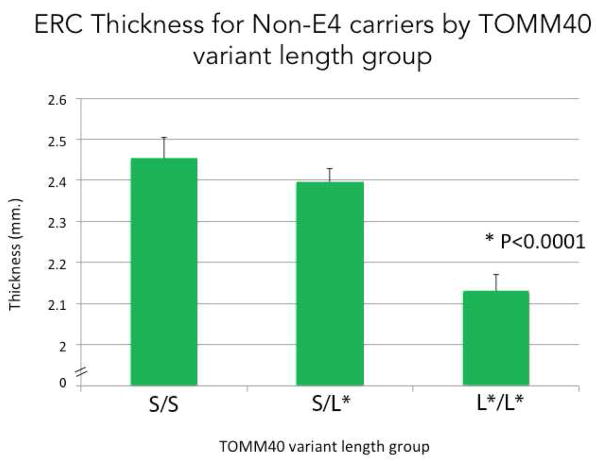
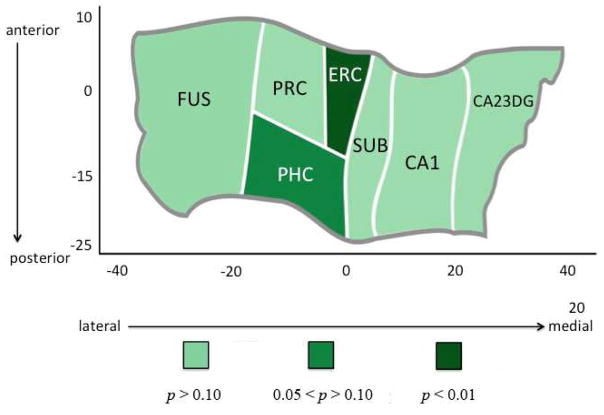
The MANCOVA revealed a significant relationship between longer poly-T lengths and thickness (F(14,54) = 3.61, p = .0003; excluding the ε2’s: (F(14,46) = 3.29, p = .001) in non-ε4 carriers. Follow-up univariate analyses indicated that ERC thickness was significantly associated with longer poly-T lengths (F(2,33) = 16.21, p <.0001; excluding the ε2’s: (F(2,30) = 14.67, p < .0001), with the L*/L* group showing significantly reduced thickness compared to both the S/L* and S/S groups (both p < .0001) (Figure 3). In addition, PHC thickness was marginally associated with increasing poly-T lengths (F(2,33) = 3.22, p =.054; excluding the ε2’s: (F(2,30) = 3.28, p =.051). No other subregions showed significant differences between increasing poly-T variant lengths in either ε4-carriers or non-carriers (Figure 4). We did not find a significant relationship between TOMM40 poly-T lengths and Z-scaled cognitive score in any of the five domains of cognitive function.
FIGURE 3. ERC Thickness for ε4-Non-Carriers by TOMM40 variant length.
Averaged ERC thickness for subjects in each of the TOMM40 summed variant length group (S/S group: 2.45mm; S/L* group: 2.40mm; L*/L* group: 2.13mm). Univariate analyses indicated that ERC thickness was significantly associated with increasing poly-T lengths (F(2,33) = 16.21, p <.0001), with the VL group showing a significantly reduced thickness compared to both the L and S groups (both p < .0001).
FIGURE 4. Hippocampal Complex Unfolding Reveals Subregional Relationship with Increasing Length of TOMM40 Poly-T Variants.
A cortical unfolding procedure was used to produce a flat map of the hippocampal complex. Regions are color coded according to the strength of the statistical association between TOMM40 poly-T variant length and cortical thickness in individual subregions within the hippocampus and surrounding neocortex. Results revealed that ERC thickness was significantly associated with increasing poly-T lengths (p <.0001), with the VL group showing a significantly reduced thickness compared to both the L and S groups (both p < .0001). In addition, PHC thickness was almost significantly associated with increasing poly-T lengths (p =.054). No other subregions showed significant differences between increasing poly-T variant lengths in either ε4-carriers or non-carriers.
4. Discussion
We show here that in older, normal control subjects who do not carry the APOEε4 variant, longer TOMM40 poly-T lengths are significantly associated with thinner ERC. Our results demonstrate an association between the TOMM40 poly-T variant and subregional morphological differences in the MTL only in subjects who do not carry the APOEε4 risk variant. The pattern of cortical thinning in subjects with low APOE-risk, but elevated TOMM40 risk closely resembles that seen in healthy, older APOEε4 carriers. To our knowledge, this is the first investigation to investigate the effect of the TOMM40 gene on brain morphology in MTL subregions. These data highlight the importance of assessing multiple risk variants in order to detect morphological differences in healthy, older adults before the possible onset of clinical symptoms. Our results also underscore the importance of investigations that assess the integrity of the ERC in healthy, older adults when assessing risk for AD, and highlight the utility of imaging tools that isolate individual subregions of the MTL, including the ERC. We suggest that, given the growth of automated hippocampal segmentation tools in recent years [47], assessing the structural integrity of the ERC plays a crucial role in assessing risk for AD, even in nondemented healthy, older control subjects.
Novel brain imaging techniques reveal structural changes that may be phenotypic in prodromal AD. For decades, the most prominent genetic marker for preclinical manifestations of the disease has been the APOEε4 variant on chromosome 19. TOMM40’s location in proximity to the APOE gene has prompted queries for whether the effects noted for this newly discovered gene are working in concert with, independently of, or instead of APOE’s effects. Because TOMM40 is in strong linkage disequilibrium with APOE on chromosome 19, the ε4 allele of APOE is almost exclusively linked to the L poly-T variant. As suggested in an article detailing the full impact of the TOM40-mediated mitochondrial protein import mechanism in aging [18], the effects of the VL/VL genotype may be associated with presymptomatic events in younger people that are masked by later pathology in advanced AD. Several studies have suggested that SNPs within TOMM40 are associated with increased risk for AD [9,48–57] or associated endophenotypes of the disease, including cognitive performance [58] and hippocampal atrophy [59] independently of APOE. However, another study failed to replicate the association between TOMM40 variants and risk for AD [60]. These results underscore the importance of further investigations into the relationship between these multiple risk variants, and suggest the need for investigations in younger, healthy subjects before the appearance of more widespread brain changes in later disease stages.
The nonspecific nature of APOE’s effects in AD was the original impetus leading investigators to postulate that other genes/proteins in the chromosomal interval containing APOE might be responsible for the wide variation in genetic risk associated with AD [8,9]. The ε4 risk variant is neither necessary nor sufficient for disease onset and genetics research alone is unlikely to definitively diagnose AD [62]. However, identifying which markers have the greatest sensitivity and specificity among those identified as markers for AD-risk will allow us to assess and follow subjects with the greatest likelihood of cognitive decline related to genetic risk for AD-onset over time, thereby strengthening the likelihood of maximizing the effect of current therapeutic interventions as well as testing novel therapies as they are developed. Additionally, within this analysis, the 4 subjects who carried at least one copy of the APOEε2 variant, were analyzed in the non-APOEε4 group as the intention was to investigate the effect of TOMM40 poly-T length in the absence of the APOEε4 variant. The data here are too small to analyze APOEε2 subjects separately, but we suggest that in future, larger datasets, the question of whether TOMM40- associated morphology differences exist in APOEε2 carriers is worthy of investigating.
It is noteworthy that the genotype and family history of AD distribution in the population studied here differs from that found in a random sampling of the general population. Typically, 20–25% of the general public carries at least one copy of the ε4 variant [63–65] while in the present study, 37% carried at least one APOE ε4 copy. Additionally, depending on genotype, 54–85% of subjects in this study reported a family history of dementia compared to 10% in the general population Our recruitment method yielded a sample of highly motivated, physically healthy subjects concerned about age-related memory problems and resulted in a sample enriched for possession of the ε4 risk variant. While the sample may not be representative of the general population, having a higher concentration of subjects with the APOEε4 variant does not explain the cortical thickness differences in ERC between the genetic groups. Additionally, as mentioned in the statistical methods section, family history was used as a covariate, ensuring any effect of that factor was not responsible for the morphology results. Finally, genotype and family history percentages reported here are similar to those reported in our lab [27,66,67] and others [68,69].
Additional limitations must also be acknowledged. The sample size is small and, unfortunately, the ethnic breakdown across the groups resulted in limited diversity for statistical analysis. The analytical method of cortical unfolding reported here is an extremely time-consuming technique. However, advances in imaging methodology, both in image acquisition and in data analysis, are expected to make more rapid analysis possible in the near future. Larger analyses should address race and ethnicity given that they are known to vary with dementia risk [70] and are also suspected to vary with TOMM40 variant length [71]. Finally, we also acknowledge that future studies will be more powerful in detecting differences in morphology associated with genetic risk using longitudinal assessment as opposed to the crosssectional analysis we report here [72].
These results demonstrate specific subregional morphological changes within the MTL related to the TOMM40 gene in the absence of the APOEε4 risk factor. Identifying relationships between gene-brain risk metrics in the absence of the APOEε4 allele promises to shed light on the question of which ε4 negative subjects are at greater risk for AD-progression. As clinical trials of novel AD treatments continue, identifying biomarkers that isolate subjects at greater risk for AD than the general population will enhance our ability to identify subjects likely to benefit from these interventions and demonstrate results from effective treatments.
High-resolution MRI combined with APOE and TOMM40 used for subregional MTL analysis.
Longer TOMM40 T-lengths correlated with thinner entorhinal cortex in APOEe3 carriers.
TOMM40 variant length is contributor to AD-like MTL pathology in absence of APOEε4.
Systematic review
Several studies implicate APOE’s neighbor on chromosome 19, TOMM40, in AD risk. In homozygous APOEε3 subjects, a phylogenetic experiment suggested that possession of longer-length poly-T repeats was associated with increased disease risk and earlier age of onset, but further structural MRI studies have shown mixed results.
Interpretation
To our knowledge, this is the first study to investigate the effect of TOMM40 on morphology in MTL subregions and show thinner entorhinal cortex in older, heterogeneous APOEε3 control subjects with longer TOMM40 poly-T lengths. We focused on subjects who do not carry the ε4 risk variant of APOE, in order to investigate the contribution of the poly-T variant length in the absence of APOE risk.
Future Directions
Our results underscore the importance of assessing the entorhinal cortex in healthy, older adults when investigating risk for AD and highlight the importance of identifying gene-brain risk metrics in the absence of APOEε4.
Acknowledgments
Supported by National Institutes of Health grants [P01-AG025831, AG13308, P50 AG 16570, MH/AG58156, MH52453; AG10123; M01-RR00865, F31AG047041], the Department of Energy [Contract DE-FC03-87-ER60615]; General Clinical Research Centers Program; the Fran and Ray Stark Foundation Fund for Alzheimer’s Disease Research; the Ahmanson Foundation; the Larry L. Hillblom Foundation; the Lovelace Foundation; the Sence Foundation; the McMahan Foundation; the Judith Olenick Elgart Fund for Research on Brain Aging; and the Parlow-Solomon Professorship on Aging. No company provided support of any kind for this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bergem AL, Engedal K, Kringlen E. The role of heredity in late-onset Alzheimer disease and vascular dementia. A twin study. Arch Gen Psychiatry. 1997;54:264–70. doi: 10.1001/archpsyc.1997.01830150090013. [DOI] [PubMed] [Google Scholar]
- 2.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–74. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 3.Waring SC, Rosenberg RN. Genome-wide association studies in Alzheimer disease. Arch Neurol. 2008;65:329–34. doi: 10.1001/archneur.65.3.329. [DOI] [PubMed] [Google Scholar]
- 4.Bertram L, Tanzi RE. The Genetics of Alzheimer’s Disease. Prog Mol Biol Transl Sci. 2012;107:79–100. doi: 10.1016/B978-0-12-385883-2.00008-4. [DOI] [PubMed] [Google Scholar]
- 5.Ashford JW, Mortimer JA. Non-familial Alzheimer’s disease is mainly due to genetic factors. J Alzheimers Dis. 2002;4:169–77. doi: 10.3233/jad-2002-4307. [DOI] [PubMed] [Google Scholar]
- 6.Mayeux R, Saunders AM, Shea S, Mirra S, Evans D, Roses AD, et al. Utility of the apolipoprotein E genotype in the diagnosis of Alzheimer’s disease. Alzheimer’s Disease Centers Consortium on Apolipoprotein E and Alzheimer’s Disease. N Engl J Med. 1998;338:506–11. doi: 10.1056/NEJM199802193380804. [DOI] [PubMed] [Google Scholar]
- 7.Roses AD. An inherited variable poly-T repeat genotype in TOMM40 in Alzheimer disease. Arch Neurol. 2010;67:536–41. doi: 10.1001/archneurol.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roses AD, Lutz MW, Amrine-Madsen H, Saunders aM, Crenshaw DG, Sundseth SS, et al. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. Pharmacogenomics J. 2010;10:375–84. doi: 10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takei N, Miyashita A, Tsukie T, Arai H, Asada T, Imagawa M, et al. Genetic association study on in and around the APOE in late-onset Alzheimer disease in Japanese. Genomics. 2009;93:441–8. doi: 10.1016/j.ygeno.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Lutz MW, Crenshaw DG, Saunders AM, Roses AD. Genetic variation at a single locus and age of onset for Alzheimer’s disease. Alzheimers Dement. 2010;6:125–31. doi: 10.1016/j.jalz.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caselli RJ, Dueck AC, Huentelman MJ, Lutz MW, Saunders AM, Reiman EM, et al. Longitudinal modeling of cognitive aging and the TOMM40 effect. Alzheimers Dement. 2012;8:490–5. doi: 10.1016/j.jalz.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maruszak A, Pepłońska B, Safranow K, Chodakowska-Żebrowska M, Barcikowska M, Zekanowski C. TOMM40 rs10524523 polymorphism’s role in late-onset Alzheimer’s disease and in longevity. J Alzheimers Dis. 2012;28:309–22. doi: 10.3233/JAD-2011-110743. [DOI] [PubMed] [Google Scholar]
- 13.Lutz MW, Crenshaw DG, Saunders AM, Roses AD. The importance of being connected. J Alzheimers Dis. 2011;24:247–51. doi: 10.3233/JAD-2010-101765. [DOI] [PubMed] [Google Scholar]
- 14.Lutz MW, Crenshaw D, Welsh-Bohmer KA, Burns DK, Roses AD. New Genetic Approaches to AD: Lessons from APOE-TOMM40 Phylogenetics. Curr Neurol Neurosci Rep. 2016;16:48. doi: 10.1007/s11910-016-0643-8. [DOI] [PubMed] [Google Scholar]
- 15.Lutz MW, Crenshaw DG, Saunders AM, Roses AD. The importance of being connected. J Alzheimers Dis. 2011;24:247–51. doi: 10.3233/JAD-2010-101765. [DOI] [PubMed] [Google Scholar]
- 16.Perry AJ, Rimmer KA, Mertens HD, Waller RF, Mulhern TD, Lithgow TGP. Structure, topology and function of the translocase of the outer membrane of mitochondria. Plant Physiol Biochem. 2008;46:265–74. doi: 10.1016/j.plaphy.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Hansson Petersen CA, Alikhani N, Behbahani H, Wiehager B, Pavlov PF, Alafuzoff I, et al. The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc Natl Acad Sci U S A. 2008;105:13145–50. doi: 10.1073/pnas.0806192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottschalk WK, Lutz MW, He YT, Saunders AM, Burns DK, Roses AD, et al. The Broad Impact of TOM40 on Neurodegenerative Diseases in Aging. J Park Dis Alzheimer’s Dis. 2014;1 doi: 10.13188/2376-922X.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherman EL, Taylor RD, Go NE, Nargang FE. Effect of mutations in Tom40 on stability of the translocase of the outer mitochondrial membrane (TOM) complex, assembly of Tom40, and import of mitochondrial preproteins. J Biol Chem. 2006;281:22554–65. doi: 10.1074/jbc.M601630200. [DOI] [PubMed] [Google Scholar]
- 20.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc Natl Acad Sci U S A. 2006;103:5644–51. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang S, ran Ma T, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc Natl Acad Sci U S A. 2005;102:18694–9. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, et al. Mitochondrial Abnormalities in Alzheimer’s Disease. J Neurosci. 2001;21:3017–23. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su B, Wang X, Zheng L, Perry G, Smith MA, Zhu X. Abnormal mitochondrial dynamics and neurodegenerative diseases. Biochim Biophys Acta. 2010;1802:135–42. doi: 10.1016/j.bbadis.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beal MF. Mitochondria take center stage in aging and neurodegeneration. Ann Neurol. 2005;58:495–505. doi: 10.1002/ana.20624. [DOI] [PubMed] [Google Scholar]
- 25.Chaturvedi RK, Flint Beal M. Mitochondrial diseases of the brain. Free Radic Biol Med. 2013;63:1–29. doi: 10.1016/j.freeradbiomed.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 26.de Leon M, Bobinski M, Convit A, Wolf O, Insausti R. Usefulness of MRI measures of entorhinal cortex versus hippocampus in AD. Neurology. 2001;56:820–1. doi: 10.1212/wnl.56.6.820. [DOI] [PubMed] [Google Scholar]
- 27.Burggren AC, Zeineh MM, Ekstrom AD, Braskie MN, Thompson PM, Small GW, et al. Reduced cortical thickness in hippocampal subregions among cognitively normal apolipoprotein E e4 carriers. Neuroimage. 2008;41:1177– 83. doi: 10.1016/j.neuroimage.2008.03.039. S1053-8119(08)00259-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velayudhan L, Proitsi P, Westman E, Muehlboeck J-S, Mecocci P, Vellas B, et al. Entorhinal cortex thickness predicts cognitive decline in Alzheimer’s disease. J Alzheimers Dis. 2013;33:755–66. doi: 10.3233/JAD-2012-121408. [DOI] [PubMed] [Google Scholar]
- 29.Zeineh MM, Engel SA, Thompson PM, Bookheimer SY. Dynamics of the hippocampus during encoding and retrieval of face-name pairs. Science (80- ) 2003;299:577–80. doi: 10.1126/science.1077775. [DOI] [PubMed] [Google Scholar]
- 30.Zeineh MM, Engel SA, Bookheimer SY. Application of cortical unfolding techniques to functional MRI of the human hippocampal region. Neuroimage. 2000;11:668–83. doi: 10.1006/nimg.2000.0561. [DOI] [PubMed] [Google Scholar]
- 31.Ekstrom AD, Bazih AJ, Suthana NA, Al-Hakim R, Ogura K, Zeineh M, et al. Advances in high-resolution imaging and computational unfolding of the human hippocampus. Neuroimage. 2009;47:42–9. doi: 10.1016/j.neuroimage.2009.03.017. S1053-8119(09)00239- 0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eldridge LL, Engel SA, Zeineh MM, Bookheimer SY, Knowlton BJ. A dissociation of encoding and retrieval processes in the human hippocampus. J Neurosci. 2005;25:3280–6. doi: 10.1523/JNEUROSCI.3420-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sicotte NL, Kern KC, Giesser BS, Arshanapalli A, Schultz A, Montag M, et al. Regional hippocampal atrophy in multiple sclerosis. Brain. 2008;131:1134–41. doi: 10.1093/brain/awn030. [DOI] [PubMed] [Google Scholar]
- 34.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–59. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 35.Small GW, Kepe V, Ercoli LM, Siddarth P, Bookheimer SY, Miller KJ, et al. PET of brain amyloid and tau in mild cognitive impairment. N Engl J Med. 2006;355:2652–63. doi: 10.1056/NEJMoa054625. 355/25/2652 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Small GW, Siddarth P, Burggren AC, Kepe V, Ercoli LM, Miller KJ, et al. Influence of cognitive status, age, and APOE-4 genetic risk on brain FDDNP positron-emission tomography imaging in persons without dementia. Arch Gen Psychiatry. 2009;66:81–7. doi: 10.1001/archgenpsychiatry.2008.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 38.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state” A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–98. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 39.Ercoli LM, Siddarth P, Kepe V, Miller KJ, Huang S-C, Cole GM, et al. Differential FDDNP PET patterns in nondemented middle-aged and older adults. Am J Geriatr Psychiatry. 2009;17:397–406. doi: 10.1097/JGP.0b013e318198750b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bilder RM, Goldman RS, Robinson D, Reiter G, Bell L, Bates JA, et al. Neuropsychology of first-episode schizophrenia: initial characterization and clinical correlates. Am J Psychiatry. 2000;157:549–59. doi: 10.1176/appi.ajp.157.4.549. [DOI] [PubMed] [Google Scholar]
- 41.Burggren A, Renner B, Jones M, Donix M, Suthana NA, Martin-Harris L, et al. Thickness in Entorhinal and Subicular Cortex Predicts Episodic Memory Decline in Mild Cognitive Impairment. Int J Alzheimers Dis. n.d doi: 10.4061/2011/956053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donix M, Linda M, Ercoli Prabha Siddarth, PhD2, Ercoli Jesse Linda M, PhD2, Siddarth Prabha, PhD2, Brown Jesse A, BA1,2, Martin-Harris Laurel, BA1,2, Burggren Alison C, PhD1,2, Miller Karen J, PhD2, Small Gary W, MD2,4, Bookh Susan Y., PD Influence of Alzheimer’s Disease Family History and Genetic Risk on Cognitive Performance in Healthy Middle-Aged and Older People. doi: 10.1053/S1067-2516(03)00401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wenham PR, Price WH, Blandell G. Apolipoprotein E genotyping by one-stage PCR. Lancet. 1991;337:1158–9. doi: 10.1016/0140-6736(91)92823-k. [DOI] [PubMed] [Google Scholar]
- 44.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97:11050–5. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeineh MM, Mazziotta JC, Thompson PM, Bookheimer SY. Hippocampal flat maps of cortical thickness and power. New York, New York: 2003. [Google Scholar]
- 46.Johnson SC, La Rue A, Hermann BP, Xu G, Koscik RL, Jonaitis EM, et al. The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOE ε3/ε3 genotype. Alzheimers Dement. 2011;7:456–65. doi: 10.1016/j.jalz.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yushkevich PA, Amaral RSC, Augustinack JC, Bender AR, Bernstein JD, Boccardi M, et al. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: towards a harmonized segmentation protocol. Neuroimage. 2015;111:526–41. doi: 10.1016/j.neuroimage.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin ER, Lai EH, Gilbert JR, Rogala AR, Afshari AJ, Riley J, et al. SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am J Hum Genet. 2000;67:383–94. doi: 10.1086/303003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu C-E, Seltman H, Peskind ER, Galloway N, Zhou PX, Rosenthal E, et al. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer’s disease: patterns of linkage disequilibrium and disease/marker association. Genomics. 2007;89:655–65. doi: 10.1016/j.ygeno.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, et al. Evidence for novel susceptibility genes for late-onset Alzheimer’s disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16:865–73. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- 51.Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 52.Naj AC, Beecham GW, Martin ER, Gallins PJ, Powell EH, Konidari I, et al. Dementia revealed: novel chromosome 6 locus for late-onset Alzheimer disease provides genetic evidence for folate-pathway abnormalities. PLoS Genet. 2010;6:e1001130. doi: 10.1371/journal.pgen.1001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamboh MI, Demirci FY, Wang X, Minster RL, Carrasquillo MM, Pankratz VS, et al. Genome-wide association study of Alzheimer’s disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S, Swaminathan S, Shen L, Risacher SL, Nho K, Foroud T, et al. Genome-wide association study of CSF biomarkers Abeta1-42, t-tau, and p-tau181p in the ADNI cohort. Neurology. 2011;76:69–79. doi: 10.1212/WNL.0b013e318204a397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naslund J, Haroutunian V, Mohs R, Davis KL, Davies P, Greengard P, et al. Correlation between elevated levels of amyloid beta-peptide in the brain and cognitive decline. Jama. 2000;283:1571–7. doi: 10.1001/jama.283.12.1571. [DOI] [PubMed] [Google Scholar]
- 56.Davies G, Harris SE, Reynolds CA, Payton A, Knight HM, Liewald DC, et al. A genome-wide association study implicates the APOE locus in nonpathological cognitive ageing. Mol Psychiatry. 2014;19:76–87. doi: 10.1038/mp.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Omoumi A, Fok A, Greenwood T, Sadovnick AD, Feldman HH, Hsiung G-YR. Evaluation of late-onset Alzheimer disease genetic susceptibility risks in a Canadian population. Neurobiol Aging. 2014;35:936, e5–12. doi: 10.1016/j.neurobiolaging.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 58.Hayden KM, McEvoy JM, Linnertz C, Attix D, Kuchibhatla M, Saunders AM, et al. A homopolymer polymorphism in the TOMM40 gene contributes to cognitive performance in aging. Alzheimers Dement. 2012;8:381–8. doi: 10.1016/j.jalz.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Potkin SG, Guffanti G, Lakatos A, Turner Ja, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PLoS One. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jun G, Vardarajan BN, Buros J, Yu C-E, Hawk MV, Dombroski BA, et al. Comprehensive search for Alzheimer disease susceptibility loci in the APOE region. Arch Neurol. 2012;69:1270–9. doi: 10.1001/archneurol.2012.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cruchaga C, Nowotny P, Kauwe JSK, Ridge PG, Mayo K, Bertelsen S, et al. Association and expression analyses with single-nucleotide polymorphisms in TOMM40 in Alzheimer disease. Arch Neurol. 2011;68:1013–9. doi: 10.1001/archneurol.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saunders AM, Schmader K, Breitner JC, Benson MD, Brown WT, Goldfarb L, et al. Apolipoprotein E epsilon 4 allele distributions in late-onset Alzheimer’s disease and in other amyloid-forming diseases. Lancet (London, England) 1993;342:710–1. doi: 10.1016/0140-6736(93)91709-u. [DOI] [PubMed] [Google Scholar]
- 63.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science (80- ) 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 64.Tanzi RE, Bertram L. New frontiers in Alzheimer’s disease genetics. Neuron. 2001;32:181–4. doi: 10.1016/s0896-6273(01)00476-7. [DOI] [PubMed] [Google Scholar]
- 65.Maruszak A, Canter JA, Styczyńska M, Żekanowski C, Barcikowska M. Mitochondrial haplogroup H and Alzheimer’s disease—Is there a connection? Neurobiol Aging. 2009;30:1749–55. doi: 10.1016/j.neurobiolaging.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 66.Bookheimer SY, Strojwas MH, Cohen MS, Saunders AM, Pericak-Vance MA, Mazziotta JC, et al. Patterns of brain activation in people at risk for Alzheimer’s disease. N Engl J Med. 2000;343:450–6. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donix M, Burggren AC, Suthana N a, Siddarth P, Ekstrom AD, Krupa AK, et al. Longitudinal changes in medial temporal cortical thickness in normal subjects with the APOE-4 polymorphism. Neuroimage. 2010;53:37–43. doi: 10.1016/j.neuroimage.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zintl M, Schmitz G, Hajak G, Klunemann H-H. ApoE Genotype and Family History in Patients With Dementia and Cognitively Intact Spousal Controls. Am J Alzheimers Dis Other Demen. 2009;24:349–52. doi: 10.1177/1533317509333040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, et al. Preclinical evidence of Alzheimer’s disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N Engl J Med. 1996;334:752–8. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- 70.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. Jama. 1997;278:1349–56. [PubMed] [Google Scholar]
- 71.Roses AD, Lutz MW, Saunders AM, Goldgaber D, Saul R, Sundseth SS, et al. African-American TOMM40’523-APOE haplotypes are admixture of West African and Caucasian alleles. Alzheimer’s Dement. 2014;10:592–601. e2. doi: 10.1016/j.jalz.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 72.Harrison TM, Mahmood Z, Lau EP, Karacozoff AM, Burggren AC, Small GW, et al. An Alzheimers Disease Genetic Risk Score Predicts Longitudinal Thinning of Hippocampal Complex Subregions in Healthy Older Adults. eNeuro. 2016;3 doi: 10.1523/ENEURO.0098-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]