Abstract
The need to provide effective and timely antimicrobial treatment to cancer patients with infections is well-recognized, but tempered by preliminary, but accumulating, evidence that antibiotic-induced microbiome dysbiosis affects cancer therapy response, non-infectious toxicities, and infectious complications. Given only a minority of empirically treated cancer patients are proven to have a true bacterial infection, it is important to consider the potential negative consequences of extensive broad-spectrum antimicrobial use on the commensal microbiota. Herein, we review the literature substantiating the dilemma oncologists face when treating suspected or documented infections with respect to the interaction between the host microbiome, antibiotics, and cancer-related clinical outcomes. We propose microbiome-based explorations that could assist oncologists in optimizing treatment strategies for cancer-related infections as well as the cancer, itself. In addition, we discuss knowledge gaps and challenges in this nascent field that must be addressed in order to deliver medically relevant translational applications. We anticipate the emerging knowledge regarding the role of the microbiota in the health of cancer patients may cause a reappraisal of the manner in which antibiotics are used in the oncologic setting and how microorganisms are viewed by oncologists.
Keywords: microbiome, cancer, antibiotics, infection, therapeutic response
Historical Beneficial Aspects of Antimicrobials in the Cancer Population
Antimicrobial therapy has markedly improved the outcome of cancer patients over the last 50 years. The potential dramatic impact of antimicrobials in oncology became clear when infections replaced hemorrhage and leukemia itself as the leading cause of death among acute leukemia patients in the 1960s [1]. By the early 1970s, the development of methicillin for penicillin-resistant Staphylococcus aureus and carbenicillin for Pseudomonas aeruginosa meant serious infections could be effectively treated, even amid persistent neutropenia [2]. As a result, neutropenic fever became an oncologic emergency demanding the rapid administration of broad-spectrum antibiotics which markedly improved the outcomes of neutropenic patients with proven infections, particularly due to Pseudomonas aeruginosa [3, 4]. Eventually, the high rates of morbidity and mortality associated with infections in hematologic malignancy patients led to large randomized controlled trials which demonstrated that prophylactic administration of a fluoroquinolone, (i.e. levofloxacin), reduced rates of neutropenic fever and confirmed infections [5]. Thus, current oncology dogma primarily considers bacteria as a threat to patient health with a low threshold for initiation of broad-spectrum antimicrobials in the preventive or therapeutic setting.
How Antibiotic-Induced Microbiome Alteration Affects the Cancer Patient
Feasible and affordable genetic means to comprehensively assay the bacteria present in a variety of sample types has paved the way for large scale investigations, such as the Human Microbiome Project [6]. In addition to 16S rRNA gene sequencing, microbiome characterization methodologies have expanded to other “-omics” approaches to include whole genome shotgun sequencing, RNAseq, and metabolomics, which more precisely delineate bacterial community structure, gene presence/expression, and metabolic activity [7]. Use of these methodologies has illuminated that the microflora have profound effects on human health such as altering cytokine profiles, influencing inflammatory immune responses, and altering metabolites [8–11].
Although it is recognized that systemically administered antimicrobials can have a dramatic impact on the composition and function of the gastrointestinal microbiome [12], recent advances have also demonstrated that antibiotic effects on the microbiome influence the response to cancer immunotherapy. Specifically, Iida et al. described tumor necrosis and immune responses to be significantly reduced in antibiotic-treated colon carcinoma and melanoma tumor-bearing mice receiving immunostimulatory CpG-oligodeoxynucleotide treatment [13]. Similarly, Vetizou et al. demonstrated that melanoma tumors in antibiotic-treated mice failed to respond to CTLA-4 blockade immunotherapy, and that the presence of Bacteriodes fragilis was critical to the anti-tumor effect [14]. Recently, it has also been discovered that specific microbiota shape innate and adaptive immune system influencing the PD-1-PD-L1 axis [15, 16], although no studies have specifically shown the effects of antibiotic treatment on the microbiota and anti-PD-L1 treatment response.
In addition to influencing immunotherapy response, antibiotic-treated animals also display significantly reduced tumor regression and survival in cytotoxic therapy scenarios, such as oxaliplatin-treated lymphoma-bearing mice [13]. Likewise, Viaud et al. observed that receipt of antibiotics with activity against Gram-positive bacteria reduced T-helper lymphocyte and lymphoma responses in mice treated with cyclophosphamide [17]. Beyond animal models, recently it was shown that patients being treated with cyclophosphamide for chronic lymphocytic leukemia and cisplatin for relapsed lymphoma who also received anti-Gram-positive antibiotics had significantly lower overall response rate and survival [18].
Furthermore, it is becoming increasingly clear that antimicrobial induced microbiome disruption is also a key factor in cancer treatment related toxicities. For example, administration of antibiotics to mice undergoing hematopoietic stem-cell transplantation (HSCT) significantly increased the severity of graft versus host disease (GHVD) and mortality [19, 20]. Consistent with murine data, investigators found receipt of antibiotics with potent anti-anaerobic activity was associated with increased GVHD risk and GVHD-related mortality following allogeneic HSCT in patients [19, 21]. Additionally, fluoroquinolone receipt, low microbial diversity, and Gammaproteobacteria-domination of fecal microbiota were predictive of pulmonary complications among HSCT recipients [22].
Antibiotic-induced microbial dysbiosis is also a crucial aspect in the cancer patient’s risk for infectious toxicities. A prime example of the “Catch-22” relationship between antimicrobial therapy and cancer care is the hematologic malignancy patient. In these patients, depletion of native commensals by antibacterial prophylaxis and empirical treatment of neutropenic fever is compounded by mucosal barrier injury from cytotoxic chemotherapy, leading to proliferation of pathogenic bacteria, translocation across disrupted intestinal epithelium, and subsequent infection (Figure 1) [23]. In leukemia and HSCT patients, receipt of particular antibiotics, such as metronidazole, was associated with decreased microbial diversity, and consequently increased intestinal domination by pathogens that commonly cause hospital-acquired and bloodstream infections [24–27]. Similarly, it was also found that broad-spectrum antibiotic receipt, specifically carbapenems, was associated with loss of bacterial diversity in both the oral and stool microbiomes of acute myeloid leukemia patients during induction chemotherapy, which was in turn correlated with higher subsequent infectious risk in the 90 days post neutrophil recovery [28]. Moreover, there is a considerable body of evidence clearly linking high rates of Clostridium difficile infections in cancer patients, particularly those undergoing HSCT, to disruption of the normal intestinal flora due a combination of repeated use of antibiotics, immune suppression, and cancer therapy [24].
Figure 1. The “Catch 22” relationship between antimicrobial therapy and cancer treatment in the oncology patient.
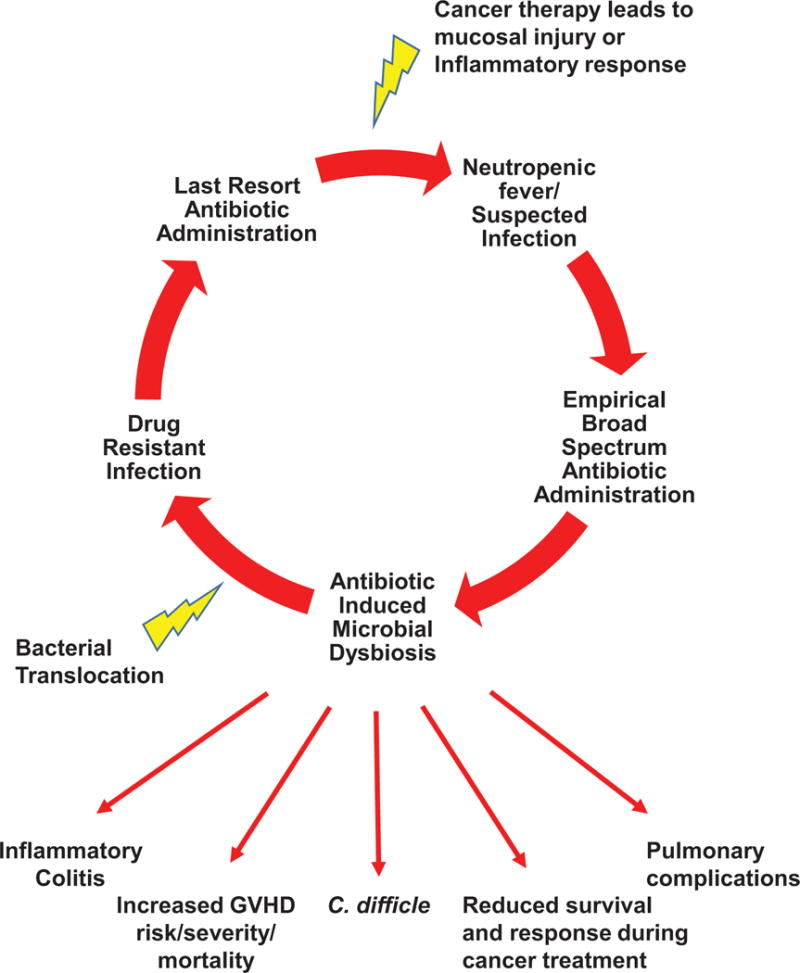
This figure depicts how depletion of native commensals by antibacterial prophylaxis and empirical treatment of neutropenic fevers or suspected infections is compounded by mucosal barrier injury from cytotoxic chemotherapy, leading to proliferation of pathogenic bacteria, translocation across disrupted intestinal epithelium, and subsequent resistant infections. This leads to a vicious cycle of re-current infectious issues and other cancer treatment related toxicities as a result of antibiotic induced microbial dysbiosis.
The microbiome as a possible prognostic or diagnostic biomarker in oncology
The studies outlined above indicate that the ability to comprehensively assess or alter human microbiota composition may be a valuable tool in improving cancer outcomes. Indeed, from a prognostic standpoint, the composition of the microbiome prior to chemotherapy has been demonstrated to be predictive of infectious outcomes for patients with acute myelogenous leukemia and lymphoma [28, 29]. Similarly, the diversity of the GI microbiome at the time of engraftment following HSCT is associated with risk of development of and mortality from GVHD [20, 30]. Finally, the abundance of the Bacteroidetes phylum was recently correlated with resistance to the development of immune-mediated colitis in melanoma patients treated with the immune check-point inhibitor, ipilimumab [31]. Thus, these data suggest the importance of developing probability indices which risk-stratify cancer patients with respect to microbiome measurements and other clinical factors, such as antimicrobial administration [32–34]. Predictive risk scores that incorporate microbiome measurements would need to include factors such as diversity metrics, absence of beneficial microbes or microbial by-products (i.e. those associated with pathogen colonization resistance, resistance against treatment complications, or anti-tumor effects) and domination by specific microbes related to infection. Through these types of examinations, one could envision the microbiome being incorporated as a baseline screening tool to predict which patients may respond better to cancer therapy, are at risk for treatment related toxicity, or are at risk for infectious complications.
Microbiome composition measurements may also assist with optimizing the choice and duration of antimicrobials in the cancer patient with respect to maintaining beneficial commensal microorganisms. For instance, we advocate for trials to assess whether rapid de-escalation of broad spectrum antimicrobials can be done safely in patients with negative cultures in the setting of asymptomatic febrile neutropenia. By merging such studies with longitudinal microbiome analyses, it could be determined whether such de-escalation helps preserve microbiome composition and if particular microbiome characteristics are associated with a need to re-initiate antimicrobials. In addition, investigating the use of more narrow spectrum antibiotics as well as shorter duration of therapy for infections are needed, as it has been suggested that the number of antibiotics and total antibiotic exposure is linked with recurrent infectious complications in leukemia patients (manuscript in press). Moreover, pharmacokinetic studies that link concentrations of antimicrobials in the intestinal lumen to effects on the microbiome are needed. Along the same lines, it will be important to assess not only the impact of the antimicrobial itself, but also its elimination (biliary vs. renal). Such data could be used to design interventions to minimize the off target effects of systemically administered antimicrobials on the commensal microbiota.
Microbiome measurements also raise the possibility to extend antimicrobial administration in oncology patients into the arena of personalized medicine. As genomic methodologies advance in terms of decreasing price and rapid availability of results, the ability to use microbiome samples to rapidly determine the scope of pathogens and antibiotic resistance genes present within an individual is becoming a real possibility [35]. By cataloging the antimicrobial resistome for each patient using metagenomic analyses, physicians could inform their therapeutic considerations for prophylaxis and infection. For example, if a particular patient were known to have intestinal domination by a pathogen resistant to standard empiric antimicrobials, oncologists could take a more individualized approach to antimicrobial initiation were that patient to develop infectious symptoms. Moreover, using microbiome measurements, other DNA-sequencing based approaches, or biomarkers, such as procalcitonin, to separate infectious from non-infectious fevers in the oncology patient would also greatly facilitate antimicrobial targeting and microbiota preservation. In addition to differentiating infectious from non-infectious fevers, it is also crucial to understand and discern colonization versus infection. For example, the unmet need to better distinguish C. difficle colonized patients from patients with C. difficle colitis is leading to a mass overdiagnosis and overtreatment [36]. It is highly likely that knowledge of microbiome interactions with the host will play an essential role in answering these needs.
The microbiome as a possible direct intervention tool
Direct manipulation of the microbiome also offers a possibility for improving cancer therapy, minimizing toxicities, and mitigating the impact of infectious diseases. For example, a recent study suggests that fecal transplantation may ameliorate steroid-resistant GVHD in HSCT recipients [37]. Similarly, fecal transplantation in mice increased responses to immunotherapy, raising the possibility that optimizing the microbiome prior to immune modulating treatment could improve response [15]. Though, randomized trials examining the efficacy of microbiome remediation are needed to fully evaluate therapeutic potential.
As antimicrobials are a dwindling resource, using the microbiome as a direct interventional tool could improve antimicrobial utilization by offering an alternative treatment strategy for infectious complications, alleviating antibiotic resistance, and preserving drug efficacy. Such microbiome based methods include fecal transplantation, targeted addition of a single or defined combination of bacterial species (probiotics), or prebiotics designed to stimulate the growth and retention of specific beneficial species in the form of dietary based intervention. It is thought that autologous fecal transplant could prevent pathogen intestinal colonization, infection, and development of antibiotic resistance [38, 39]. Consequently, if cancer hospitals begin to bank patient feces prior to cancer treatment, a patient’s native fecal microbiota could be implanted either continuously throughout treatment or administered after broad-spectrum antimicrobial treatment to counteract the microbiome damage potentially caused by antimicrobial treatment or chemotherapy.
However, one concern with using the administration of specific bacterial cocktails or fecal transplant in the immunocompromised patient is the risk for infection, as there have been numerous reports of septicemia associated with use of probiotic therapy such as Lactobacillus bacteremia or Saccharomyces fungemia [40, 41]. Thus, prebiotic administration or dietary intervention may be more desirable toxicity mitigation strategies. Recent examinations have suggested the beneficial impact of fiber on the microbiome as it relates to inflammation and mucosal barrier injury, particularly in that specific fibers increase the number of butyrate-producing bacteria [42, 43]. The short-chain fatty acid butyrate is importantly involved in adaptive immune responses, such as colonic T cell differentiation [44–46]. These data indicate bypassing the microbiota and providing bacterial metabolites, such as short-chain fatty acids, is an alternative possibility. Additionally, these studies suggest the importance of performing microbiome examination in tandem with metabolic and immunology research in order to improve intervention strategies which specifically target the host microbiota.
Critical cancer-microbiome knowledge gaps
It is important to remember the era of cancer-microbiome research is relatively nascent and thus fundamental questions remain unanswered. For example, how useful are single microbiome measurements given the microbiome inter- and intra-patient variability, particularly when ill? Although the majority of oncology patients lose microbial diversity during chemotherapy, inter-patient changes are highly variable with some patients maintaining a relatively preserved microbiota while others exhibit microbiome domination by one or two pathogens [25, 28]. Gaining knowledge regarding the factors that drive such drastically different microbiota trajectories is essential to designing and targeting microbiota preservation strategies. In addition to differences among individuals, more information is needed regarding variance in local microbiota composition at the intestinal mucosa versus what is present in stool samples.
Moreover, the integration of more advanced approaches, such whole genomic sequencing and metabolomics, are needed to potentially uncover mechanisms by which the microbiome can impact on clinical outcomes. For example, significant progress is being made towards culturing the entire intestinal bacterial microbiome using methods such as “culturomics”, to not only improve upon the identification of viable species within the gut, but also to capture the functional biodiversity [47]. Moreover, elucidating the role that the mycobiome and virome play in immune responses, cancer-therapy response, cancer treatment toxicities, and infectious complications will also need to be incorporated in future research, as these areas remain mostly unexplored. This effort, however, will need to include improving sequencing methods and databases for fungi and viruses.
It is also crucial to improve our statistical methodologies so that the complex nature of microbiome data, particularly with regard to longitudinal sampling, can be incorporated into clinical models. Statistical challenges include developing strategies to look for associations in high-dimensional data, a problem that is also being addressed by other types of big data (i.e. exome, proteomics, transcriptomics, etc.). Some challenges are unique to the microbiome, which features the additional layer of evolutionary relationships and potential interactions between bacteria, fungi, and viruses. The further development of biostatistical methods that can identify statistically meaningful relationships among networks integrating high dimensional microbiome data with complex variables such as gene function, metabolites, antibiotic administration, diet, and patient outcomes are key to conceiving dependable interventions.
Conclusion
The dramatic impact of the commensal microbiota on the health of the cancer patient is increasing in appreciation. As profound effects of antibiotics on the human microbiome have been demonstrated, it is imperative that antibiotic administration and stewardship strategies in patients with malignancy be considered within the context of the microbiome. Many possible future avenues of investigation exist that could potentially aid physicians in treating cancer-related infections while limiting collateral damage to the microbiota (Table 1). As more exploratory work is done to understand the microbiome’s role in cancer and cancer treatment related toxicities, carefully designed animal models and interventional trials will be critical to moving beyond basic association or biomarker studies in order to determine the mechanisms by which the microbiome modulates patient outcomes. The integration of microbiome-based approaches into the clinical arena offers a tremendous new opportunity to improve outcomes across the cancer care continuum.
Table 1.
Translational microbiome-based research strategies and interventions to support the management of infectious diseases and antimicrobial administration among high-risk cancer patients.
| Infectious Disease Management Objective | Microbiome Research Strategy |
|---|---|
| Risk stratification of patients for infection or colonization with antibiotic resistant pathogens prior to cancer treatment |
|
|
|
| Personalization of antimicrobial administration and infection control decisions for optimal patient outcomes |
|
|
|
|
|
| Infection prevention or microbiome synergism with antimicrobial therapy during cancer treatment |
|
Translational Relevance.
Antimicrobial therapy is critical to the health of cancer patients. However, initial clinical studies in patients and laboratory based investigations in murine models have demonstrated that disruption of the microbiome induced by antimicrobials impacts chemo- and immunotherapy response as well as treatment-related toxicities. Equally alarming is the vicious cycle of treating ever increasing multi-drug resistant infections with broad spectrum antibiotics that further deplete the commensal microflora. Consequently, cancer clinicians face a challenging and unique dilemma when managing infections in cancer patients. It is imperative that oncologists improve their antibiotic prophylaxis and treatment strategies with consideration of microbiome research. This perspective reviews the literature substantiating the interplay of antibiotics, the microbiome, and cancer, while offering possible avenues of investigation that could help physicians treat infections while maintaining the beneficial impact of the microbiota. Additionally, we discuss how manipulation of the microbiome could assist in optimizing cancer treatment outcomes.
Acknowledgments
We would like to thanks Dr. Cathy Eng, Dr. Elizabeth Mittendorf, and Chelcy Brumlow, M.S. for their contribution of making suggestions for this manuscript.
Financial Support: Jessica Galloway-Peña is supported by the Odyssey Program and CFP Foundation at The University of Texas MD Anderson Cancer Center. Samuel A. Shelburne is funded through The University of Texas MD Anderson Multi-Disciplinary Research Program and other funds through MD Anderson Cancer Center. Robert R. Jenq is funded through the NHLBI R01HL124112 and the Cancer Prevention and Research Institute of Texas Recruitment of Rising Stars Program RR160089.
References
- 1.Hersh EM, Bodey GP, Nies BA, Freireich EJ. Causes of death in acute leukemia: a ten-year study of 414 patients from 1954–1963. JAMA. 1965;193:105–109. doi: 10.1001/jama.1965.03090020019005. [DOI] [PubMed] [Google Scholar]
- 2.Bodey GP. The changing face of febrile neutropenia-from monotherapy to moulds to mucositis. Fever and neutropenia: the early years. J Antimicrob Chemother. 2009;63(Suppl 1):i3–13. doi: 10.1093/jac/dkp074. [DOI] [PubMed] [Google Scholar]
- 3.Freifeld AG, Bow EJ, Sepkowitz KA, Boeckh MJ, Ito JI, Mullen CA, et al. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the infectious diseases society of america. Clin Infect Dis. 2011;52(4):e56–93. doi: 10.1093/cid/cir073. [DOI] [PubMed] [Google Scholar]
- 4.Schimpff S, Satterlee W, Young VM, Serpick A. Empiric therapy with carbenicillin and gentamicin for febrile patients with cancer and granulocytopenia. N Engl J Med. 1971;284(19):1061–1065. doi: 10.1056/NEJM197105132841904. [DOI] [PubMed] [Google Scholar]
- 5.Bucaneve G, Micozzi A, Menichetti F, Martino P, Dionisi MS, Martinelli G, et al. Levofloxacin to prevent bacterial infection in patients with cancer and neutropenia. N Engl J Med. 2005;353(10):977–987. doi: 10.1056/NEJMoa044097. [DOI] [PubMed] [Google Scholar]
- 6.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noecker C, McNally CP, Eng A, Borenstein E. High-resolution characterization of the human microbiome. Transl Res. 2016 doi: 10.1016/j.trsl.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDermott AJ, Huffnagle GB. The microbiome and regulation of mucosal immunity. Immunology. 2014;142(1):24–31. doi: 10.1111/imm.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishio J, Honda K. Immunoregulation by the gut microbiota. Cell Mol Life Sci. 2012;69(21):3635–3650. doi: 10.1007/s00018-012-0993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535(7610):65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 11.Blottiere HM, de Vos WM, Ehrlich SD, Dore J. Human intestinal metagenomics: state of the art and future. Curr Opin Microbiol. 2013;16(3):232–239. doi: 10.1016/j.mib.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. 2014;124(10):4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabe H, Nordstrom I, Andersson K, Lundell AC, Rudin A. Staphylococcus aureus convert neonatal conventional CD4(+) T cells into FOXP3(+) CD25(+) CD127(low) T cells via the PD-1/PD-L1 axis. Immunology. 2014;141(3):467–481. doi: 10.1111/imm.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, Enot DP, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342(6161):971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pflug N, Kluth S, Vehreschild JJ, Bahlo J, Tacke D, Biehl L, et al. Efficacy of antineoplastic treatment is associated with the use of antibiotics that modulate intestinal microbiota. Oncoimmunology. 2016;5(6):e1150399. doi: 10.1080/2162402X.2016.1150399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shono Y, Docampo MD, Peled JU, Perobelli SM, Velardi E, Tsai JJ, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Trans Med. 2016;8(339):339ra371. doi: 10.1126/scitranslmed.aaf2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209(5):903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2014;20(5):640–645. doi: 10.1016/j.bbmt.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris B, Morjaria SM, Littmann ER, Geyer AI, Stover DE, Barker JN, et al. Gut microbiota predict pulmonary infiltrates after allogeneic hematopoietic cell transplantation. Am J Respir Crit Care Med. 2016;194(4):450–463. doi: 10.1164/rccm.201507-1491OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taur Y, Pamer EG. Microbiome mediation of infections in the cancer setting. Genome Med. 2016;8(1):40. doi: 10.1186/s13073-016-0306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taur Y, Pamer EG. The intestinal microbiota and susceptibility to infection in immunocompromised patients. Curr Opin Infect Dis. 2013;26(4):332–337. doi: 10.1097/QCO.0b013e3283630dd3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55(7):905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120(12):4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Vliet MJ, Tissing WJ, Dun CA, Meessen NE, Kamps WA, de Bont ES, et al. Chemotherapy treatment in pediatric patients with acute myeloid leukemia receiving antimicrobial prophylaxis leads to a relative increase of colonization with potentially pathogenic bacteria in the gut. Clin Infect Dis. 2009;49(2):262–270. doi: 10.1086/599346. [DOI] [PubMed] [Google Scholar]
- 28.Galloway-Pena JR, Smith DP, Sahasrabhojane P, Ajami NJ, Wadsworth WD, Daver NG, et al. The role of the gastrointestinal microbiome in infectious complications during induction chemotherapy for acute myeloid leukemia. Cancer. 2016;122(14):2186–2196. doi: 10.1002/cncr.30039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montassier E, Al-Ghalith GA, Ward T, Corvec S, Gastinne T, Potel G, et al. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med. 2016;8(1):49. doi: 10.1186/s13073-016-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124(7):1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. doi: 10.1038/ncomms10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tosh PK, McDonald LC. Infection control in the multidrug-resistant era: tending the human microbiome. Clin Infect Dis. 2012;54(5):707–713. doi: 10.1093/cid/cir899. [DOI] [PubMed] [Google Scholar]
- 33.Halpin AL, McDonald LC. Editorial Commentary: The dawning of microbiome remediation for addressing antibiotic resistance. Clin Infect Dis. 2016;62(12):1487–1488. doi: 10.1093/cid/ciw187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halpin AL, de Man TJ, Kraft CS, Perry KA, Chan AW, Lieu S, et al. Intestinal microbiome disruption in patients in a long-term acute care hospital: A case for development of microbiome disruption indices to improve infection prevention. Am J Infect Control. 2016;44(7):830–836. doi: 10.1016/j.ajic.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325(5944):1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamboj M, Sheahan A, Sun J, Taur Y, Robilotti E, Babady E, et al. Transmission of Clostridium difficile during hospitalization for allogeneic stem cell transplant. Infect Control Hosp Epidemiol. 2016;37(1):8–15. doi: 10.1017/ice.2015.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kakihana K, Fujioka Y, Suda W, Najima Y, Kuwata G, Sasajima S, et al. Fecal microbiota transplantation for patients with steroid-resistant/dependent acute graft-versus-host disease of the gut. Blood. 2016 doi: 10.1182/blood-2016-05-717652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khanna S, Tosh PK. A clinician’s primer on the role of the microbiome in human health and disease. Mayo Clin Proc. 2014;89(1):107–114. doi: 10.1016/j.mayocp.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Crum-Cianflone NF, Sullivan E, Ballon-Landa G. Fecal microbiota transplantation and successful resolution of multidrug-resistant-organism colonization. J Clin Microbiol. 2015;53(6):1986–1989. doi: 10.1128/JCM.00820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cesaro S, Chinello P, Rossi L, Zanesco L. Saccharomyces cerevisiae fungemia in a neutropenic patient treated with Saccharomyces boulardii. Support Care Cancer. 2000;8(6):504–505. doi: 10.1007/s005200000123. [DOI] [PubMed] [Google Scholar]
- 41.Land MH, Rouster-Stevens K, Woods CR, Cannon ML, Cnota J, Shetty AK. Lactobacillus sepsis associated with probiotic therapy. Pediatrics. 2005;115(1):178–181. doi: 10.1542/peds.2004-2137. [DOI] [PubMed] [Google Scholar]
- 42.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 46.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


