Abstract
Purpose
Expression of the ΔN isoform of p63 (ΔNp63) is a diagnostic marker highly specific for lung squamous cell carcinoma (SCC). We previously found that Syntaxin Binding Protein 4 (STXBP4) regulates ΔNp63 ubiquitination, suggesting that STXBP4 may also be a SCC biomarker. To address this issue, we investigated the role of STXBP4 expression in SCC biology and the impact of STXBP4 expression on SCC prognosis.
Experimental design
We carried out a clinicopathological analysis of STXBP4 expression in 87 lung SCC patients. Whole transcriptome analysis using RNA-seq was performed in STXBP4-positive and STXBP4-negative tumors of lung SCC. Soft agar assay and xenograft assay were performed using overexpressing or knockdown SCC cells.
Results
Significantly higher levels of STXBP4 expression were correlated with accumulations of ΔNp63 in clinical lung SCC specimens (Spearman’s rank correlation ρ=0.219). Notably, STXBP4-positive tumors correlated with three important clinical parameters: T factor (P<0.001), disease stage (P=0.030) and pleural involvement (P=0.028). Whole transcriptome sequencing followed by pathway analysis indicated that STXBP4 is involved in functional gene networks that regulate cell growth, proliferation, cell death and survival in cancer. Platelet-Derived Growth Factor Receptor alpha (PDGFRα) was a key downstream mediator of STXBP4 function. In line with this, shRNA mediated STXBP4 and PDGFRA knockdown suppressed tumor growth in soft agar and xenograft assays.
Conclusions
STXBP4 plays a crucial role in driving SCC growth and is an independent prognostic factor for predicting worse outcome in lung SCC. These data suggest that STXBP4 is a relevant therapeutic target for patients with lung SCC.
Keywords: TP63, STXBP4, PDGFRα, Lung Squamous Cell Carcinoma (Lung SCC), p40
Introduction
Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all cases of lung cancer, and is mainly sub-classified into adenocarcinoma (AC) and squamous cell carcinoma (SCC) (1). Current treatment strategies for NSCLC include chemotherapy, depending on the histological tumor type, and targeted agents for patients whose tumors carry a specific targetable genomic alteration. Although there have been significant advances in the treatment of lung SCC, further improvements in prognosis are dependent upon the identification of SCC specific molecules or genomic alterations that can be used as therapeutic biomarkers and/or targets (2).
Several immunohistochemical markers have been investigated for their utility in distinguishing lung SCC from lung AC, including TTF-1, napsin A and CK5/6 and the ΔN isoform of p63 (ΔNp63) (3-5). The latter is a highly specific marker for lung SCC and genomic regions containing the TP63 gene are frequently amplified in a variety of SCCs, including lung, head and neck, bladder and cervical cancers (4, 6-8). Although these findings suggest that ΔNp63 is a lung SCC oncogene, the pathologic relevance of p63 in tumorigenesis remains unclear (9, 10).
Alternative splicing of the TP63 gene generates transcripts encoding two opposing classes of proteins: one containing the transactivation domain (TAp63) and the other lacking the domain (ΔNp63) (11-13). Early studies showed that ΔNp63 acts as a dominant-negative transcriptional repressor to inhibit p53- or TAp63-mediated transcription in vitro and in vivo, consistent with a potential oncogenic role for the ΔNp63 isoform (12, 14). However, the ΔNp63 isoform also has transcriptional activity that is independent of the second transactivation domain (15).
ΔNp63 is regulated in a coordinated manner by two scaffold proteins, Syntaxin Binding Protein 4 (STXBP4) and Receptor of activated kinase C1 (RACK1; encoded by the GNB2L1), which bind to ΔNp63 (16, 17). STXBP4, originally identified as a glucose transporter, is localized on human chromosome 17q22 and plays a role in the translocation of transport vesicles from the cytoplasm to the plasma membrane (18, 19). While ΔNp63 plays a role in maintaining the viability and proliferative capacity of basal epithelial cells, STXBP4 is a positive regulator of ΔNp63 stability and is also crucial for keratinocyte proliferation (16, 20).
In this report, we focused on STXBP4 and its oncogenic function in lung SCC, with a particular emphasis on the interactions between STXBP4 and p63. We also addressed the relevance of STXBP4 expression to patient prognosis. Initially, we assessed the expression of STXBP4 and ΔNp63 in SCC tumors by immunohistochemistry, and found that positive STXBP4 expression signified worse Overall Survival (OS) and Progression-Free Survival (PFS). We further performed a genome-wide transcriptome analysis (RNA-seq) using Next Generation Sequencing (NGS) and found that Platelet-Derived Growth Factor Receptor α (PDGFRα) was positively correlated with the expression of STXBP4. In line with this, shRNA mediated depletion of PDGFRα suppressed the growth of a lung SCC cell line in soft agar and xenograft tumor assays, similar to the findings obtained when the expression of STXBP4 or ΔNp63 were knocked down. Taken together, our data address the physiological role and diagnostic potential of STXBP4 in lung SCC, and suggest that PDGFRα may be a key mediator of STXBP4-mediated oncogenic activity.
Materials and Methods
Cell Culture
The human lung SCC cell lines, RERF-LC-Sq1 and EBC-1 were obtained from the Japanese Collection of Research Bioresources (JCRB). The cell lines were last authenticated by short tandem repeat (STR) analysis on December 22, 2015 (RERF-LC-Sq1), or on June 10, 2016 (EBC-1). RERF-LC-Sq1 cells were cultured in RPMI1640 with 10% fetal bovine serum (FBS), and EBC-1 cells were cultured in Eagle’s minimal essential medium (EMEM) with 10% FBS at 37°C in a 5% CO2 incubator.
Patients
Human tissue specimens were surgically resected from 87 lung SCC patients at Gunma University Hospital and its affiliated hospitals between August 2003 and December 2010 (21). The main eligibility criteria were as follows: age 20 to 85 years; performance status based on Eastern Cooperative Oncology Group ≤ 2; estimated life expectancy ≥ 3 months; adequate hepatic, cardiac, renal, and bone marrow functions. The study was conducted in compliance with the Declaration of Helsinki, and was approved by the Institutional Review Board of the participating hospitals and institutions. All patients provided written informed consent before registration. Tumor samples were stored at −80°C until use.
Immunohistochemistry
Immunohistochemical analysis was performed on formalin-fixed and paraffin-embedded SCC sections. The sections were de-paraffinized, blocked in PBS containing 5% FBS for 1 hr, and incubated overnight with diluted primary antibodies at 4°C in a humidified chamber. Staining reactions were developed using Vectastain universal ABC Kit (Vector Laboratories, Burlingame, CA) and then DAB Kit (Vector Laboratories) for immunohistochemistry. Meyer’s hematoxylin (IHC world, Woodstock, MD) was used as a nuclear counterstain. STXBP4, p63, ΔNp63 levels were assessed by immunohistochemical staining and scored using a semi-quantitative method: 1 ≤ 10%, 2 = 10-25%, 3 = 25-50%, 4 = 51-75% and 5 ≥ 75% of positive cells. The tumors in which the stained cancer cells were scored as 3, 4, or 5 were defined as STXBP4-positive; 1 and 2 were defined as STXBP4-negative.
We used antibodies specific for p63 (4A4) (Santa Cruz Biotechnology, Dallas, TX) and STXBP4 (Abcam, Cambridge, MA). Rabbit polyclonal ΔNp63 antibody was previously described (16). CD147 (Santa Cruz Biotechnology) and mTOR (Cell Signaling Technology, Danvers, MA) immunohistochemical staining was performed according to the procedures described in a previous report (22). The following diluted antibodies were used: p63 (1:100 dilution); ΔNp63 (1:100 dilution); STXBP4 (1:100 dilution); CD147 (1:100 dilution); mTOR (1:80 dilution). Highly cellular areas of the sections were evaluated for Ki-67 expression. All epithelial cells with nuclear staining of any intensity were defined as high expression. Approximately 1,000 nuclei were counted on each slide. Proliferative activity was assessed as the percentage of Ki-67-stained nuclei (Ki-67 labeling index) in the sample. The median value of the Ki-67 labeling index was evaluated, and tumor cells with greater than the median value were defined as high expressors. The sections were assessed using light microscopy in a blind fashion by at least two of the authors.
Plasmids and antisense oligonucleotides
Human cDNAs encoding FLAG-tagged or HA-tagged ΔNp63α, STXBP4 and PDGFRα were cloned into the LPCX retroviral expression vector (Takara Bio, Shiga, Japan). The sequences of the above constructs were verified using DNA sequencing. For siRNA experiments, 19 nucleotide siRNA duplexes with 3’dTdT overhangs were synthesized by Dharmacon (GE Dharmacon, Lafayette, CO). The siRNA oligonucleotide sequences for Luciferase control (LUC), ΔNp63, STXBP4 and PDGFRα, are described in the Supplementary Information. For siRNA transfection, RERF-LC-Sq1 or EBC-1 cells were transfected with 50 nM siRNA using DharmaFECT 1 siRNA transfection reagent (GE Dharmacon) according to the manufacturer’s instruction. For shRNA experiments, the shRNAs for Luciferase (LUC), ΔNp63, STXBP4 and PDGFRα oligonucleotides were cloned into the pLKO.1 puro lentivirus expression vector between Age I site and Eco RI site. The sequences of the above constructs were verified using DNA sequencing. The target sequences of the shRNA oligonucleotides are described in Supplementary Information.
Immunoblotting analysis
Immunoblotting analysis was performed as previously described (23). In short, cells were solubilized with lysis buffer (20 mM sodium phosphate [pH 7.0], 125 mM NaCl, 30 mM sodium pyrophosphate, 0.1% NP-40, 5 mM EDTA, 10 mM sodium fluoride, 0.1 mM Na3VO4 and 1 mM phenylmethylsulfonyl fluoride) supplemented with Complete protease inhibitor cocktail (Roche, Penzberg, Germany), and homogenized by passage through a 20G needle. The eluates were then concentrated and separated by SDS-PAGE. Transfer to nitrocellulose membranes and screening using rabbit polyclonal antibodies for ΔNp63 and STXBP4 were carried out as previously described (24). We used antibodies specific for p63 (4A4), ΔNp63α, STXBP4, Phospho-PDGFRα (Tyr849) (Cell Signaling Technology), PDGFRα (Abcam), phospho-p38MAPK (Thr180/Tyr182) (Cell Signaling Technology), p38MAPK (Cell Signaling Technology) and β-Actin (Sigma-Aldrich, St. Louis, MO).
Genome-wide transcriptome analysis (RNA-seq) and real-time RT-PCR
Total RNA was prepared from surgically resected samples using a RNeasy Mini kit (Qiagen, Hilden, Germany) after homogenizing with Mixer Mill MM400 (Qiagen). RNA quality was assessed using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). High quality RNA (RNA integrity numbers > 7.0) from six STXBP4-positive and six STXBP4-negative samples were used for genome-wide transcriptome analysis (RNA-seq experiments). mRNAs were captured using a Dynabeads mRNA DIRECT Micro Purification Kit (Thermo Fisher Scientific, Waltham, MA). The mRNA was then used to generate sequencing libraries of barcoded fragments using the Ion Total RNA-Seq Kit v2 (Thermo Fisher Scientific) following the manufacturer’s instructions. Libraries were sequenced on an Ion Proton System using four libraries per Ion PI Chip v2, Ion PI Template OT2 200 kit v3 and Ion PI Sequencing 200 kit v3 (Thermo Fisher Scientific). BAM files generated by the Ion Proton System were converted to FASTQ files using bam2fastq software (v1.1.0, https://gsl.hudsonalpha.org/information/software/bam2fastq), and reads shorter than 21 nucleotides were removed. Quantitation of each gene was undertaken as previously described (25). Briefly, the reads were aligned to the UCSC reference human genome 19 (hg19) using a combination of Tophat2 (v2.0.11, http://ccb.jhu.edu/software/tophat/index.shtml), and the Bowtie2 (2.2.2.0, http://bowtie-bio.sourceforge.net/index.shtml) pipelines. The read counts were obtained using Partek Genomics Suite software (http://www.partek.com/). Differentially expressed genes were detected using edgeR software (26) and genes with a FDR < 0.50 (p < 0.01) were analyzed by Ingenuity Pathway Analysis (Qiagen).
For real-time RT-PCR, relative RNA quantities were measured by Universal Probe Library set (Roche) with KAPA Master mix (KAPA Biosystems, Wilmington, MA) on a StepOne real-time PCR system (Thermo Fisher Scientific). The Universal Probe Library Human ACTB Gene Assay (Roche) was used for an endogenous normalization control. Sequence detection software was utilized for data analysis, and relative fold induction was determined by the comparative threshold cycle method using standard curves, which were generated by plotting the observed Ct values against the standard dilutions of a positive control sample. In all experiments, the average of three independent reactions is shown with error bars indicating standard deviation. Gene expression data were downloaded from the Gene Expression Omnibus database (GSE84339).
Subcutaneous xenografts
A total of 5 × 106 lentivirally transduced or retrovirally expressed cells were injected subcutaneously into nude mice (BALB/c-nu/nu, CLEA Japan, Tokyo, Japan) and tumor size was measured after 20 days (RERF-LC-Sq1) or 14 days (EBC-1). All animal procedures were performed with the approval of the Animal Ethics Committee of Gunma University.
Anchorage-independent growth
RERF-LC-Sq1 cells were transduced with lentiviruses carrying shRNAs for Luciferase (LUC), ΔNp63, STXBP4 or PDGFRα. For soft agar assays, the cells were grown in triplicate for 12 days, after which anchorage-independent growth was quantified with a CytoSelect-96 kit (Cell Biolabs, San Diego, CA).
Statistical analysis
Probability values (P-value) < 0.05 indicated a statistically significant difference. Fisher’s exact test was used to examine the association of two categorical variables. The correlation between different variables was analyzed using the nonparametric Spearman’s rank test. Follow-up for the 87 patients was conducted using the patient medical records. The Kaplan-Meier method was used to estimate survival as a function of time, and survival differences were analyzed by the log-rank test. The day of surgery was defined as the starting day for measuring postoperative survival. OS was determined as the time from tumor resection to death from any cause. PFS was defined as the time between tumor resection and first disease progression or death. Multivariate analyses were performed using a stepwise Cox proportional hazards model to identify independent prognostic factors. Statistical analysis was performed using JMP 8 (SAS) software.
Results
Survival outcomes according to STXBP4 and p63 expression
The clinicopathological features of the 87 patients included in this study are shown in Table 1. The median age of the patients was 72 (range 56 to 84), the majority of patients were male (92.0 %), and former or current smokers (98.9 %). All patients received radical surgery with evidence of pathological stage IA/B in 54.0 %, stage IIA/B in 26.4 %, and stage IIIA in 18.4 % of patients. Pleural involvement, lymphatic permeation and venous invasion were observed in 41 patients (47.1%), 47 patients (54.0%), and 40 patients (46.0%), respectively.
Table 1.
Patient's demographics according to STXBP4 expression
| Variables | STXBP4 |
ΔNp63 |
p63 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| High No. (%) |
Low No. (%) |
P-value | Positive No. (%) |
Negative No. (%) |
P-value | Positive No. (%) |
Negative No. (%) |
P-value | |
| Age | 0.42 | 0.051 | 0.22 | ||||||
| ≤ 65yr | 9 (10.3) | 9 (10.3) | 10 (11.5) | 8 ( 9.2) | 16 (18.4) | 2 ( 2.3) | |||
| > 65 yr | 43 (50.5) | 26 (29.9) | 20 (23.0) | 49 (56.3) | 51 (58.6) | 18 (20.7) | |||
| Sex | 0.70 | 0.69 | < 0.99 | ||||||
|
| |||||||||
| Male | 47 (54.0) | 33 (37.9) | 27 (31.0) | 53 (60.9) | 61 (70.1) | 19 (21.8) | |||
| Female | 5 ( 5.7) | 2 ( 2.3) | 3 ( 3.4) | 4 ( 4.6) | 6 ( 4.6) | 1 ( 3.4) | |||
|
| |||||||||
| Differentiation | 0.080 | 0.034 | 0.24 | ||||||
| Well or Moderately | 43 (49.4) | 23 (26.4) | 27 (31.0) | 39 (44.8) | 53 (60.9) | 13 (15.0) | |||
| Poorly | 9 (10.3) | 12 (13.8) | 3 ( 3.4) | 18 (20.7) | 14 (16.1) | 7 ( 8.0) | |||
|
| |||||||||
| T factor | < 0.001 | 0.33 | < 0.001 | ||||||
| T1 | 8 (9.2) | 19 (21.8) | 7 ( 8.0) | 20 (23.0) | 27 (31.0) | 0 ( 0.0) | |||
| T2-3 | 44 (50.6) | 16 (18.4) | 23 (26.4) | 37 (42.5) | 40 (46.0) | 20 (23.0) | |||
|
| |||||||||
| N factor | > 1 | > 1 | 0.28 | ||||||
| N0 | 36 (41.4) | 25 (28.7) | 21 (24.1) | 40 (46.0) | 49 (56.3) | 12 (13.8) | |||
| N1-2 | 16 (18.4) | 10 (11.5) | 9 (10.3) | 17 (19.5) | 18 (20.7) | 8 ( 9.2) | |||
|
| |||||||||
| Disease stage | 0.030 | 0.37 | 0.20 | ||||||
| I | 23 (26.4) | 24 (27.6) | 14 (16.1) | 33 (37.9) | 39 (44.8) | 8 ( 9.2) | |||
| II-III | 29 (33.3) | 11 (12.6) | 16 (18.4) | 24 (27.6) | 28 (32.2) | 12 (13.8) | |||
|
| |||||||||
| Pleural Involvement | 0.028 | 0.66 | 0.29 | ||||||
| Positive | 30 (34.5) | 11 (12.6) | 13 (14.9) | 28 (32.2) | 30 (34.5) | 11 (13.8) | |||
| Negative | 22 (25.3) | 24 (27.6) | 17 (19.5) | 29 (33.3) | 37 (42.5) | 9 (10.3) | |||
|
| |||||||||
| Lymphatic permeation | 0.83 | > 1 | 0.61 | ||||||
| Positive | 29 (33.3) | 18 (20.7) | 16 (18.4) | 31 (35.6) | 35 (40.2) | 12 (13.8) | |||
| Negative | 23 (26.4) | 17 (19.5) | 14 (16.1) | 26 (29.9) | 32 (36.8) | 8 ( 9.2) | |||
|
| |||||||||
| Vascular invasion | 0.39 | > 1 | > 1 | ||||||
| Positive | 26 (29.9) | 14 (16.1) | 14 (16.1) | 26 (29.9) | 31 (35.6) | 9 (10.3) | |||
| Negative | 26 (29.9) | 21 (24.1) | 16 (18.4) | 31 (35.6) | 36 (41.4) | 11 (12.6) | |||
P-values were obtained by Fisher's exact test.
Clinical stage at the time of initial diagnosis was determined according to the seventh edition of General Rule for Clinical and Pathological Record of Lung Cancer (2010), the Japan Lung Cancer Society.
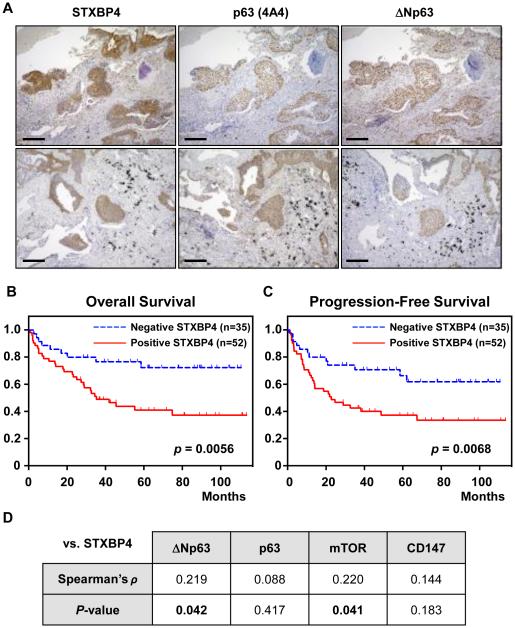
Frequently, lung SCCs exhibit simultaneous up-regulation of both TAp63 and ΔNp63, and ΔNp63 in particular, is a putative diagnostic marker for pulmonary SCC (10). To address the clinical significance of STXBP4 expression, we investigated whether high expression of this gene correlates with ΔNp63 status. We found that 59.8 % (52/87) of all patients were STXBP4-positive, and STXBP4 expression was detected in those tumors that showed an accumulation of p63 (Fig. 1A).
Figure 1.
STXBP4 expression is correlated with p63 expression and poor prognosis in lung SCC. (A) Representative immunohistochemical staining of a lung SCC. STXBP4 immunostaining demonstrates a nuclear and cytoplasmic pattern with a score of 5. Scale bars are 200 μm. (B, C) Kaplan-Meier analysis of Overall Survival (OS) and Progression Free Survival (PFS) defined according to STXBP4 expression. A statistically significant difference in OS and PFS was observed between the STXBP4-positive patients and those with low STXBP4 expression [OS, p = 0.0056(A); PFS, p = 0.0068 (B)]. P-values were obtained by log-rank test. (D) Spearman’s rank correlation was performed based on the expression levels of STXBP4 and ΔNp63.
Statistical correlation analysis between STXBP4 expression and clinicopathological features revealed that pathological local tumor factor stage (Disease stage), pathological tumor-node-metastasis (TNM) stage, and pleural involvement as a local invasion factor, were correlated with STXBP4-positivity (Table 1). On the other hand, age, gender, and pathological differentiation were not correlated with STXBP4 expression. The expression of ΔNp63 was significantly correlated with age and differentiation status, but not with clinical stage of disease. Remarkably, patient survival was significantly associated with T factor, disease stage, pleural involvement, and STXBP4 expression, as assessed by univariate analysis. Multivariate analysis confirmed that STXBP4 expression and disease stage were independent prognostic factors in lung SCC patients with poor OS and PFS (Table 2).
Table 2.
Univariate and multivariate survival analysis in all patients
| Variables | Overall Survival |
Progression-Free Survival |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate |
Multivariate |
Univariate |
Multivariate |
||||||
| 5-yrs rate (%) |
p-value | HR (95% CI) |
p-value | 5-yrs rate (%) |
p- value |
HR (95% CI) |
p-value | ||
| Age | 0.853 | 0.504 | 0.965 | 0.452 | |||||
| ≤ 65yr | 49 | 1.32 (0.60 - 3.21) |
44 | 1.34 (0.64 - 3.06) |
|||||
| > 65 yr | 55 | 51 | |||||||
|
| |||||||||
| Sex | 0.579 | 0.690 | 0.215 | 1.90 | |||||
| Male | 55 | 1.25 (0.36 - 3.26) |
51 | 1.90 (0.63 - 4.55) |
|||||
| Female | 38 | 21 | |||||||
|
| |||||||||
| Differentiation | 0.206 | 0.447 | |||||||
| WD | 68 | 54 | |||||||
| MD/PD | 50 | 48 | |||||||
|
| |||||||||
| T factor | 0.011 | 0.011 | |||||||
| T1 | 75 | 75 | |||||||
| T2-3 | 44 | 44 | |||||||
|
| |||||||||
| N factor | 0.207 | 0.011 | |||||||
| No | 57 | 56 | |||||||
| N1-2 | 43 | 29 | |||||||
|
| |||||||||
| Disease Stage | 0.006 | 0.033 | 0.002 | 0.002 | |||||
| I | 70 | 2.17 (1.06 - 4.24) |
67 | 2.94 (1.49 - 5.67) |
|||||
| II - III | 42 | 29 | |||||||
|
| |||||||||
| Lymphatic permeation |
0.448 | 0.182 | |||||||
| Positive | 50 | 44 | |||||||
| Negative | 62 | 54 | |||||||
|
| |||||||||
| Vascular invasion |
0.239 | 0.365 | |||||||
| Positive | 49 | 48 | |||||||
| Negative | 57 | 51 | |||||||
|
| |||||||||
| Pleural involvement |
0.014 | 0.049 | |||||||
| Positive | 41 | 41 | |||||||
| Negative | 65 | 56 | |||||||
|
| |||||||||
| STXBP4 | 0.005 | 0.028 | 0.006 | 0.040 | |||||
| Positive | 41 | 37 | |||||||
| Negative | 72 | 2.24 ( 1.09 - 5.10) |
66 | 2.02 (1.03 - 4.24) |
|||||
|
| |||||||||
| ΔNp63 | 0.362 | 0.458 | |||||||
| Positive | 51 | 45 | |||||||
| Negative | 61 | 54 | |||||||
|
| |||||||||
| p63 | 0.523 | 0.111 | |||||||
| Positive | 53 | 51 | |||||||
| Negative | 54 | 36 | |||||||
Kaplan-Meier analysis of OS and PFS according to STXBP4 expression revealed a statistically significant difference in OS and PFS between the patients who were STXBP4-positive compared with those who were STXBP4-negative (Fig. 1B and 1C). The five-year survival rate and median survival time for all patients were 50.2 % and 38.3 months (0.75 to 111.5 months), respectively. The median PFS and OS (21.2 months versus 52.2 months; P < 0.05) were shorter in STXBP4-positive patients compared with STXBP4-negative patients (Fig. 1B and 1C). STXBP4-positive patients showed poor OS (log-rank P < 0.01) and PFS (log-rank P < 0.01) compared with those with STXBP4-negative patients. Interestingly, STXBP4 levels significantly predicted outcome in patients with tumors expressing high ΔNp63 levels (Supplementary Fig. 1A and 1B), while the complementary analysis showed that ΔNp63 levels did not significantly predict OS (P = 0.35) and PFS (P = 0.54) in the STXBP4 high-expressing group (Supplementary Fig. 1C and 1D). Thus, these results indicate that STXBP4 could be an independent prognostic marker for predicting poor outcome in lung SCC.
We observed significantly higher levels of STXBP4 expression in those tumors that showed an accumulation of ΔNp63 (Spearman’s ρ = 0.219; P < 0.05), while among all p63 isoforms, no significant correlations were observed (P > 0.5) (Fig. 1D). Interestingly, high mammalian target of rapamycin (mTOR), a major controller of growth and is often deregulated in cancer (27), was significantly correlated with STXBP4-positivity (Spearman’s ρ = 0.220; P < 0.05), while other tumor markers, including CD147, a member of the immunoglobulin superfamily involved in angiogenesis (P < 0.5), and Ki-67, a general marker for cell division (P < 0.1), were not significantly correlated (Fig. 1D).
Transcriptional profiling and functional screening to identify possible downstream mediators of STXBP4
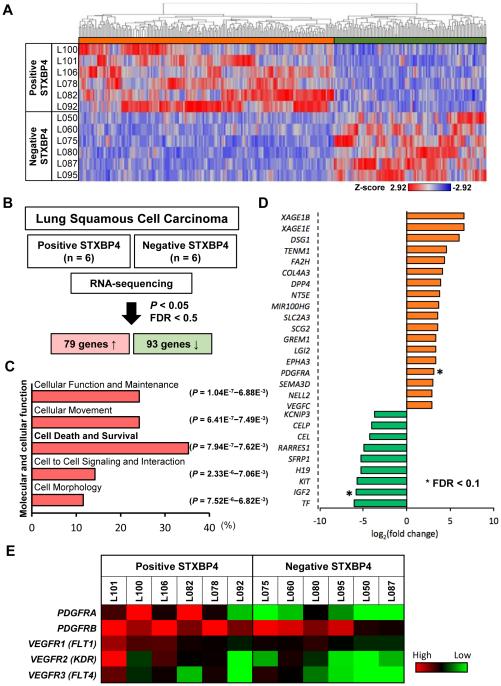
Hierarchical cluster analysis after alignment of a total of 15,346 genes to the reference sequence, showed that STXBP4-positive and STXBP4-negative tumors had distinctly different gene expression profiles (Fig. 2A). Among the differentially expressed genes (P-value < 0.05, False Discovery Rate (FDR) < 0.5), we identified a total of 172 genes that were either significantly up-regulated (79 genes) or down-regulated (93 genes) in the STXBP4-positive tumors. These candidate genes potentially represent a network involved in STXBP4-mediated biology (Fig. 2B). To address this possibility in more detail, we carried out Ingenuity Pathway Analysis (IPA), which revealed that more than 30% of the affected genes were classified in the functional class of “Cell Death and Survival”. This finding supported our experimental observations that STXBP4 could be linked to the poor prognosis of lung SCC (Fig. 2C).
Figure 2.
Gene expression profiling of clinical samples from lung SCC patients. (A) A cluster diagram of RNA-seq data from six pairs of STXBP4-positive and STXBP4-negative samples. The color bars represent relative expression levels: Red indicates higher than average expression and blue indicates lower than average expression. (B) 79 significantly differentially expressed upregulated genes and 93 downregulated genes (P < 0.05 and FDR < 0.5) were identified. (C) Functional analysis of differentially expressed genes was performed by Ingenuity Pathway Analysis. (D) The genes are listed in descending order of normalized expression. *FDR < 0.1. (E) A cluster diagram of PDGFR and VEGFR expression from RNA-seq analysis. The color bars represent relative expression levels: red indicates higher than average expression and blue indicates lower than average expression.
Additionally, other significant functional classes identified by IPA, including “Cellular Movement” and “Cell to Cell Signaling and Interaction”, may be relevant to the correlation of STXBP4-positivity with local tumor progression related to local tumor size (T factor) and disease stage (Fig. 2C). The canonical pathway analysis characterized two signaling pathways as the functional relationship of STXBP4-positivity. “Cellular Movement” and “Cell Morphology”, have been predicted as the most significantly activated canonical pathways (Supplementary Fig. 2).
IPA revealed that STXBP4-positivity was also correlated with the expression of growth factor receptors and components of downstream pathways. Among these genes listed in descending order of normalized expression, PDGFRA was a significant up-regulated gene (FDR < 0.1) (Fig. 2D and 2E), and a most relevant candidate for addressing the growth of STXBP4-positive lung SCC cells (Supplementary Fig. 3). PDGFRα is a receptor tyrosine kinase that is a critical regulator of growth and proliferation of certain cell types during embryonal development (28, 29). In subsequent experiments described later in this study, PDGFRα proved to be a key mediator of STXBP4 oncogenic activity.
STXBP4 regulates PDGF-PDGFR signaling in lung SCC
PDGF and PDGFR isoforms have important functions in the regulation of growth and survival of certain cell types (28, 29), and upregulation of PDGF-PDGFR signaling drives tumor cell growth. Indeed, the oncogenic properties of mutated or amplified PDGFRα have been studied in several tumor types, and PDGFRβ has been linked to not only tumor angiogenesis via paracrine effects, but also cancer metastasis (30, 31).
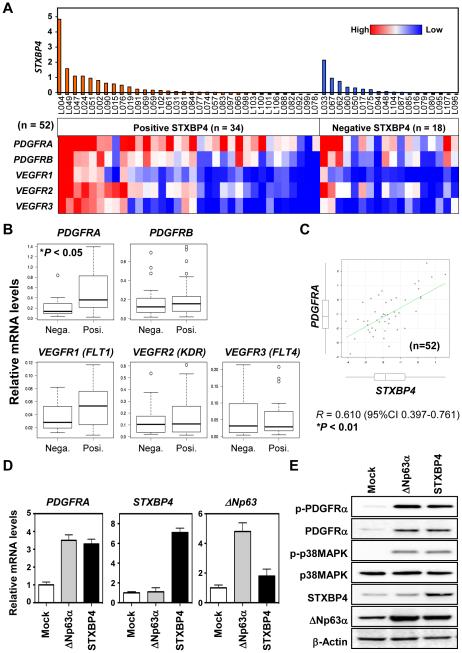
We measured mRNA expression levels by real-time RT-PCR in a total of 52 available samples from lung SCC patients for which high quality RNA was available (RIN > 2.0). The mRNA levels of STXBP4 were also correlated with ΔNp63 mRNA levels in those 52 samples (Supplementary Fig. 4A). Interestingly, we observed that PDGFRA expression was significantly up-regulated in STXBP4-positive lung SCC samples compared with STXBP4-negative samples (P < 0.05) (Fig. 3A and 3B). On the other hand, PDGFRB, VEGFR1 (FLT1), VEGFR2 (KDR) and VEGFR3 (FLT4) were consistently, but not significantly, up-regulated in STXBP4-positive lung SCC samples. STXBP4-positive samples defined by immunohistochemistry also had high STXBP4 mRNA expression levels compared with STXBP4-negative samples, and interestingly, PDGFRA mRNA levels were also significantly correlated with STXBP4 mRNA levels (Fig. 3C). Additionally, analyses of The Cancer Genome Atlas (TCGA) datasets of lung SCC (n = 488), supported in part, our finding that PDGFRA mRNA levels are significantly correlated with STXBP4 mRNA levels (P = 0.015) (Supplementary Fig. 4B).
Figure 3.
PDGFRA expression is significantly upregulated in STXBP4-positive samples from lung SCC Patients. (A) A cluster diagram of relative expression of PDGFRs and VEGFRs by real-time RT-PCR analysis. Tumor specimens were collected from lung SCC patients with surgical resection. A total of 52 high RNA integrity number (RIN > 2.0) STXBP4-positive samples (n=34) and STXBP4-negative samples (n=18), were used for transcriptome profiling by real-time RT-PCR. (B) PDGFRA mRNA was significantly upregulated in STXBP4-positive lung SCC samples. Relative mRNA levels of PDGFRs and VEGFRs of the STXBP4-positive samples (n=35) and STXBP4-negative samples (n=19); *P < 0.05. (C) Scatter plot of relative mRNA expression levels of PDGFRA and STXBP4. (D) STXBP4 or ΔNp63α induces PDGFRA expression in a lung SCC cell line, EBC-1. The cells were retrovirally transduced with empty vector control (Mock), ΔNp63α or STXBP4. The mRNA levels of PDGFRA, STXBP4 or ΔNp63 were determined by real-time RT-PCR. (E) EBC-1 cells transduced as in (D), were subjected to immunoblotting using antibodies for phospho-PDGFRα (p-PDGFRα), PDGFRα, phospho-p38MAPK (p-p38MAPK), p38MAPK, STXBP4, ΔNp63 or β-Actin.
To confirm the enhanced expression levels of PDGFRA in STXBP4 high expressing tumors, the lung SCC cell line, EBC-1, was transduced with STXBP4 and ΔNp63α retroviruses. As shown in Fig. 3D and 3E, the STXBP4 transduced stable clones showed high induction levels of both PDGFRA mRNA level (Fig. 3D) and PDGFRα protein levels (Fig. 3E), consistent with our findings in 52 resected patient lung SCC samples (Fig. 3B and 3C). Correspondingly, high expressing STXBP4 cells had elevated ΔNp63 protein levels, but not mRNA levels. Additionally, the relative increase in PDGFRA mRNA expression is more marked in STXBP4 low-expressing EBC-1 cells compared with STXBP4 high-expressing RERF-LC-Sq1 cells (Supplementary Fig. 5A and 5B). The results indicate that STXBP4 regulates PDGFRα expression in lung SCC, most likely in a ΔNp63-dependent manner, and also suggest that STXBP4 may serve as a new SCC biomarker.
STXBP4-depletion represses lung SCC tumor growth in vivo
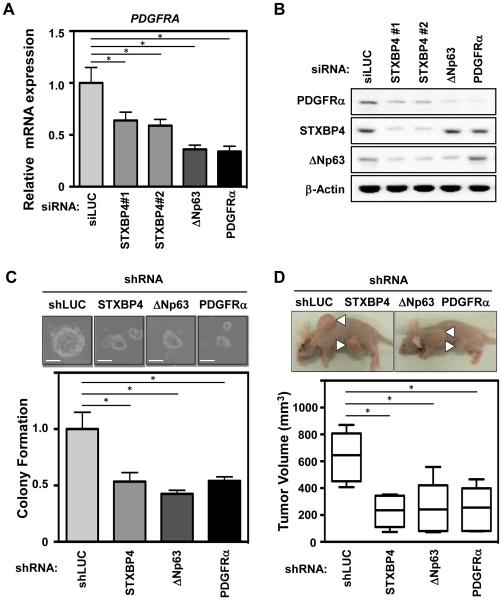
We next examined the oncogenic role of STXBP4 in regulating the expression of PDGFRα in a lung SCC cell line using a loss-of-function approach. Two independent siRNAs against STXBP4 (STXBP4#1 and STXBP4#2) were transfected into the lung SCC cell line, RERF-LC-Sq1, and a siRNA targeting luciferase (siLUC) was used as a control. The STXBP4 knockdown cells showed low expression of STXBP4, which correlated with down-regulation of both PDGFRA mRNA (Fig. 4A) and PDGFRα protein levels (Fig. 4B), consistent with our findings in resected patient lung SCC samples.
Figure 4.
STXBP4-depletion inhibits lung SCC tumorigenesis and modulates PDGF signaling in vivo. (A) The lung SCC cells, RERF-LC-Sq1, were treated with siRNAs for Luciferase (siLUC) as a control, STXBP4#1, STXBP4#2, ΔNp63 or PDGFRα. Total RNAs were quantified by real-time RT-PCR analysis and the induction levels of PDGFRA were determined by the relative Ct method. (B) RERF-LC-Sq1 cells depleted as in (A), were subjected to immunoblotting using anti-PDGFRα, STXBP4, ΔNp63 or β-Actin antibodies. (C) The growth of RERF-LC-Sq1 cells after shRNA mediated PDGFRα, STXBP4 or ΔNp63 knockdown was monitored by soft agar colony formation assays. Standard deviations (SD) are plotted. *P < 0.05. (D) Representative images of xenografts from subcutaneously transplanted with lentivirally shRNA transduced Luciferase as a control (shLUC), Stxbp4, ΔNp63 or PDGFRα knockdown RERF-LC-Sq1 cells (n = 6 for each knockdown). The results of six independent injections of knockdown cells are shown. Twenty days after implantation, the length (L) and width (W) of the tumor mass were measured, and the tumor volume (TV) was calculated using the equation: TV = (L × W2)/2. *P < 0.05.
In order to evaluate the functional relevance of STXBP4 and PDGFRα expression during tumor formation, we monitored the colony formation of RERF-LC-Sq1 cells lentivirally transduced with STXBP4, ΔNp63 or PDGFRα shRNAs. As shown in Fig. 4C, PDGFRα, STXBP4 or ΔNp63 knockdown in RERF-LC-Sq1 cells, led to decreased anchorage-independent colony formation in soft agar. Subcutaneous transplantation of PDGFRA or STXBP4 knockdown clones into immunodeficient mice resulted in suppressed tumor formation compared with control luciferase shRNA xenografts (Fig. 4D). Additionally, the knockdown effect of STXBP4 and suppression of tumorigenesis are more marked in high STXBP4 expressing RERF-LC-Sq1 cells compared with low STXBP4 expressing EBC-1 cells (Fig. 4 and Supplementary Fig. 6). These data suggest that downregulation of STXBP4 decreases PDGFRα expression and suppresses tumor formation.
Overall, our results indicate that STXBP4 has oncogenic activity both in vitro and in vivo, and further suggest that STXBP4 could be a critical driver of tumor propagation through regulating the PDGFRα pathway.
Discussion
We demonstrated that STXBP4 expression in clinical specimens was closely associated with T factor (P < 0.001), disease stage (P = 0.030), and pleural involvement (P = 0.028). Furthermore, univariate and multivariate analysis indicated that STXBP4 expression was an independent prognostic factor for OS and PFS. While p63 is essential for normal epidermal stratification and the proliferative potential of epithelial stem cells, ΔNp63 is thought to maintain the proliferative potential of basal regenerative cells, including stem cells, in skin, thymus, breast, prostate, and urothelial stratified epithelium (20, 32-36). STXBP4 can physically interact with ΔNp63 and is indispensable for stabilizing ΔNp63, which is consistent with a putative diagnostic role for STXBP4 in lung SCC.
Polymorphisms of STXBP4/COX11 (rs6504950; AA/AG-genotype) were associated with a significantly decreased risk of carcinogenesis in a meta-analysis of breast cancer patients (37). Although functional assessments of these polymorphisms were not undertaken, and the extent of loss of STXBP4 function in tumors was not studied, the data suggested that STXBP4 could play a role in carcinogenesis and tumor progression in breast cancer patients. Our study provides diagnostic role of STXBP4 alongside ΔNp63 in lung SCC.
The pathological function of STXBP4 in human cancers remains unclear. However, STXBP4 can physically interact with p63 and is indispensable for stabilizing ΔNp63 even in normal conditions (16). Consistent with STXBP4 localization in both the nucleus and cytoplasm, it has been suggested that nuclear STXBP4 has p63-mediated functions, and that cytoplasmic STXBP4 could facilitate other functions in a p63-independent manner (16). In fact, our data indicated that STXBP4 induction partially increased tumor growth even in the absence of elevated ΔNp63 (Supplementary Fig. 7A), and that PDGFRA induction also partially increased tumor growth even in STXBP4 knockdown cells (Supplementary Fig. 7B). Thus, STXBP4 may contribute to the susceptibility and severity of cancer in a p63-dependent and independent manner. Indeed, amplification and overexpression of p63 has frequently been observed in a variety of SCCs, including lung cancers and head and neck cancers (8, 38). However, p63 expression is decreased during progression to invasion and metastasis of lung, breast and bladder cancer, and loss of p63 expression is associated with worse prognosis in some cases (35, 39, 40). It could be the balance between the TA isotype (tumor suppressive) and ΔN isotype (oncogenic), as well as the tissue context, which is critical for proliferation and differentiation in both epithelial stem cells and cancer stem cells.
Global transcriptome profiling using next-generation sequencing technologies has become more common for comprehensive gene expression analysis to explore novel regulators and target genes in different types of cancers. In this report, genome-wide transcriptome analysis identified mediators of STXBP4 activity, including PDGFRα, which contributes to cell growth and metastasis in a p63-dependent manner. PDGF family proteins consist of several disulfide-bonded, dimeric isoforms (PDGF AA, PDGF AB, PDGF BB, PDGF CC and PDGF DD) that bind in a specific pattern to two related receptor tyrosine kinases, PDGFRα and PDGFRβ (41). PDGFRα homodimers bind to all PDGF isoforms except those containing PDGF D (42). A number of different signaling pathways, including mTOR (Fig. 1D) and MAPK (Fig. 3E), are initiated by activated PDGF receptors, and stimulate cell growth, actin reorganization, migration, and differentiation (43, 44).
PDGF receptors are expressed at low levels in normal lung epithelial cells, however, increased PDGFRα expression has been reported in lung cancer. PDGFRβ expression is observed mainly in stromal cells, but also in the sarcomatoid type of NSCLC (45). Based on recent evidence, inhibition of the p53/NF-Y complex by mutant gain of function p53 enhances PDGFRβ expression and promotes metastasis in a subset of pancreatic cancers (31). In addition, the interaction of mutant p53 with p63 regulates the expression of p63 target genes to enhance invasion and metastasis (46). Hence, the oncogenic activity of mutant p53 is a consequence of the physical association between mutant p53 and the p53 family members, p63 and p73.
Treatment strategies for lung cancer are based on the assumption that an individual patient's cancer is purely of one subtype. Since many cancers are heterogeneous and relatively resistant to chemotherapy or radiation, there is strong interest in molecular-targeted therapies based on tumor biology. In particular, targeted agents that inhibit the epidermal growth factor receptor (EGFR) or anaplastic lymphoma kinase (ALK) are approved for the treatment of NSCLC harboring genetic alterations in the genes encoding these proteins (47). EGFR inhibitors, such as erlotinib and gefitinib, are only effective against NSCLCs with EGFR mutations, which occur almost exclusively in lung AC. Similarly, the recently identified EML4-ALK rearrangement, which predicts susceptibility to the targeted agent crizotinib, also occurs only in lung AC. Unfortunately, therapeutic advances in the treatment of lung SCC have lagged behind those for AC (48). Therefore, the capacity to distinguish between lung AC and SCC is particularly important for the effective use of novel targeted therapies to treat patients with these NSCLC subtypes.
Inhibition of the PDGFRα signaling pathway by treatment with a neutralizing PDGFRα antibody, MEDI-575, had minimal effect on tumor cell proliferation in preclinical models of NSCLC (49). Lung SCC histology also identified patients at a higher risk of bleeding during treatment with bevacizumab, a monoclonal anti-VEGF antibody (50). Thus, more studies are required to determine whether specific inhibition of PDGF receptors, without inhibition of VEGF receptors, is of any benefit for lung cancer patients. These issues highlight the growing importance of accurate identification of NSCLC subtypes for assigning patients to appropriate histology-based therapies and the triage of tissue for appropriate molecular studies.
Supplementary Material
Translational Relevance.
ΔNp63 is a diagnostic marker highly specific for lung squamous cell carcinoma (SCC), but the regulation of p63 protein stability and the pathologic relevance of p63 in tumorigenesis remains unclear. We report here for the first time that Syntaxin Binding Protein 4 (STXBP4) expression increases the oncogenic potential of ΔNp63, and is STXBP4 an independent negative prognostic marker for predicting poor outcome in lung SCC. Transcriptional analysis (RNA-seq) using Next Generation Sequencing in STXBP4-positive and STXBP4-negative lung SCC indicated that Platelet-Derived Growth Factor Receptor α (PDGFRα) is a key downstream mediator of STXBP4 function. The data suggest that STXBP4 is a new diagnostic marker in lung SCC, and STXBP4 might be a relevant therapeutic target for the treatment of patients with this disease.
Acknowledgements
We thank Dr. Arito Yamane for instrument settings of Ion Proton system, and Gombodorj, N., Umezawa, S., Suto, Y., Nakamura, M., Ito, M., Ichihara, A. and Horigome, E. for expert technical assistances. We also thank Drs. Vincenzo Castronovo, Andrei Turtoi and Akeila Bellahcene in University of Liege for valuable discussions. This work was supported, in part, by the Promotion Plan for the Platform of Human Resource Development for Cancer, New Paradigms - Establishing Center for Fostering Medical Researchers of the Future Programs by Ministry of Education, Culture, Sports, Science and Technology of Japan, and Gunma University Initiative for Advanced Research (GIAR). This work was also supported, in part, by The Yasuda Medical Foundation and by NIH grant CA87497.
Footnotes
Disclosure of Potential Conflicts of Interest: M. Nishiyama receives commercial research support from Yakult Honsha Co. Ltd. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clinic proceedings. 2008;83(5):584–94. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Recondo G, Jr., Recondo G, Sr., Galanternik F, Greco M, de la Vega M, Canton ED, et al. Immunotherapy for Non-Small Cell Lung Cancer - Finally a Hint of Hope. Rev Recent Clin Trials. 2016 doi: 10.2174/1574887111666160330121349. [DOI] [PubMed] [Google Scholar]
- 3.Mukhopadhyay S, Katzenstein AL. Subclassification of non-small cell lung carcinomas lacking morphologic differentiation on biopsy specimens: Utility of an immunohistochemical panel containing TTF-1, napsin A, p63, and CK5/6. Am J Surg Pathol. 2011;35(1):15–25. doi: 10.1097/PAS.0b013e3182036d05. [DOI] [PubMed] [Google Scholar]
- 4.Massion PP, Taflan PM, Jamshedur Rahman SM, Yildiz P, Shyr Y, Edgerton ME, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 2003;63(21):7113–21. [PubMed] [Google Scholar]
- 5.Sniezek JC, Matheny KE, Westfall MD, Pietenpol JA. Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope. 2004;114(12):2063–72. doi: 10.1097/01.mlg.0000149437.35855.4b. [DOI] [PubMed] [Google Scholar]
- 6.Candi E, Dinsdale D, Rufini A, Salomoni P, Knight RA, Mueller M, et al. TAp63 and DeltaNp63 in cancer and epidermal development. Cell Cycle. 2007;6(3):274–85. doi: 10.4161/cc.6.3.3797. [DOI] [PubMed] [Google Scholar]
- 7.Mills AA. p63: oncogene or tumor suppressor? Curr Opin Genet Dev. 2006;16(1):38–44. doi: 10.1016/j.gde.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Di Como CJ, Urist MJ, Babayan I, Drobnjak M, Hedvat CV, Teruya-Feldstein J, et al. p63 expression profiles in human normal and tumor tissues. Clin Cancer Res. 2002;8(2):494–501. [PubMed] [Google Scholar]
- 9.Nonaka D. A study of DeltaNp63 expression in lung non-small cell carcinomas. Am J Surg Pathol. 2012;36(6):895–9. doi: 10.1097/PAS.0b013e3182498f2b. [DOI] [PubMed] [Google Scholar]
- 10.Bishop JA, Teruya-Feldstein J, Westra WH, Pelosi G, Travis WD, Rekhtman N. p40 (DeltaNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod Pathol. 2012;25(3):405–15. doi: 10.1038/modpathol.2011.173. [DOI] [PubMed] [Google Scholar]
- 11.Melino G, Lu X, Gasco M, Crook T, Knight RA. Functional regulation of p73 and p63: development and cancer. Trends Biochem Sci. 2003;28(12):663–70. doi: 10.1016/j.tibs.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2(3):305–16. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 13.Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15(11):1363–7. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- 14.McKeon F. p63 and the epithelial stem cell: more than status quo? Genes Dev. 2004;18(5):465–9. doi: 10.1101/gad.1190504. [DOI] [PubMed] [Google Scholar]
- 15.Duijf PH, Vanmolkot KR, Propping P, Friedl W, Krieger E, McKeon F, et al. Gain-of-function mutation in ADULT syndrome reveals the presence of a second transactivation domain in p63. Hum Mol Genet. 2002;11(7):799–804. doi: 10.1093/hmg/11.7.799. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Peart MJ, Prives C. Stxbp4 regulates DeltaNp63 stability by suppression of RACK1-dependent degradation. Mol Cell Biol. 2009;29(14):3953–63. doi: 10.1128/MCB.00449-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fomenkov A, Zangen R, Huang YP, Osada M, Guo Z, Fomenkov T, et al. RACK1 and stratifin target DeltaNp63alpha for a proteasome degradation in head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle. 2004;3(10):1285–95. doi: 10.4161/cc.3.10.1155. [DOI] [PubMed] [Google Scholar]
- 18.Min J, Okada S, Kanzaki M, Elmendorf JS, Coker KJ, Ceresa BP, et al. Synip: a novel insulin-regulated syntaxin 4-binding protein mediating GLUT4 translocation in adipocytes. Mol Cell. 1999;3(6):751–60. doi: 10.1016/s1097-2765(01)80007-1. [DOI] [PubMed] [Google Scholar]
- 19.Saito T, Okada S, Yamada E, Ohshima K, Shimizu H, Shimomura K, et al. Syntaxin 4 and Synip (syntaxin 4 interacting protein) regulate insulin secretion in the pancreatic beta HC-9 cell. J Biol Chem. 2003;278(38):36718–25. doi: 10.1074/jbc.M305114200. [DOI] [PubMed] [Google Scholar]
- 20.Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol. 2003;23(7):2264–76. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimizu K, Kaira K, Tomizawa Y, Sunaga N, Kawashima O, Oriuchi N, et al. ASC amino-acid transporter 2 (ASCT2) as a novel prognostic marker in non-small cell lung cancer. Br J Cancer. 2014;110(8):2030–9. doi: 10.1038/bjc.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaira K, Sunose Y, Arakawa K, Sunaga N, Shimizu K, Tominaga H, et al. Clinicopathological significance of ASC amino acid transporter-2 expression in pancreatic ductal carcinoma. Histopathology. 2015;66(2):234–43. doi: 10.1111/his.12464. [DOI] [PubMed] [Google Scholar]
- 23.Rokudai S, Aikawa Y, Tagata Y, Tsuchida N, Taya Y, Kitabayashi I. Monocytic leukemia zinc finger (MOZ) interacts with p53 to induce p21 expression and cell-cycle arrest. J Biol Chem. 2009;284(1):237–44. doi: 10.1074/jbc.M805101200. [DOI] [PubMed] [Google Scholar]
- 24.Rokudai S, Laptenko O, Arnal SM, Taya Y, Kitabayashi I, Prives C. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc Natl Acad Sci U S A. 2013;110(10):3895–900. doi: 10.1073/pnas.1300490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8(9):1765–86. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 27.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heldin CH. Targeting the PDGF signaling pathway in tumor treatment. Cell communication and signaling : CCS. 2013;11:97. doi: 10.1186/1478-811X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai Y. Platelet-derived growth factor receptor tyrosine kinase inhibitors: a review of the recent patent literature. Expert opinion on therapeutic patents. 2010;20(7):885–97. doi: 10.1517/13543776.2010.493559. [DOI] [PubMed] [Google Scholar]
- 30.Cao R, Bjorndahl MA, Religa P, Clasper S, Garvin S, Galter D, et al. PDGF-BB induces intratumoral lymphangiogenesis and promotes lymphatic metastasis. Cancer Cell. 2004;6(4):333–45. doi: 10.1016/j.ccr.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 31.Weissmueller S, Manchado E, Saborowski M, Morris JPt, Wagenblast E, Davis CA, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell. 2014;157(2):382–94. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 33.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–8. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 34.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102(27):9625–30. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbieri CE, Tang LJ, Brown KA, Pietenpol JA. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 2006;66(15):7589–97. doi: 10.1158/0008-5472.CAN-06-2020. [DOI] [PubMed] [Google Scholar]
- 36.Higashikawa K, Yoneda S, Tobiume K, Taki M, Shigeishi H, Kamata N. Snail-induced down-regulation of DeltaNp63alpha acquires invasive phenotype of human squamous cell carcinoma. Cancer Res. 2007;67(19):9207–13. doi: 10.1158/0008-5472.CAN-07-0932. [DOI] [PubMed] [Google Scholar]
- 37.Tang L, Xu J, Wei F, Wang L, Nie WW, Chen LB, et al. Association of STXBP4/COX11 rs6504950 (G>A) polymorphism with breast cancer risk: evidence from 17,960 cases and 22,713 controls. Arch Med Res. 2012;43(5):383–8. doi: 10.1016/j.arcmed.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi K, Wu L, Caballero OL, Hibi K, Trink B, Resto V, et al. Frequent gain of the p40/p51/p63 gene locus in primary head and neck squamous cell carcinoma. Int J Cancer. 2000;86(5):684–9. doi: 10.1002/(sici)1097-0215(20000601)86:5<684::aid-ijc13>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 39.Karni-Schmidt O, Castillo-Martin M, Shen TH, Gladoun N, Domingo-Domenech J, Sanchez-Carbayo M, et al. Distinct expression profiles of p63 variants during urothelial development and bladder cancer progression. Am J Pathol. 2011;178(3):1350–60. doi: 10.1016/j.ajpath.2010.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26(36):5169–83. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 41.Deuel TF, Silverman NJ, Kawahara RS. Platelet-derived growth factor: a multifunctional regulator of normal and abnormal cell growth. BioFactors. 1988;1(3):213–7. [PubMed] [Google Scholar]
- 42.Bergsten E, Uutela M, Li X, Pietras K, Ostman A, Heldin CH, et al. PDGF-D is a specific, protease-activated ligand for the PDGF beta-receptor. Nat Cell Biol. 2001;3(5):512–6. doi: 10.1038/35074588. [DOI] [PubMed] [Google Scholar]
- 43.Ostman A, Heldin CH. Involvement of platelet-derived growth factor in disease: development of specific antagonists. Advances in cancer research. 2001;80:1–38. doi: 10.1016/s0065-230x(01)80010-5. [DOI] [PubMed] [Google Scholar]
- 44.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22(10):1276–312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donnem T, Al-Saad S, Al-Shibli K, Andersen S, Busund LT, Bremnes RM. Prognostic impact of platelet-derived growth factors in non-small cell lung cancer tumor and stromal cells. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2008;3(9):963–70. doi: 10.1097/JTO.0b013e3181834f52. [DOI] [PubMed] [Google Scholar]
- 46.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137(1):87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 47.Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. The New England journal of medicine. 2014;371(23):2167–77. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 48.Socinski MA, Stinchcombe TE, Moore DT, Gettinger SN, Decker RH, Petty WJ, et al. Incorporating bevacizumab and erlotinib in the combined-modality treatment of stage III non-small-cell lung cancer: results of a phase I/II trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30(32):3953–9. doi: 10.1200/JCO.2012.41.9820. [DOI] [PubMed] [Google Scholar]
- 49.Laing N, McDermott B, Wen S, Yang D, Lawson D, Collins M, et al. Inhibition of platelet-derived growth factor receptor alpha by MEDI-575 reduces tumor growth and stromal fibroblast content in a model of non-small cell lung cancer. Molecular pharmacology. 2013;83(6):1247–56. doi: 10.1124/mol.112.084079. [DOI] [PubMed] [Google Scholar]
- 50.Johnson DH, Fehrenbacher L, Novotny WF, Herbst RS, Nemunaitis JJ, Jablons DM, et al. Randomized phase II trial comparing bevacizumab plus carboplatin and paclitaxel with carboplatin and paclitaxel alone in previously untreated locally advanced or metastatic non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22(11):2184–91. doi: 10.1200/JCO.2004.11.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.