To the Editor
Phosphoglucomutase 3 (PGM3) deficiency is an autosomal recessive syndrome with immunologic phenotypes ranging from a hyper-IgE syndrome to severe combined immunodeficiency (SCID). To date, PGM3 deficiency has been described in twenty-nine individuals from thirteen families, with the majority (23 individuals from 9 families) presenting with a hyper-IgE phenotype (1–5). Hypomorphic PGM3 mutations result in altered glycan expression, making this deficiency a congenital disorder of glycosylation (CDG). Among individuals with partially preserved immune function and associated serum IgE elevations, the phenotype is complex and includes severe atopic disease, Th17-associated autoimmunity, and infectious susceptibility, in addition to connective tissue and neurologic abnormalities. PGM3 converts N-acetylglucosamine-6-phosphate (GlcNAc-6-P) to GlcNAc-1-P. The fundamental defect in PGM3 deficiency appears to arise from inadequate production of GlcNAc-1-P. The resulting reduction in substrate for the subsequent enzymatic step in the hexosamine biosynthetic pathway causes a secondary deficiency of uracil diphosphate (UDP)-GlcNAc in primary dermal fibroblasts (1) and in peripheral blood mononuclear cells (PBMCs) (Fig 1A).
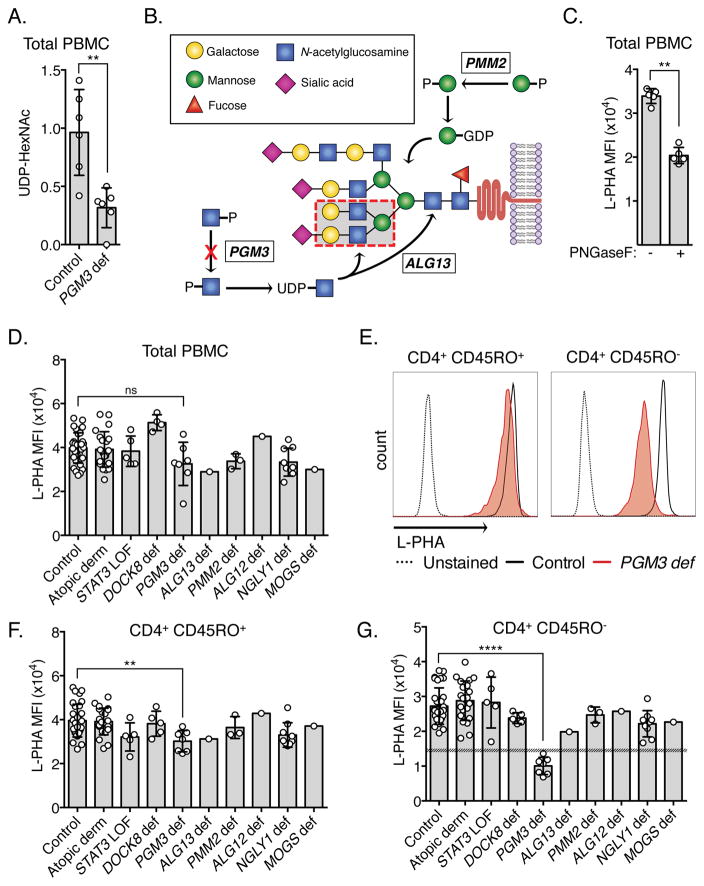
Figure 1. Reduced naïve CD4 N-glycan complexity identifies PGM3 deficiency from other congenital glycosylation and hyper-IgE disorders.
(A) Intracellular UDP-HexNAc in PGM3 deficient (n = 6) and control PBMCs (n = 6). Unpaired t-test; **P<0.01. (B) Schematic of complex N-glycan formation with L-PHA binding site shown as a red dashed box. (C) L-PHA staining intensity of PBMCs before and after PNGAseF treatment (n = 5). Mann-Whitney test; **P<0.01. (D) L-PHA staining intensity of PBMCs from controls (n = 32), individuals with atopic dermatitis (AD, n = 22), DOCK8 deficiency (n = 4), STAT3 loss-of-function (STAT3 LOF, n = 5), PGM3 deficiency (n = 7), ALG13 deficiency (n = 1), PMM2 deficiency (n = 3), ALG12 deficiency (n = 1), NGLY1 deficiency (n = 9), and MOGS deficiency (n = 1). Representative (E) and combined (F, G) L-PHA staining of CD45RO+ and CD45RO− CD4+ cells. Shaded area in (G) delineates the upper 99% confidence interval (CI) of PGM3 deficient cells and the closest lower 99% CI (STAT3 LOF). Mann-Whitney test; **P<0.01, ****P<0.0001.
UDP-GlcNAc is fundamental to normal N-linked glycosylation of proteins. It is incorporated into the core structure of all N-linked glycoproteins and is a key component of branching structures of complex N-glycans (Fig 1B). N-glycan complexity has been shown to have dramatic effects on T cell function including altering the activation threshold of the TCR and promoting CTLA-4-mediated growth arrest (6, 7). Diminished intracellular concentrations of UDP-GlcNAc seen in PGM3 deficiency have been associated with altered N-glycosylation of serum proteins but alterations in cellular glycan expression have not yet been characterized (1).
Complete loss of function in PGM3 is embryonically lethal in mice (8). To date, all individuals identified with PGM3 deficiency express mutant protein, thus limiting the ability to screen for this disorder using conventional protein detection assays. Furthermore, functional enzymatic activity assays are complicated and require specialized expertise. To get around these obstacles, we exploited predicted glycan differences to develop a lectin-based flow cytometric assay that can quantify expression of complex branched N-glycans and successfully identify PGM3 deficient individuals (see the Online Repository Methods for a complete description of assays).
N-glycan branching was measured using a fluorescein-conjugated plant lectin, L-phytohemagglutinin (L-PHA), which has specificity for 2,6-branched complex N-glycans with bisecting N-acetylglucosamine (GlcNAc) (Fig 1B) (9). The dependence of L-PHA staining on N-glycan expression was confirmed by treatment of peripheral blood mononuclear cells (PBMCs) with PNGaseF – an enzyme that hydrolyzes nearly all N-glycans – resulting in a significant reduction in staining intensity (Fig 1C).
We examined PBMCs isolated from PGM3 deficient individuals and controls, as well as from other disorders with significant elevations in IgE (atopic dermatitis, DOCK8 deficiency, and dominant negative STAT3 mutations resulting in loss of function) and from patients with congenital disorders of glycosylation (CDGs) (caused by autosomal recessive loss of function mutations in ALG13, PMM2, ALG12, or MOGS) and deglycosylation (NGLY1). Patient ages, allergic disease prevalence, serum IgE, and absolute eosinophil counts are listed in Table I. Phosphomannomutase-2 (PMM2) facilitates conversion of phosphorylated mannose in a manner similar to PGM3, potentially limiting mannose substrate for all N-glycans, but not specifically limiting N-glycan branching or complexity (E1). Asparagine-linked glycosylation homologs 13 and 12 (ALG13/ALG12) assist in the addition of sugars to the N-glycan core structure, and therefore are predicted to affect total N-glycosylation expression (E2). Mannosyl-oligosaccharide glucosidase (MOGS) and N-glycanase 1 (NGLY1) are glycosidases involved in glycoprotein quality control, and defects may result in accumulation of high-mannose type glycans for the former (E3, E4).
Table I.
Clinical and laboratory data from screened individuals
| Diagnosis (N) | Age (yr) Median, (range) | Serum IgE (IU/mL) Median, (range) | Eosinophils (cells/μL) Median, (range) | Allergy N (%) | Clinical Features (N) |
|---|---|---|---|---|---|
| PGM3 deficiency (N = 7) | 33 (4–38) | 27,200 (1,927–35,445) | 900 (200–3,600) | 7 (100%) | AD (7), food allergy (5), asthma (1), S. aureus* (5) |
|
| |||||
| Atopic dermatitis (N = 22) | 16 (6–52) | 20,689 (5,129–185,033) | 1,545 (500–4,550) | 22 (100%) | AD (22), food allergy (20), asthma (18), S. aureus (17) |
| DOCK8 deficiency (N = 4) | 27 (22–31) | 7,408 (1,162–31,403) | 2,735 (560–6,750) | 4 (100%) | AD (4), food allergy (4), asthma (1), |
| STAT3 LOF (N = 5) | 19 (12–57) | 3,670 (3,219–29,611) | 630 (530–2,220) | 5 (100%) | AD (5), food allergy (2), asthma (1), S. aureus (5) |
| ALG13 deficiency (N = 1) | 9 | 9 | 80 | none | - |
| PMM2 deficiency (N = 3) | 4 (3–5) | 8 (0–27) | 110 (0–170) | 2 (100%) | AD (2) - mild |
| ALG12 deficiency (N = 1) | 20 | 2 | 60 | none | - |
| NGLY1 deficiency (N = 9) | 8 (3–18) | 26 (2–92) | 230 (40–1,260) | 1 (11%) | AD (1) - mild |
| MOGS deficiency (N = 1) | 17 | 9 | 300 | 1 (100%) | food allergy (1) – egg, unconfirmed |
Susceptibility to skin and soft tissue infection with Staphylococcus aureus (S. aureus).
Despite having reduced UDP-GlcNAc pools, L-PHA staining in PGM3-deficient PBMCs en masse displayed no discernable difference from controls or other genetic disorders (Fig 1D). However, examining distinct leukocyte populations (for gating strategy see Fig E2), significant but relatively modest reductions were observed in multiple cell populations, most significantly within the CD8+ CD45RO− and CD45RO+ pools, with the notable exception of CD19+ B cells. While statistically significant, L-PHA staining intensity of these PGM3 deficient cell populations overlapped with one or more control population, therefore none of these differences was large enough to discern every individual with PGM3 deficiency clearly from the other disorders (Fig 1E, F; E1). However, closer inspection of CD4+ CD45RO− cells, revealed a marked reduction in N-glycan complexity in PGM3 deficiency compared to all other samples, clearly distinguishing PGM3 deficiency from all other disorders of hyper-IgE as well as all other CDGs tested (Fig 1E, G). These differences appeared specific to detection of 2,6-branching complex N-glycans since staining with concanavalin A (Con A), a lectin which binds alpha-linked mannose residues present on all N-glycans, failed to demonstrate such a defect (Fig E3). Despite the marked reductions seen in CD4+ CD45RO− cells, the complex N-glycan expression in the CD4+ CD45RO+ population appeared relatively unperturbed. We hypothesize the differences seen between these populations is related to metabolic changes that are known to occur with lymphocyte activation.
We have developed a lectin-based flow cytometric assay that can clearly identify PGM3 deficiency by examining extracellular L-PHA staining of CD4+ CD45RO− T cells. It is possible that other glycosylation disorders could result in a similar glycan pattern, and follow-up sequencing remains necessary. However, we predict that similar glycosylation defects in lymphocytes will likely be associated with clinical phenotypes resembling PGM3 deficiency. Thus, this and other lectin-based flow cytometry may also aid in screening for and identifying CDGs associated with immune dysregulation. This assay is easy to perform with low cost, using a lectin conjugate and conjugated antibodies standard to flow cytometry labs, and can help stratify individuals for subsequent targeted or genomic sequencing.
Hematopoietic stem cell transplantation (HSCT) has demonstrated efficacy in PGM3 deficiency-associated SCID. Because of the high morbidity associated with the hyper-IgE phenotype, HSCT may also be considered for this presentation in the future. Lectin-based flow cytometry-screening could aid in early diagnosis without delaying HSCT in this clinical context.
Supplementary Material
Acknowledgments
Funding: This research was supported by the Intramural Research Programs of the NIH, National Cancer Institute, Center for Cancer Research, of the National Institute of Allergy and Infectious Diseases, and of the National Institute for Diabetes and Digestive and Kidney Diseases. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government
We thank the patients, their families, and the healthy volunteers for participating in this study, as well as the clinical staff of the LAD, LCID, and UDP/NHGRI for their efforts and contributions. We also thank Michael Demetriou for sharing his expertise.
Abbreviations used
- PGM3
Phosphoglucomutase-3
- CDG
congenital disorder of glycosylation
- SCID
severe combined immunodeficiency
- GlcNAc
N-acetylglucosamine
- UDP
uracil diphosphate
- HSCT
hematopoietic stem cell transplant
- DOCK8
dedicator of cytokinesis 8
- STAT3
signal transducer and activator of transcription 3
- PMM2
phosphomannomutase-2
- ALG13
asparagine-linked glycosylation homolog 13
- ALG12
asparagine-linked glycosylation homologs 12
- MOGS
mannosyl-oligosaccharide glucosidase
- NGLY1
N-glycanase 1
- L-PHA
L-phytohemagglutinin
- PBMC
peripheral blood mononuclear cell
- MFI
mean fluorescence intensity
- Con A
concanavalin A
Footnotes
Conflicts of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang Y, Yu X, Ichikawa M, Lyons JJ, Datta S, Lamborn IT, et al. Autosomal recessive phosphoglucomutase 3 (PGM3) mutations link glycosylation defects to atopy, immune deficiency, autoimmunity, and neurocognitive impairment. J Allergy Clin Immunol. 2014;133(5):1400–9. 9 e1–5. doi: 10.1016/j.jaci.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stray-Pedersen A, Backe PH, Sorte HS, Morkrid L, Chokshi NY, Erichsen HC, et al. PGM3 mutations cause a congenital disorder of glycosylation with severe immunodeficiency and skeletal dysplasia. Am J Hum Genet. 2014;95(1):96–107. doi: 10.1016/j.ajhg.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sassi A, Lazaroski S, Wu G, Haslam SM, Fliegauf M, Mellouli F, et al. Hypomorphic homozygous mutations in phosphoglucomutase 3 (PGM3) impair immunity and increase serum IgE levels. J Allergy Clin Immunol. 2014;133(5):1410–9. 9 e1–13. doi: 10.1016/j.jaci.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernth-Jensen JM, Holm M, Christiansen M. Neonatal-onset T(−)B(−)NK(+) severe combined immunodeficiency and neutropenia caused by mutated phosphoglucomutase 3. J Allergy Clin Immunol. 2016;137(1):321–4. doi: 10.1016/j.jaci.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Ali M, Ben-Mustapha I, Mekki N, Mellouli F, Khemiri M, Bejaoui M, et al. A founder mutation in PGM3 is responsible for a severe form of hyper-IgE like syndrome in Tunisian patients. J Clin Immunol. 2016;36(3):235–334. [Google Scholar]
- 6.Demetriou M, Granovsky M, Quaggin S, Dennis JW. Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature. 2001;409(6821):733–9. doi: 10.1038/35055582. [DOI] [PubMed] [Google Scholar]
- 7.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129(1):123–34. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 8.Greig KT, Antonchuk J, Metcalf D, Morgan PO, Krebs DL, Zhang JG, et al. Agm1/Pgm3-mediated sugar nucleotide synthesis is essential for hematopoiesis and development. Mol Cell Biol. 2007;27(16):5849–59. doi: 10.1128/MCB.00802-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cummings RD, Kornfeld S. Characterization of the structural determinants required for the high affinity interaction of asparagine-linked oligosaccharides with immobilized Phaseolus vulgaris leukoagglutinating and erythroagglutinating lectins. J Biol Chem. 1982;257(19):11230–4. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.