Abstract
Protein-protein interactions are essential for almost all intracellular and extracellular biological processes. Regulation of protein-protein interactions is one strategy to regulate cell fate in a highly selective manner. Specifically, peptides are ideal candidates for inhibition of protein-protein interactions because they can mimic a protein surface to effectively compete for binding. Additionally, peptides are synthetically accessible and can be stabilized by chemical modifications. In this review, we survey screening and rational design methods for identifying peptides to inhibit protein-protein interactions, as well as methods for stabilizing peptides to effectively mimic protein surfaces. In addition, we discuss recent applications of peptides to regulate protein-protein interactions for both basic research and therapeutic purposes.
Graphical abstract
Introduction
Protein-protein interactions (PPIs) are essential for various biological processes, including cell proliferation, cell division, signal transduction, transcription, translation, and programmed cell death. PPIs also play a critical role in various diseases and pathological conditions such as neurodegeneration, cardiovascular diseases, and cancer [1]. The interactome, the complete list of the molecular interactions in a cell, has been predicted to contain about 650,000 PPIs. Because PPIs mediates critical molecular communications, PPI-modifying drugs provide a massive therapeutic potential. However, targeting PPIs with small molecules, which are traditionally used to modulate a single enzyme function, is challenging for two main reasons. Primarily, the binding surfaces between proteins are usually large (1,500–3,000 Å2) and involve many polar and hydrophobic interactions. In addition, binding surfaces are typically flat, without a defined binding pocket for binding of a small-molecule drug. Targeting PPIs with small molecules has produced a mix of both exciting success stories and frustrating challenges, and has been discussed in several recent reviews [1–3]. Peptides and peptidomimetics (modified peptides), on the other hand, are perfect candidates to target PPIs. Interestingly, it has been estimated that 15–40% of all PPIs are mediated by a short linear peptide [2]. Peptides can be rationally designed based on the natural sequences that mediates PPI in the proteins, and therefore can mask a critical part of the binding surface; furthermore, peptidomimetics can be chemically modified to stabilize the bioactive conformation mimicking the 3D protein structure. Additionally, peptides and peptidomimetics can modulate intracellular targets by crossing the cell membrane independently (e.g. cyclosporine) or via conjugation to cell-penetrating vehicle peptides [4]. In this review we will discuss common approaches for developing peptides to investigate and modulate PPIs, as well as examples of therapeutic application of peptides as PPI regulators.
Peptides derived through high-throughput methods
Peptides to modulate PPIs are often discovered via high-throughput screening. Because many natural small molecules have evolved to interact with proteins, and vice versa, screening of natural product libraries for PPI inhibitors is often used. Natural macrocyclic peptides in particular are large and flexible enough to modulate protein surfaces, yet their cyclic structure provides some rigidity, increased resistance to proteolysis and cell-permeability, therefore also making excellent drug candidates [5]. For example, screening of a 2070-compound macrocycle library identified robotnikinin, a macrocyclic peptide that disrupts the interaction between Patched and Sonic hedgehog proteins in the hedgehog signal transduction pathway [6].
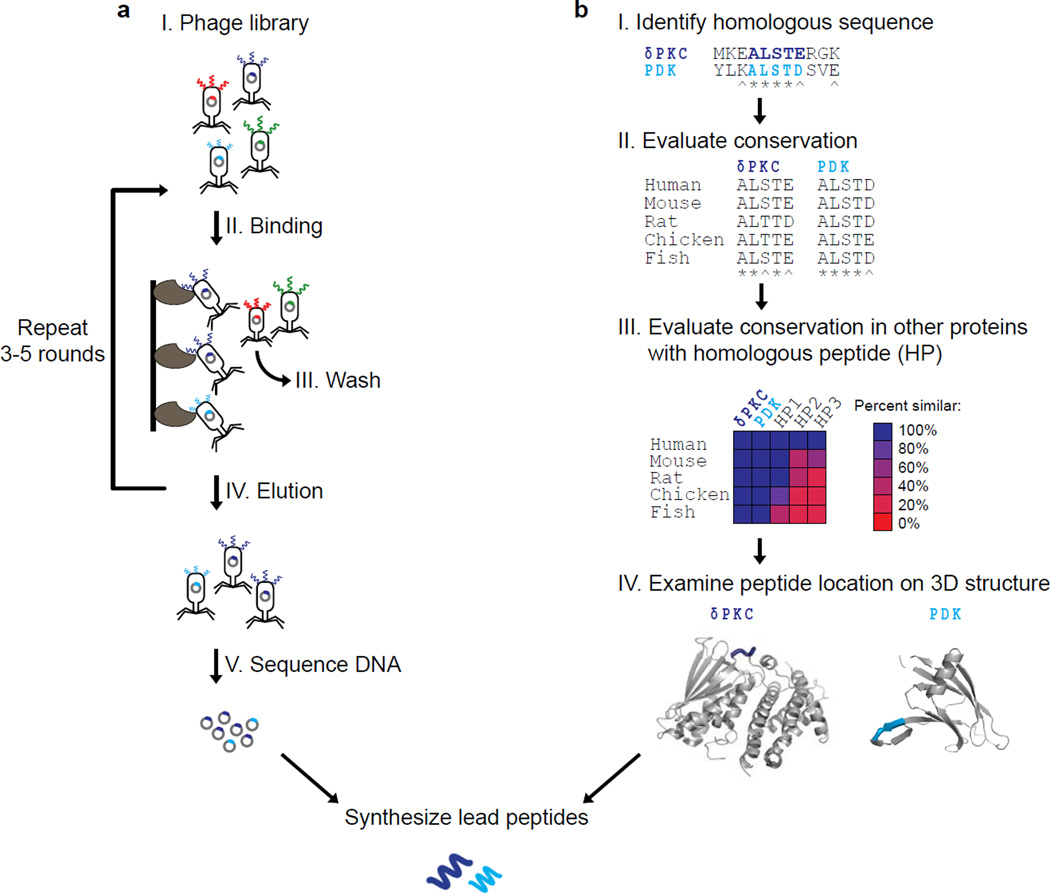
Protein-protein interactions are more often probed via selection methods such as phage display [7] in which bacteriophages (viruses consisting of DNA or RNA encapsulated by coat proteins) are engineered to uniformly display a library of random-sequence peptides fused to the outward-facing terminus of one of several phage coat proteins (Fig. 1a, I) [7]. In this selection process, called biopanning [7], the protein target is immobilized and the phage display library is introduced for binding (Fig. 1a, II), then the unbound phage is washed away (Fig. 1a, III) and the bound phage is eluted (Fig. 1a, IV). This process is repeated for 3–5 rounds, to enrich for the strongest binders. The selected phage DNA is sequenced to identify the amino acid sequence that provides strong binding to the target (Fig. 1a, V). Phage display is faster, less expensive, and requires less complicated data analysis techniques than high-throughput screening of small molecule libraries [8].
Figure 1. Workflows for peptide identification via phage display selection or rational design.
(a) In phage display, bacteriophage is engineered to uniformly display a library of peptides on the surface (I). The library is introduced to the immobilized protein of interest (II), the unbound phage is washed away (III), and the bound phage is eluted (IV). This process is repeated for 3–5 rounds to enrich for the strongest binders. Finally, the phage DNA is sequenced (V) to identify the selected peptide sequences, and these lead peptides are synthesized for further characterization. (b) In a rational design approach for developing peptide inhibitors of PPIs, the proteins of interest are first aligned to identify short sequences of homology (I). These sequences are checked for evolutionary conservation (II); other proteins sharing this homologous peptide (HP) sequence are also checked for evolutionary conservation (III). A sequence that is specific to the desired PPI should be conserved in the proteins of interest, but not in other proteins. The peptide sequence is then mapped onto the three-dimensional structures of the proteins of interest (PDK (PDB: 1JM6), C2 domain of δPKC (PDB:1BDY)) (IV), if available. Finally, the peptides are synthesized for further characterization. * denotes identity, ^ denotes homology.
Phage display was developed in 1985 [9] and is still the most commonly used display method [10]. However, display can also be done in yeast [11] or bacteria [12]. If complicated folding patterns or post-translational modifications are desired, insect or mammalian cells can also be used for display [13]. Due to limits of transformation efficiency, commercial phage display libraries typically have a diversity of 108–1010[7]; to evade this limitation, acellular display methods have also been developed, in which DNA is transcribed and translated in vitro, then is covalently linked to the translated peptide via a covalent linker. These libraries have no upper limit on peptide diversity [14].
Phage display has been used to characterize protein-protein interactions in several ways. Traditionally, phage display has been used to map epitope regions by displaying protein fragments and panning for the epitope against the immobilized monoclonal antibody [15]. Peptide phage display is an increasingly common strategy to characterize known PPIs, discover new PPIs, and identify candidate peptides for PPI inhibition [16]. For example, a random-sequence peptide display library was used to characterize the interface and binding modes of the interaction between p300 and HIF-1α, and several of the identified peptides inhibited the interaction between p300 and HIF-1α with an IC50 of <5 µM [17]. In another study, peptide phage display was used to identify the PY binding motif (LPxY or PPxY) as a strong interactor with the oxidoreductase WWOX; the authors then validated several PY-containing proteins as novel binding partners of WWOX, including the E3 ubiquitin ligase ITCH [18].
Although high-throughput screening methods have proven effective for identifying peptides that regulate PPIs, there are drawbacks. The process and resources needed for synthesizing and maintaining hundreds of thousands of compounds or phages, as well as for optimizing high-throughput assays, are costly and time-consuming. As protein interfaces become better understood through X-ray crystal structures of protein complexes and computational methods, rationally designing of peptides inhibitors of PPIs without conducting large screens is enabled.
Rational design of peptides to inhibit PPIs
Rational design is an attractive approach to efficiently develop peptides to selectively inhibit PPIs. Interactions between short linear sequences in proteins were shown to be crucial for cell signaling, and the interactions are often transient, allowing for rapid changes in response to varying stimuli [19]. Our laboratory has developed peptides derived from proteins interfaces that act as competitive inhibitors of PPIs demonstrating high selectivity and bioactivity [20–23]. Focusing on protein kinase C (PKC) and its scaffold protein receptor for activated C kinase (RACK) [22, 24–26] we developed peptides that act as specific PKC regulators and studied them in several animal models including cancer [27], cardiovascular diseases [28] and neurodegenerative disease [29] as well as in humans [30, 31]. Recently we developed a series of peptides derived from δPKC to selectively inhibit the docking and phosphorylation of individual substrates, described below; these peptides are highly selective inhibitors of the PPIs between δPKC and one substrate [32–34].
Several protein kinases have docking site for the substrate that is outside of the active/catalytic site of the enzyme. The docking site may recognize a short peptide motif in the target protein substrates, distinct from the phosphoacceptor site on the substrate [35]. The docking site providing further selectivity for the substrate without compromising the stereochemical requirements for efficient catalysis at the active site [36]. Docking is particularly prevalent in serine/threonine kinases and has been characterized for a number of protein kinase families including c-Jun N-terminal kinases (JNKs), cyclin-dependent kinases (CDKs), and mitogen-activated protein kinases (MAPKs) (for review: [35]). For example, Lee et al. identified a six amino acid docking site at the C-terminal Src kinase (Csk), and a peptide mimicking the docking site inhibited Csk phosphorylation of Src (IC50 = 21 µM) by inhibiting the Src-Csk interaction, but only moderately inhibited Csk kinase activity against other substrates [37].
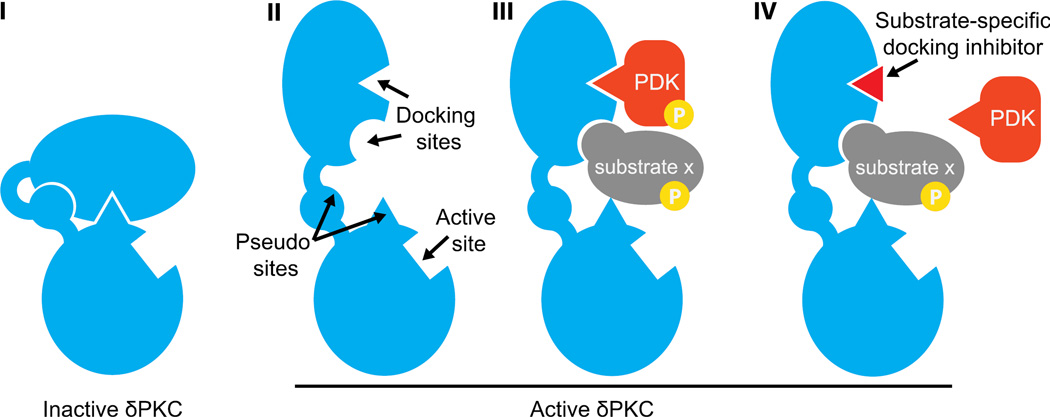
We hypothesized that the substrate-specific docking site on the kinase may be masked by an intramolecular interaction in the inactive kinase conformation, and activation of the kinase results in a conformational change revealing this docking site (Fig. 2). Furthermore, we hypothesize that in some cases this intramolecular interaction site on the kinase mimics the PKC-binding site on the substrate, and therefore will have similar sequence. Based on this, we developed a general procedure to design novel selective protein-protein interactions inhibitors (Fig. 1b). First, the two proteins that are known to interact are aligned to identify short homologous domains as candidate sequences for peptide development (Fig. 1b, I). Next, the identified candidate sequences should be checked for evolutionary conservation, as it has been shown that conserved sequences are likely to be important for protein functions such as mediation of protein-protein interactions (Fig. 1b, II) [38, 39]. Finally, to ensure the selectivity of the peptide, other proteins containing a homologous sequence are identified and conservation of this sequence is assessed (Fig. 1b, III). To reduce likelihood of off-target peptide interactions, the candidate sequence should not be conserved in these other proteins. While this method uses sequence-based design, the protein structure can also be used to guide the design of the peptide, if available.
Figure 2. A rationally designed selective inhibitor of single substrate phosphorylation.
δPKC is in equilibrium between multiple active and inactive conformations. In the inactive δPKC (I), each docking site interacts with a pseudo site in δPKC, which mimics the docking site for δPKC on the substrate. A shift in equilibrium between the inactive (I) and active (II) conformations of δPKC exposes the catalytic site and substrate-specific docking sites (docking sites for PDK and substrate×on δPKC, II). Docking of each substrate to the kinase induces proximity to the catalytic site, leading to substrate phosphorylation (P, III). A rationally designed peptide (red triangle) corresponding to the PDK docking site is a competitive inhibitor for docking to and the resulting phosphorylation of PDK by δPKC, without affecting docking and phosphorylation of other δPKC substrates (e.g., substrate x, IV).
Several peptides have been developed using this procedure and these peptides demonstrated high bioactivity and selectivity. For example, we developed a peptide based on short homologous regions between δPKC and its substrate, pyruvate dehydrogenase kinase (PDK). We rationally designed a peptide corresponding to the PDK docking site on δPKC, which we termed pseudo-PDK or ψPDK peptide. We were encouraged to find that the homologous sequences from which ψPDK was derived were located on the surface of both kinase and substrate, which is consistent with the sequence being involved in protein-protein interaction (Fig. 1b, IV). We found that ψPDK peptide selectively inhibits PDK interaction with δPKC and the resulting δPKC-mediated phosphorylation of PDK with an in vitro Kd of ~50 nM. When used in rats, ψPDK reduces cardiac injury with an IC50 of ~5 nM. Moreover, ψPDK peptide does not affect the phosphorylation of other δPKC substrates under the same experimental conditions, even at 1 µM concentration, demonstrating that PDK phosphorylation alone is critical for δPKC-mediated injury by heart attack [32].
The rational approach we identified is not unique for kinases and substrates; adopting the same approach to target proteins outside of the PKC family, we identified peptide P110 that selectively inhibits excessive mitochondrial fission by inhibiting PPI between dynamin related protein 1 (Drp1) and Fis1 [40]. We also rationally designed a peptide (p4d) which exhibits leishmanicidal and trypanocidal activity in vitro and in in vivo [41–43]. A summary of peptides derived using this rational approach, along with biological insights learned, is in Table 1.
Table 1.
List of the δPKC substrates that are discussed in the text, the peptides derived from them, and their function.
| Protein | Peptide | Kd (in vitro) | IC50 (ex vivo) | Biological activity | Ref |
|---|---|---|---|---|---|
| Pyruvate dehydrogenase kinase (PDK) |
ALSTER | 50 nM | ~5 nM | ψPDK bound to δPKC in vitro, inhibited binding between δPKC and PDK, and also selectively inhibited PDK phosphorylation in vitro and ex vivo. |
[32] |
| Glyceraldehyde -3-Phosphate Dehydrogenase (GAPDH) |
GIVEGL | n.d. | n.d. | ψGAPDH bound to δPKC in vitro, inhibited binding between δPKC and GAPDH, and also selectively inhibited GAPDH phosphorylation in vitro and ex vivo. |
[33] |
| Myristoylated alanine-rich C- kinase substrate (MARCKS) |
KAAEEP | 3.3 nM | n.d. | ψMARCKS bound to δPKC in vitro, and selectively inhibited MARCKS phosphorylation ex vivo. |
[34] |
| Dynaminrelated protein 1 (Drp1) |
YTDFDE | 2.9 nM | ~100 nM | ψDrp1 bound to δPKC in vitro, and selectively inhibited Drp1 phosphorylation ex vivo. |
[34] |
| Insulin receptor substrate 1 (IRS1) |
FRPRSKS | 2.9 nM | ~350 nM | ψIRS1 bound to δPKC in vitro, and selectively inhibited IRS1 phosphorylation ex vivo. |
[34] |
n.d., not determined.
Peptidomimetics as secondary structure mimics
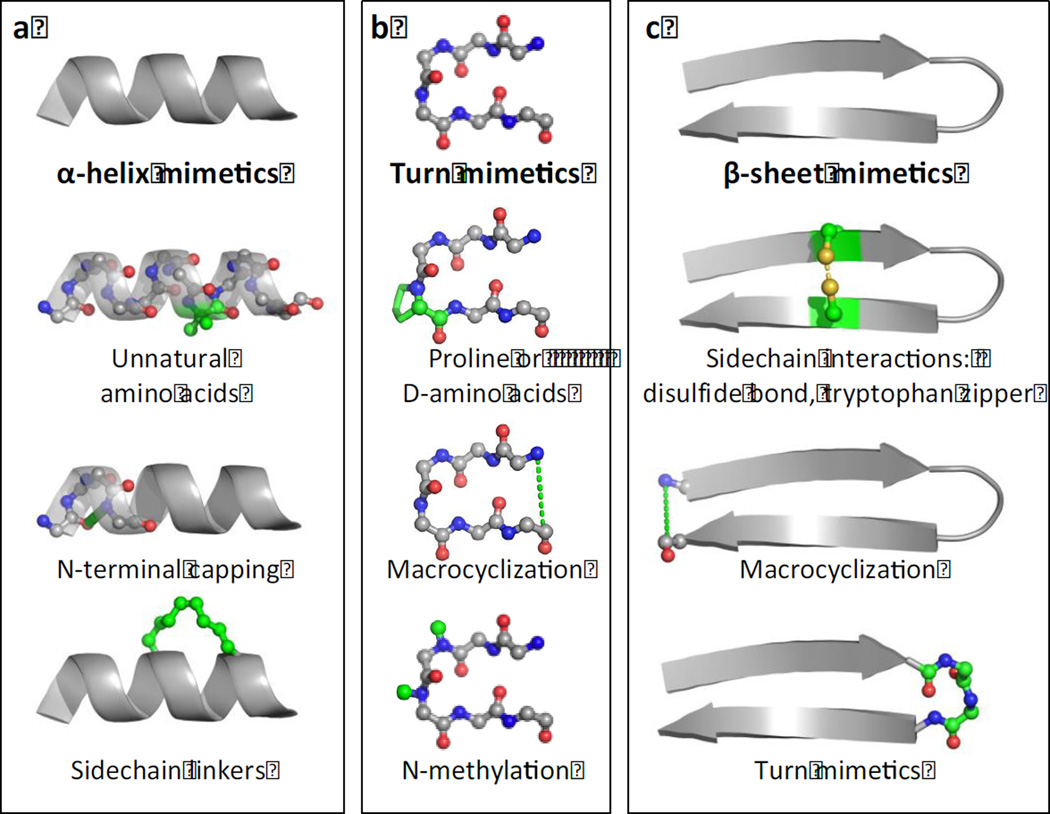
Once the region mediating the PPI is known, a peptide mimicking that region is an excellent starting point for developing inhibitors or tools to probe the interaction. Because short peptides are often flexible and unstructured, potency of inhibitors can be increased by stabilizing or inducing the desired secondary structure [44]. Protein-protein interfaces can involve all three main protein structural motifs: alpha-helices, B-strands, and loop or turn regions; peptides mimicking these motifs can be stabilized in different ways (Fig. 3).
Figure 3. Methods of stabilizing peptide mimetics of alpha-helices, turns, and beta-hairpins.
(a) alpha-helices can be stabilized by using unnatural amino acids, N-terminal capping, or sidechain linkers such as stapling. (b) Turn mimetics may be stabilized by using proline or D-amino acids, macrocyclization, or N-methylation of the backbone amide. (c) Beta-sheet mimetics, specifically beta-hairpins, can be stabilized by sidechain interactions such as disulfide bonds or tryptophan zippers, macrocyclization, or by using turn mimetics to induce hairpin formation.
In many cases, the interaction between two proteins is primarily facilitated by a single alpha-helix that includes at least two “hotspot” residues [44]. Short peptide mimics of alpha-helices can be stabilized by modifying the amino acid sequence to include natural or unnatural residues with a high helix-forming propensity, such as alpha-aminoisobutyric acid [44]. Alpha-helices can also be stabilized covalently by N-terminal capping, in which the natural hydrogen bond between the carboxyl group of the N-terminal amino acid and the amide group of the i+3 amino acid is replaced with a covalent bond, therefore seeding helix formation. Alternatively, a covalent linker can be introduced between i and i+3, i+4, or i+7 sidechains, constraining the distance between the two amino acids to stabilize the helix [45]. These linkers can be all-hydrocarbon, thiol-based, lactam, or triazole linkers [45]. In particular, stapled peptides, which are made by introducing a hydrocarbon linker between two hydrophobic residues, have found many applications both in vitro and in vivo. In many cases, stapling improves proteolytic resistance, cell penetration, half-life, and affinity [46]. For example, Glas et al. designed a helical peptide to inhibit the interaction between human 14-3-3 protein and a bacterium virulence factor exoenzyme S (exoS) [47]. The peptide was derived from an 11-amino-acid sequence in exoS, called ESp, which was known to mediate binding. A hydrocarbon staple between two hydrophobic residues in i and i+3 positions increased the affinity of the peptide to 14-3-3 by almost 30-fold. Crystal structures showed that the stapled peptide interacted with 14-3-3 in a manner very similar to ESp.
Turn regions, defined as stretches of 2–6 amino acids that have a hydrogen bond between the i and i+n backbones, are highly represented at weak and transient protein-protein interfaces. Like alpha-helices, turn mimetics can be stabilized by modifying the peptide sequence to include turn-stabilizing amino acids such as proline, which produces a kink in the peptide backbone, or D-amino acids. Unlike alpha-helices, the N- and C-termini of turn regions are often close in proximity, and so macrocyclization is a common strategy to stabilize turn mimetics. N-methylation of backbone amides also can reduce peptide flexibility by increasing steric hindrance, therefore stabilizing the turn conformation.
Beta-sheets can be partially mimicked by beta-hairpin peptides, two short antiparallel beta strands connected by a turn region [45]. Beta-hairpins can be stabilized in their turn region by methods described above. A beta-hairpin can also be stabilized by inducing proximity between the two beta-strands, therefore seeding formation of the ordered hydrogen-bonding pattern between the two strands. This can be done by head-to-tail macrocyclization, by creating a disulphide bond between the two strands, or by pi-stacking between tryptophan amino acids on each strand (tryptophan zipper) [45].
Alpha-helices, beta-sheets, and turn regions can all be mimicked using novel chemical scaffolds as well. This field is quickly growing, and has been reviewed elsewhere [45, 48].
Peptide target PPIs as therapeutics
There are several examples of peptides and peptidomimetics targeting PPIs that have been in clinical trials. For example, ALRN-6924 (Aileron) is the first potent and specific re-activator of p53, a tumor suppressor protein, which binds and inhibits two of the p53 suppressor proteins, MDM2 and MDMX. p53 represents one of the most sought after oncology drug targets due to its central role in preventing the initiation and progression of many tumors. ALRN-6924 has been evaluated as an anti-tumor therapy in patients with advanced solid tumors or lymphomas [49]. Another example is the use of Arg-Gly-Asp (RGD)-based peptidomimetics to inhibit the interaction between integrins, mainly αvβ3 and αvβ5, and their substrates. The most successful example of such a strategy is the cyclic peptide cilengitide (Merck-Serono), c(RGDf(NMe)V) [50]. Cilengitide demonstrates increased potency and specificity towards endothelial cells, and has also been tested in vivo in several phase II trials of different cancers [51]. Another example is KAI-9803 (KAI Pharmaceuticals), a small peptide that specifically inhibits the interaction between δPKC and its intracellular receptor, RACK. δPKC activation during reperfusion initiates the molecular processes for cell death which ultimately leads to inflammation and damage in the heart or the brain during a stroke. KAI-9803 was tested in a phase II clinical trial to assess safety and efficacy in patients with acute myocardial infarction undergoing reperfusion via balloon angioplasty and appeared efficacious in the patients that have complete blood vessel occlusion [30].
Conclusion
Peptides are ideal chemical tools for inhibiting PPIs; rational design and screens have identified peptides that bind with exquisite specificity and affinity to their targets, therefore having relatively few off-target effects [52, 53]. However, peptides have few disadvantages as therapeutic entities, mainly instability and lack of intracellular penetration compared to small molecules. As discussed above, instability can be addressed using local chemical manipulation such as modified or unnatural amino acids, or general modification like cyclization, which leads to conformational restraint and significant enhancement of stability. Cell penetrating peptides, short positively charged peptides, can be used to deliver peptides into cells in a safe manner [4, 54, 55]. Because of these methods of circumventing the therapeutic disadvantages of peptides, peptides are gaining increasing attention as therapeutics.
Currently, there are more than 60 approved peptide medicines on the market and this is expected to grow significantly, with approximately 140 peptide drugs currently in clinical trials [56]. In addition, there is an increasing number of new peptide drugs entering clinical studies, from 1.2 per year (1970s) to 16.8 per year since 2000 [57]. Four peptide drugs have reached global sales over US$ 1 billion, and the global peptide drug market has been predicted to reach US$25.4 billion in 2018. Finally, this market share is growing much faster than that of other pharmaceuticals, and success rates for bringing biologics to market are now about twice that of small molecule drugs [56]. In the future, the growing popularity of peptides as therapeutics may bring on a new era of drug discovery.
Highlights.
Protein-protein interactions are essential for almost all cell processes
Peptides are ideal candidates for targeting protein-protein interactions (PPIs)
We survey screening and rational design methods for identifying peptides to inhibit PPIs
Peptides increasing popularity as therapeutics may bring on a new era of drug discovery
Acknowledgments
The work was supported by National Institutes of Health grant HL52141 to D.M.-R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Fischer G, Rossmann M, Hyvönen M. Alternative modulation of protein-protein interactions by small molecules. Curr Opin Biotechnol. 2015;35:78–85. doi: 10.1016/j.copbio.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.London N, Raveh B, Schueler-Furman O. Druggable protein-protein interactions-from hot spots to hot segments. Curr Opin Chem Biol. 2013;17(6):952–959. doi: 10.1016/j.cbpa.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 3. Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: Progressing toward the reality. Chem Biol. 2014;21(9):1102–1114. doi: 10.1016/j.chembiol.2014.09.001. ** In this review the authors summarize the progress of new protein-protein interaction inhibitors that have advanced to clinical trials within the past decade. The authors also offer their prospects for the future of inhibitors that target protein-protein interations in drug discovery.
- 4.Lonn P, Dowdy SF. Cationic ptd/cpp-mediated macromolecular delivery: Charging into the cell. Expert Opin Drug Deliv. 2015;12(10):1627–1636. doi: 10.1517/17425247.2015.1046431. [DOI] [PubMed] [Google Scholar]
- 5.Driggers EM, Hale SP, Lee J, Terrett NK. The exploration of macrocycles for drug discovery-an underexploited structural class. Nat Rev Drug Discov. 2008;7(7):608–624. doi: 10.1038/nrd2590. [DOI] [PubMed] [Google Scholar]
- 6.Bauer RA, Wurst JM, Tan DS. Expanding the range of 'druggable' targets with natural product-based libraries: An academic perspective. Curr Opin Chem Biol. 2010;14(3):308–314. doi: 10.1016/j.cbpa.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamzeh-Mivehroud M, Alizadeh AA, Morris MB, Church WB, Dastmalchi S. Phage display as a technology delivering on the promise of peptide drug discovery. Drug Discov Today. 2013;18(23–24):1144–1157. doi: 10.1016/j.drudis.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Willats WG. Phage display: Practicalities and prospects. Plant Mol Biol. 2002;50(6):837–854. doi: 10.1023/a:1021215516430. [DOI] [PubMed] [Google Scholar]
- 9.Smith GP. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 10.Omidfar K, Daneshpour M. Advances in phage display technology for drug discovery. Expert Opin Drug Discov. 2015;10(6):651–669. doi: 10.1517/17460441.2015.1037738. [DOI] [PubMed] [Google Scholar]
- 11.Kondo A, Ueda M. Yeast cell-surface display-applications of molecular display. Appl Microbiol Biotechnol. 2004;64(1):28–40. doi: 10.1007/s00253-003-1492-3. [DOI] [PubMed] [Google Scholar]
- 12.van Bloois E, Winter RT, Kolmar H, Fraaije MW. Decorating microbes: Surface display of proteins on escherichia coli. Trends Biotechnol. 2011;29(2):79–86. doi: 10.1016/j.tibtech.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Beerli RR, Bauer M, Buser RB, Gwerder M, Muntwiler S, Maurer P, Saudan P, Bachmann MF. Isolation of human monoclonal antibodies by mammalian cell display. Proc Natl Acad Sci U S A. 2008;105(38):14336–14341. doi: 10.1073/pnas.0805942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothe A, Hosse RJ, Power BE. In vitro display technologies reveal novel biopharmaceutics. FASEB J. 2006;20(10):1599–1610. doi: 10.1096/fj.05-5650rev. [DOI] [PubMed] [Google Scholar]
- 15.Beghetto E, Gargano N. Lambda-display: A powerful tool for antigen discovery. Molecules (Basel, Switzerland) 2011;16(4):3089–3105. doi: 10.3390/molecules16043089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu CH, Liu IJ, Lu RM, Wu HC. Advancement and applications of peptide phage display technology in biomedical science. J Biomed Sci. 2016;23:8. doi: 10.1186/s12929-016-0223-x. * This review article summarizes recent trends in application of peptide phage display for drug discovery and development. In particular, the authors detail recent advances in using peptide phage display to identify new protein-protein interactions, and to map protein-protein interaction domains.
- 17.Kyle HF, Wickson KF, Stott J, Burslem GM, Breeze AL, Tiede C, Tomlinson DC, Warriner SL, Nelson A, Wilson AJ, Edwards TA. Exploration of the hif-1alpha/p300 interface using peptide and adhiron phage display technologies. Mol Biosyst. 2015;11(10):2738–2749. doi: 10.1039/c5mb00284b. [DOI] [PubMed] [Google Scholar]
- 18. Abu-Odeh M, Bar-Mag T, Huang H, Kim T, Salah Z, Abdeen SK, Sudol M, Reichmann D, Sidhu S, Kim PM, Aqeilan RI. Characterizing ww domain interactions of tumor suppressor wwox reveals its association with multiprotein networks. J Biol Chem. 2014;289(13):8865–8880. doi: 10.1074/jbc.M113.506790. ** Here, the authors investigate the interaction between the WW domain of the tumor suppressor WWOX and its potential binding partners. Binding partners were identified by pulldown of GST-WW fusions and subsequent LC-MS/MS. From these binding partners, a binding motif was identified which was corroborated by random peptide phage display. This study illustrates the power of phage display coupled with alternative techniques for characterizing protein-protein interfaces.
- 19.Blikstad C, Ivarsson Y. High-throughput methods for identification of protein-protein interactions involving short linear motifs. Cell communication and signaling : CCS. 2015;13:38. doi: 10.1186/s12964-015-0116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mochly-Rosen D, Khaner H, Lopez J, Smith BL. Intracellular receptors for activated protein kinase c. Identification of a binding site for the enzyme. J Biol Chem. 1991;266(23):14866–14868. [PubMed] [Google Scholar]
- 21.Souroujon MC, Mochly-Rosen D. Peptide modulators of protein-protein interactions in intracellular signaling. Nat Biotechnol. 1998;16(10):919–924. doi: 10.1038/nbt1098-919. [DOI] [PubMed] [Google Scholar]
- 22.Qvit N, Mochly-Rosen D. Highly specific modulators of protein kinase c localization: Applications to heart failure. Drug Discov Today Dis Mech. 2010;7(2):e87–e93. doi: 10.1016/j.ddmec.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mochly-Rosen D, Das K, Grimes KV. Protein kinase c, an elusive therapeutic target? Nat Rev Drug Discov. 2012;11(12):937–957. doi: 10.1038/nrd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Churchill EN, Qvit N, Mochly-Rosen D. Rationally designed peptide regulators of protein kinase c. Trends Endocrinol Metab. 2009;20(1):25–33. doi: 10.1016/j.tem.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams DR, Ron D, Kiely PA. Rack1, a multifaceted scaffolding protein: Structure and function. Cell communication and signaling : CCS. 2011;9:22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The rack1 scaffold protein: A dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62(6):1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- 27.Kim J, Mochly-Rosen D. Protein kinase c in cancer signaling and therapy. Humana Press; 2010. Regulation of pkc by protein-protein interactions in cancer; pp. 79–103. [Google Scholar]
- 28.Inagaki K, Chen L, Ikeno F, Lee FH, Imahashi K, Bouley DM, Rezaee M, Yock PG, Murphy E, Mochly-Rosen D. Inhibition of delta-protein kinase c protects against reperfusion injury of the ischemic heart in vivo. Circulation. 2003;108(19):2304–2307. doi: 10.1161/01.CIR.0000101682.24138.36. [DOI] [PubMed] [Google Scholar]
- 29.Qi X, Disatnik MH, Shen N, Sobel RA, Mochly-Rosen D. Aberrant mitochondrial fission in neurons induced by protein kinase c delta under oxidative stress conditions in vivo. Mol Biol Cell. 2011;22(2):256–265. doi: 10.1091/mbc.E10-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bates E, Bode C, Costa M, Gibson CM, Granger C, Green C, Grimes K, Harrington R, Huber K, Kleiman N, Mochly-Rosen D, et al. Intracoronary kai-9803 as an adjunct to primary percutaneous coronary intervention for acute st-segment elevation myocardial infarction. Circulation. 2008;117(7):886–896. doi: 10.1161/CIRCULATIONAHA.107.759167. [DOI] [PubMed] [Google Scholar]
- 31.Lincoff AM, Roe M, Aylward P, Galla J, Rynkiewicz A, Guetta V, Zelizko M, Kleiman N, White H, McErlean E, Erlinge D, et al. Inhibition of delta-protein kinase c by delcasertib as an adjunct to primary percutaneous coronary intervention for acute anterior st-segment elevation myocardial infarction: Results of the protection ami randomized controlled trial. Eur Heart J. 2014;35(37):2516–2523. doi: 10.1093/eurheartj/ehu177. [DOI] [PubMed] [Google Scholar]
- 32. Qvit N, Disatnik MH, Sho J, Mochly-Rosen D. Selective phosphorylation inhibitor of delta protein kinase c–pyruvate dehydrogenase kinase protein–protein interactions: Application for myocardial injury in vivo. J Am Chem Soc. 2016;138(24):7626–7635. doi: 10.1021/jacs.6b02724. ** Here, a general approach to develop novel inhibitors of protein-protein interactions is presented. Based on a rational approach, the authors developed short peptides that inhibit the protein-protein interactions between a kinase and only one of its many susbtrates. The authors demostrated the efficacy and selectivity of the peptide in various systems including in vitro, ex vivo and in vivo.
- 33.Qvit N, Joshi AU, Cunningham AD, Ferreira JC, Mochly-Rosen D. Gapdh (glyceraldehyde-3-phosphate dehydrogenase) protein-protein interaction inhibitor reveals a non-catalytic role for gapdh oligomerization in cell death. J Biol Chem. 2016;291(26):13608–13621. doi: 10.1074/jbc.M115.711630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qvit N, Kornfeld OS, Mochly-Rosen D. Engineered substrate- specific delta pkc isoform antagonists to enhance cardiac therapeutics. Angewandte Chemie International Edition in English. 2016 doi: 10.1002/anie.201605429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8(7):530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: The role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 37.Lee S, Lin X, Nam NH, Parang K, Sun G. Determination of the substrate-docking site of protein tyrosine kinase c-terminal src kinase. Proc Natl Acad Sci U S A. 2003;100(25):14707–14712. doi: 10.1073/pnas.2534493100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257(2):342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 39.Caffrey DR, Somaroo S, Hughes JD, Mintseris J, Huang ES. Are protein-protein interfaces more conserved in sequence than the rest of the protein surface? Protein Sci. 2004;13(1):190–202. doi: 10.1110/ps.03323604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi X, Qvit N, Su YC, Mochly-Rosen D. A novel drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. J Cell Sci. 2013;126(Pt 3):789–802. doi: 10.1242/jcs.114439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qvit N, Crapster JA. Peptides that target protein-protein interactions as an anti-parasite strategy. Chim Oggi Chem Today. 2014;32(6):62–66. [Google Scholar]
- 42. Qvit N, Schechtman D, Peña DA, Berti DA, Soares CO, Miao Q, Liang L, Baron LA, Teh-Poot C, Martínez-Vega P, Ramirez-Sierra MJ, et al. Scaffold proteins lack and track as potential drug targets in kinetoplastid parasites: Development of inhibitors. Int J Parasitol Drugs Drug Resist. 2016;6:74–84. doi: 10.1016/j.ijpddr.2016.02.003. ** Here, the athours use a general approach to target parasitic disases. A library of peptides that were designed based on sequence homology between a scaffold protein of the parasite and its mammalian homologues was screened for activity in two parasites: Leishamnia and Trypanosoma cruzi. One peptide showed low micromolar activity in both parasites. Next, using structure-activity relationship studies, a cyclic peptide with improved stability and increased activity was indentifed and demonstrated promising results in two animal studies.
- 43.Qvit N, Kornfeld OS. Development of a backbone cyclic peptide library as potential antiparasitic therapeutics using microwave irradiation. J Vis Exp. 2016;107:e53589. doi: 10.3791/53589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rezaei Araghi R, Keating AE. Designing helical peptide inhibitors of protein-protein interactions. Curr Opin Struct Biol. 2016;39:27–38. doi: 10.1016/j.sbi.2016.04.001. ** This review summarizes strategies for stabilizing helical peptides with chemical modifications, with a focus on sidechain crosslinking chemistry, specifically for application to protein-protein interactions. The authors highlight the importance of computational modeling, improved high-throughput screen methods, and basic understanding of sequence-structure-function relationships for effectively inhibiting protein-protein interactions with helical peptides.
- 45. Pelay-Gimeno M, Glas A, Koch O, Grossmann TN. Structure-based design of inhibitors of protein-protein interactions: Mimicking peptide binding epitopes. Angew Chem Int Ed Engl. 2015;54(31):8896–8927. doi: 10.1002/anie.201412070. * This review article elegantly describes and illustrates strategies for stabilizing and synthesizing peptidemimetics mimicking alpha helices, beta sheets, and turns. The authors further discuss past examples of using peptidomimetics for inhibiting protein-protein interactions.
- 46.Verdine GL, Hilinski GJ. Stapled peptides for intracellular drug targets. Methods Enzymol. 2012;503:3–33. doi: 10.1016/B978-0-12-396962-0.00001-X. [DOI] [PubMed] [Google Scholar]
- 47. Glas A, Bier D, Hahne G, Rademacher C, Ottmann C, Grossmann TN. Constrained peptides with target-adapted cross-links as inhibitors of a pathogenic protein-protein interaction. Angew Chem Int Ed Engl. 2014;53(9):2489–2493. doi: 10.1002/anie.201310082. ** This study targets the interaction between bacterium virulence factor ExoS and human protein 14-3-3. The authors designed and tested several macrocyclic peptidomimetics based on the known interaction motif ESp. This study is an excellent illustration of systematic and targeted peptidomimetic strategies to optimize a peptide inhibitor of a protein-protein interaction for possible therapeutic application.
- 48.Jayatunga MK, Thompson S, Hamilton AD. Alpha-helix mimetics: Outwards and upwards. Bioorg Med Chem Lett. 2014;24(3):717–724. doi: 10.1016/j.bmcl.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 49. Burgess A, Chia KM, Haupt S, Thomas D, Haupt Y, Lim E. Clinical overview of mdm2/x-targeted therapies. Front Oncol. 2016;6:7. doi: 10.3389/fonc.2016.00007. * In this review the authors give a nice overview of the current lead compounds that target either MDM2- or MDMX in various stages of development, from late pre-clinical studies to early phase clinical trials.
- 50.Mannhold R, Kubinyi H, Folkers G, Dömling A. Protein-protein interactions in drug discovery. John Wiley & Sons; 2013. [Google Scholar]
- 51.Bradley DA, Daignault S, Ryan CJ, Dipaola RS, Cooney KA, Smith DC, Small E, Mathew P, Gross ME, Stein MN, Chen A, et al. Cilengitide (emd 121974, nsc 707544) in asymptomatic metastatic castration resistant prostate cancer patients: A randomized phase ii trial by the prostate cancer clinical trials consortium. Invest New Drugs. 2011;29(6):1432–1440. doi: 10.1007/s10637-010-9420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qvit N. Development and therapeutic applications of oligonucleotides and peptides. Chim Oggi Chem Today. 2011;29(2):4–7. [Google Scholar]
- 53.Mochly-Rosen D, Qvit N. Peptide inhibitors of protein-protein interactions. Chim Oggi Chem Today. 2010;28(1):14–16. [Google Scholar]
- 54. Herce HD, Garcia AE, Cardoso MC. Fundamental molecular mechanism for the cellular uptake of guanidinium-rich molecules. J Am Chem Soc. 2014;136(50):17459–17467. doi: 10.1021/ja507790z. ** Cell pentrating peptides (CPPs) are short peptides that facilitate cellular uptake of various cargos. Since large macromolecules have no ability to enter cells and require delivery vehicles, CPPs show great potential to deliver otherwise undeliverable macromolecular therapeutics into cells for experimentation in cell culture and in vivo. Moreover, clinical data from over 25 completed Phase I and Phase II clinical trials confirm the safety of CPP delivery in vivo. Nevertheless, the mechanism by which CPPs efficiently cross the plasma membrane remains a fundamental open question. In this study the author address this question using computational, in vitro and live-cell approaches.
- 55.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: Delivery of a biologically active protein into the mouse. Science. 1999;285(5433):1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 56.Fosgerau K, Hoffmann T. Peptide therapeutics: Current status and future directions. Drug Discov Today. 2015;20(1):122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 57.Roy S, Ghosh P, Sekhar Roy N, Mazumder A, Roy K, Kumar Manna A, Mallick S, Ahmed I. Peptide based molecules as protein-protein interaction inhibitors: Tools for chemical genetics and therapy. Curr Chem Biol. 2012;6(2):145–163. [Google Scholar]