Summary
During epithelial to mesenchymal transition (EMT), cells lining the tissue periphery break up their cohesion to migrate within the tissue. This dramatic reorganization involves a poorly characterized reorientation of the apico-basal polarity of static epithelial cells into the front-rear polarity of migrating mesenchymal cells. To investigate the spatial coordination of intracellular reorganization with morphological changes, we monitored centrosome positioning during EMT in vivo, in developing mouse embryos and mammary gland, and in vitro, in cultured 3D cell aggregates and micro-patterned cell doublets. In all conditions, centrosomes moved from their off-centered position next to intercellular junctions toward extra-cellular matrix adhesions on the opposite side of the nucleus, resulting in an effective internal polarity reversal. This move appeared supported by controlled microtubule network disassembly. Sequential release of cell confinement using dynamic micropatterns, and modulation of microtubule dynamics, confirmed that centrosome repositioning was responsible for further cell disengagement and scattering.
Introduction
Epithelium provides a selective barrier for controlled directional transport in duct containing organs such as airway, intestinal track or secretory tubule glands. Cells of epithelial origins are internalized at specific developmental stages to subsequently form internal tissues (Acloque et al., 2009). As they do so, some epithelial cells convert into mesenchymal cells, which migrate into and populate the underlying interstitial tissues. This topological tissue remodeling, during which peripheral cells become internal cells, is accompanied by dramatic intra-cellular reorganization (Lamouille et al., 2014). Epithelial cells disassemble the tight junctions they formed with their neighbors and that were ensuring the selective permeability. As they move inside, they lose the contact-free edge they had toward the outer medium and get fully surrounded by cells and extra-cellular matrix (Acloque et al., 2009). Their secretory and endocytic functions, which were directed by the presence of this contact-free interface get redistributed toward adhesive edges. Therefore the epithelial-to-mesenchymal transition (EMT) not only involves reorganization of cell position and acquisition of a migratory phenotype but also implies a reorientation of cell function and polarity (Godde et al., 2010; Huang et al., 2012).
Cell polarity is an intrinsic bias in internal cell organization which is spatially adapted to extra-cellular cues and which direct cell functions (Bornens, 2008). In epithelia, intercellular junctions form a diffusion barrier between distinct plasma membrane domains and thus define the apical pole, toward the outer medium, and the basal pole, toward the extra-cellular matrix (ECM) that cells are attached to (Rodriguez-Boulan and Macara, 2014). The spatial segregation of cell-matrix adhesion and intercellular junctions physically separate the signaling and anchoring proteins they are associated with (Burute and Thery, 2012). The centrosome is off-centered toward the actin-rich apical pole (Hebert et al., 2012) and microtubule orientation along the apico-basal axis directs intra-cellular trafficking (Akhtar and Streuli, 2013). Together, the segregation of the two types of adhesions and the orientation of the microtubule network defines the apico-basal orientation of epithelial cell polarity from ECM toward the contact free edge. In mesenchymal cells, intercellular junctions are much weaker and do not define membrane domains as in epithelial cells. Instead, it is the cell migration machinery that directs cell polarity (Etienne-Manneville, 2013). Here also, mutual exclusion of signaling pathways segregate actin network polymerization at the advancing cell edge and actin contraction at the retracting edge and thereby define the mesenchymal front-rear polarity axis. Centrosome positioning toward the cell front and the associated asymmetric microtubule network organization is pivotal for the establishment and maintenance of the front-rear polarity axis of migrating cells (Etienne-Manneville, 2013; Luxton and Gundersen, 2011). Thus, during EMT the apico-basal polarity axis is converted into a front-rear axis (Godde et al., 2010; Nelson, 2009; Xu et al., 2009). The mechanism involved in the remodeling of cell internal polarity during this conversion has not yet been specifically addressed.
Simple geometrical considerations suggest that during EMT the epithelial polarity toward the contact-free edge is literally inverted toward the underlying basement membrane that mesenchymal cells digest as they move inward (Figure S1). Several examples of such polarity reversals have been observed within simplified epithelia in vitro, in response to conditions that are close to the changes that occur during EMT. Thus, modifying the composition of the extra-cellular matrix surrounding epithelial cyst, or modulation of integrins activation state, can induce the relocalization of polarity surface markers from the outside surface to the internal lumen and vice versa (Akhtar and Streuli, 2013; Nitsch and Wollman, 1980; Ojakian and Schwimmer, 1994; Rodriguez-Fraticelli et al., 2012; Wang et al., 1990; Yu et al., 2005). While the characterization of the mechanism supporting the inversion of surface markers and membrane protrusions has been addressed (Bryant et al., 2014; Scarpa et al., 2015), the reorganization of intracellular organization has not yet been studied. Interestingly, analyses of intermediate stages of developing chick auditory system and neural network, as well as observations of branching kidney tubules and inversion of thyroid follicles in culture have suggested the existence of concerted repositioning of internal organelles such as nucleus, Golgi apparatus, lysosomes and centrosome (Carney and Couve, 1989; Das and Storey, 2014; Nitsch and Wollman, 1980; Pollack et al., 1998; Yu et al., 2003). The synchrony between centrosome repositioning and epithelial cell migration within the underlying mesenchyme led to the hypothesis that internal polarity reversal is instrumental in the initiation of cell migration (Carney and Couve, 1989).
This hypothesis is supported by the observation of nucleus-centrosome axis orientation toward ECM rather than intercellular junctions in cells that are prompt in scattering (Desai et al., 2009). However epithelial transition to cell migration can occur without Golgi apparatus and centrosome repositioning (Anstrom and Raff, 1988; Revenu et al., 2014). Indeed, all cells don’t migrate with the centrosome in front of the nucleus (Higginbotham and Gleeson, 2007; Luxton and Gundersen, 2011; Pouthas et al., 2008; Tang and Marshall, 2012). Centrosome locates behind the nucleus of migrating lymphocytes (Takesono et al., 2010) and its position in neutrophils (Yoo et al., 2012) and migrating fibroblasts (Yvon et al., 2002) is debated. Some of the observed differences certainly come from the strong effect of cell microenvironment. Indeed, subtle variations of microenvironment geometry can reverse centrosome position in a given migrating cell type (Pouthas et al., 2008), making difficult to compare in vitro and in vivo conditions. Furthermore the mixed contributions of cell-cell contact, the process of migration itself and the multiple mechanisms regulating, together or separately, the position of nucleus and of centrosome add to the confusion (Luxton and Gundersen, 2011; Tang and Marshall, 2012). In the end, whether centrosome position is a cause or a consequence of cell migration could not be clarified. New methodological approaches appeared required to test whether internal polarity is actually reversed during EMT and whether it promotes or follows the cell migration process.
Here we use in-vitro models of mammary gland development (Debnath et al., 2003) and kidney tubulogenesis (Pollack et al., 1998) to study cell polarity during epithelial morphogenesis. In order to distinguish the effect of cell neighbors, cell migration, cell spreading and adhesion remodeling on cell polarization, we use micropatterns to control the shape and position of cells forming doublets. We show that nucleus-centrosome axis reorientation occurs in the few hours following mammary gland cells stimulation with transforming growth factor beta (TGF-β) and kidney cells stimulation with either TGF-β or hepatocyte growth factor (HGF). Centrosome repositioning at the cell center appeared driven by a partial disassembly of the microtubule network and the release of Partitioning defective protein Par3 from intercellular junctions. Finally, we show that polarity reversal by centrosome repositioning occurs prior to cell scattering and was necessary for cell dissociation during EMT.
Results
Polarity reversal during EMT in mouse development
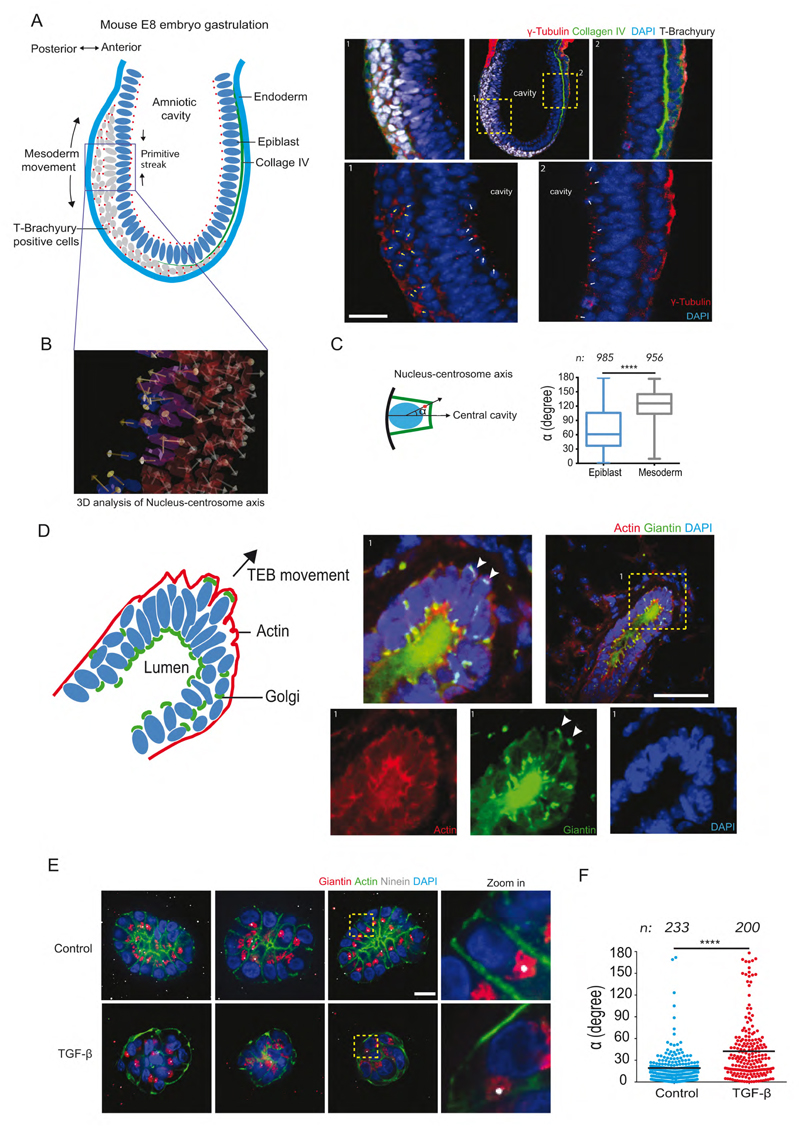
Several key developmental stages involve EMT and thus could display the polarity reversal we hypothesize. The first EMT event occurs at gastrulation, 5-6 days post fertilization. The primitive streak forms at the future posterior end of the embryo wherein a subset of epiblast cells differentiate into primary mesenchyme and ingress between the epiblast and endoderm layer (Acloque et al., 2009; Tam and Behringer, 1997). Epiblast cells are marked by expression of nuclear T-Brachyury along with breakdown of Collagen IV (Figure 1A). Using γ-tubulin as a marker for centrosome, we investigated polarity of cell populations destined for different cell fates. The centrosome in epiblast cells was localized close to the amniotic cavity, resulting in a nucleus-centrosome axis oriented toward the cavity. The epiblast cells undergoing EMT showed higher T-Brachyury expression and were positioned farther from the cavity. In these cells, centrosomes appeared relocated away from the cavity and the nucleus-centrosome axis pointed toward the endoderm layer (Figure 1B, C). This supported our hypothesis that upon the onset of EMT, when epiblast cells move inward to form the primitive streak, the nucleus-centrosome axis gets inverted.
Figure 1. Evidence of polarity reversal at various stages of mouse development and within 3D organotypic cell culture.
(A) Scheme representing germ layers of E8 mouse embryo, site of primitive streak formation and deduced nucleus-centrosome orientations from images in inset 1,2. Inset 1: Posterior end of embryo stained for T-Brachyury (white), γ-tubulin (red) and DAPI (blue). Arrows indicate cells expressing T-Brachyury with nucleus-centrosome axis (white arrows) orientated away from amniotic cavity (marked c). Inset2: Anterior end of E8 mouse showing cells without T-Brachyury and nucleus-centrosome axis pointing toward the amniotic cavity. Scale bar represents 20 μm.
(B) 3D analysis of nucleus-centrosome vectors in embryo using 3D image analysis software.
(C) Quantification of angle α contained by normal to cell base and nucleus-centrosome vector in mesoderm (T-brachyury positive) and Epiblast (T-brachyury negative) cells.
(D) Scheme representing nucleus-Golgi apparatus axis of cells in growing terminal end bud of female mouse mammary gland at 6-7 weeks of age. Inset 1: Merged image of terminal end bud stained for Golgi apparatus (green), F-actin (red) and nucleus (blue). Images of separate channels are below. Scale bar represents 50 μm.
(E) Examples of control and TGF-β treated MCF10A 3D cultures (Day7) labeled for Golgi apparatus (green), centrosome (white), F-actin (green) with zoomed and cropped image showing nucleus-centrosome orientation on the right. Scale bar represents 20 μm.
(F). Scatter plots show quantification of angle α for control and TGF-β treated MCF10A cells. n represents total number of cells quantified from Control (N=23) and TGF-β (N=25) acini. Two tailed-non-parametric Mann-Whitney test was used, ****: p<0.0001.
Later, at puberty, the ductal network within the mouse mammary gland expands by invading the surrounding fat pad. At this stage, specialized structures called terminal end buds appear at the end of the primary ducts (Hinck and Silberstein, 2005). Cell rearrangement and the collective migration of cells out of the bud is based on partial EMT (as the cells did not transit up to individual migration) (Ewald et al., 2008; Godde et al., 2010). Cell polarity axis orientation was inferred from the orientation of the Golgi apparatus with respect to the nucleus. In luminal cells, the Golgi apparatus was positioned toward the duct (Figure 1D) as in the case of lactating acini (Akhtar and Streuli, 2013). Interestingly, a few cells at the tip of the growing terminal end buds showed complete inversion of the nucleus-Golgi axis (Figure 1D and Movie S1). Other cells near the tip displayed a mispositioned Golgi apparatus suggestive of intermediate stages of polarity reversal (Figure 1D).
We thus found indications of nucleus-centrosome axis inversion at two distinct stages of mouse development, supporting our working hypothesis of polarity reversal occurring during EMT. Investigating the mechanics of polarity inversion in vivo at single cell resolution remains technically challenging. In order to obtain further insights into this process we used simpler and more accessible working systems to study the induction and consequences of those polarity reversals where parameters of interest could be better controlled.
Polarity reversal in 3D mammary gland cell culture
Self-organized mammary acini in 3D gels recapitulate numerous features of native tissue, including epithelial cell polarization (Debnath et al., 2003) and constitute a robust system amenable to induction of morphogenesis by the addition of growth factors (Debnath et al., 2003; Montesano et al., 2007; Seton-Rogers et al., 2004). We used MCF10A 3D cultures as a model of mammary gland acini to investigate polarity changes that may occur upon induction of EMT by TGF-β (Xu et al., 2009; Zhang et al., 2014) (Figure S2A-C). After seven days culture in 3D gels of laminin-rich basement membrane (commercialized as Matrigel), MCF10A show acini-like structures with enriched apical actin indicating the site of a future lumen (Figure 1E). Nontreated cysts showed a regular arrangement of nuclei in a single layer and were placed equidistant from the acinus center (Figure 1E). This regular geometric organization was lost in cysts treated with TGF-β1 (5 ng/ml) for 5 days. The severe disorganization of cell arrangement in cysts was also associated with misorientation of the polarity axes (Figure 1E, F). We found a similar disorganization of acini assembled from MDCK cells of kidney origin upon treatment with HGF (Figure S2D-G). Thus, cell mispositioning and polarity axes disorientation are closely connected in these 3D architectures. This complex interplay between cell shape, position and polarity raised a few central questions that are difficult to address in 3D culture systems. In particular, polarity misorientation could result from defective internal polarization mechanism or from a correct polarization in a perturbed context due to mispositioning of neighboring cells. Furthermore, cell migration is known to actively regulate both epithelial (Wang et al., 2013) and mesenchymal (Luxton and Gundersen, 2011) cell polarities, suggesting that cell motility could also actively direct polarity reorientation in these 3D cysts.
Polarity reversal in micropatterned cells
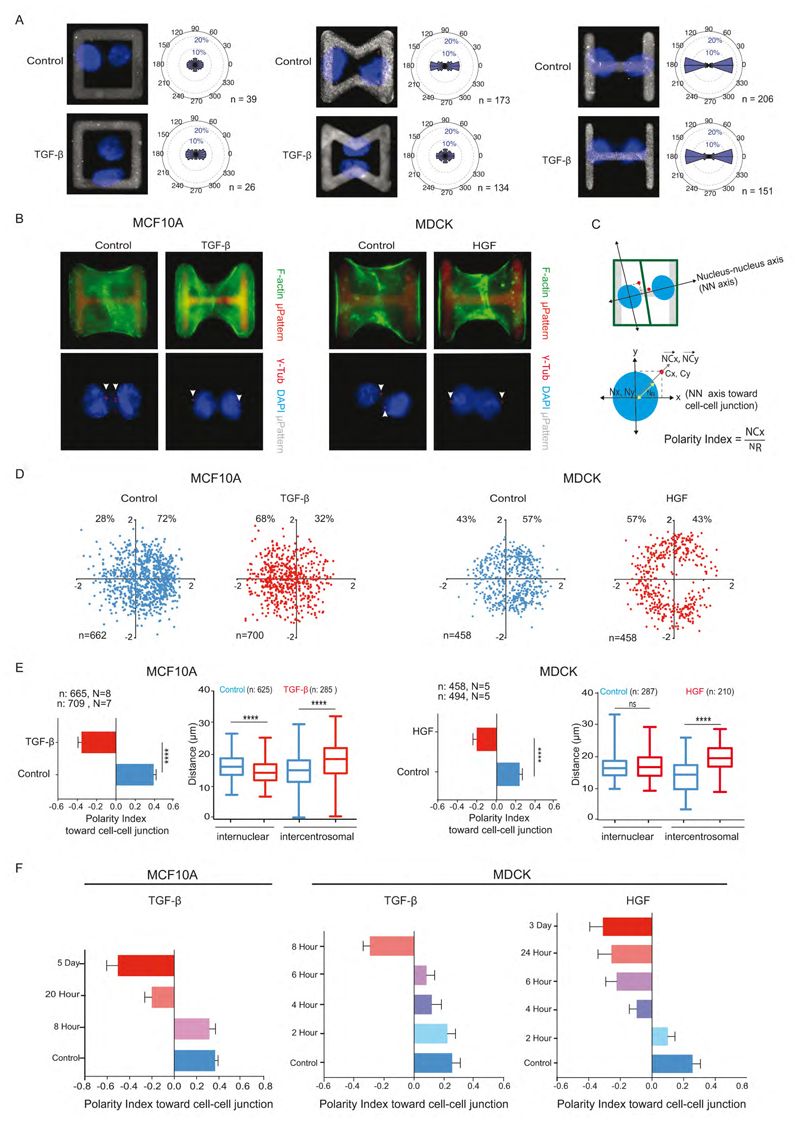
It appeared necessary to dissect the role of cell polarization, cell positioning and cell motion in tissue reorganization during EMT. This prompted us to work on a simpler but more controlled cell culture model, which can still recapitulate important aspects of morphogenesis (Théry, 2010). We first aimed at eliminating the variable effect due to the presence of multiple neighbors and their movement by adapting a minimal tissue model of 2 cells confined on micropattern geometry. To prevent the possible effect of cell movement on polarity reversal we looked for micropattern geometries that could block cell migration. Restricting micropattern size is not sufficient to prevent cell movement because cells can exchange their positions and rotate within the micropatterned area (Tseng et al., 2012). Square-shaped micropatterns could not prevent the rotation of normal epithelial cells and were even less able to constrain that of TGF-β -induced mesenchymal cells (Figure 2A, left). Bowtie-shaped micropatterns stabilized the position of epithelial cells but could not prevent mesenchymal cell motion (Figure 2A, middle). H-shaped micropattern could block both epithelial (Tseng et al., 2012) and mesenchymal cell motion, placing them in similar and thus comparable conditions (Figure 2A, right). Hence H-shaped micropatterns were used in further experiments to compare epithelial and mesenchymal cell polarity.
Figure 2. Polarity reversal is an early feature of EMT.
(A) Images of nuclei (blue) of MCF10A cell doublets on square (i), bowtie (ii) and H-shaped (iii) microcropatterns (grey). Graphs represent angular distribution of nucleus-nucleus axis (NN axis) orientation of cell doublets. n indicates number of cells.
(B) MCF10A and MDCK cell doublets on H-shaped micropattern were stained for F-actin (green) (top) or centrosome (red) and DNA (blue) (bottom).
(C) Axes system defined by NN axis (X axis) passing through center of nuclei of cell doublets and an axis perpendicular to NN axis (Y axis). Normalized nucleus-centrosome vector coordinates (NCx, NCy) were calculated by subtracting coordinates of centrosome (Cx, Cy) from Nucleus (Nx, Ny) and normalized by the nucleus distance (NR).
(D) Scatter plot of normalized NC vector. The total number of cells and the respective proportions (%) on positive and negative x-axis are indicated.
(E) Horizontal histograms show the quantification of polarity index, i.e normalized X coordinate of NC vector. N indicates the number of independent experiments, whereas n indicates the total number of single cells. Vertical box plots show the quantification of inter-nuclear and inter-centrosome distance.
(F) Polarity index toward cell-cell junction in control (blue) after varying the duration of TGF-β treatment and HGF treatment to MCF10A and MDCK cells.
Two tailed-non-parametric Mann-Whitney tests were used. ****: p<0.0001. Error bars indicate SEM. See also Figure S2 and S3.
Single MCF10A or MDCK cells were plated on H-shaped micropatterns and fixed 24 hour later to give them enough time to divide once and form daughter cell doublets (Figure 2B). Nucleus-centrosome vector orientations were measured with respect to the nucleus-nucleus axis pointing toward the intercellular junction (Figure 2C). Nucleus-centrosome distances were normalized with respect to nucleus size (Figure 2C). Thus, positive coordinates corresponded to nucleus-centrosome axes pointing toward adjacent cells and large values to highly eccentric centrosome positions. Both epithelial MCF10A (cultured in defined medium) and MDCK cells (cultured in classical growth medium with serum) displayed marked polarization toward the intercellular junction (Figure 2D). Strikingly, MCF10A cells pretreated with TGF-β for 5 days and MDCK cells pretreated with HGF for 3 days both displayed the opposite polarity orientation (Figure 2D), although contacting cells were still interacting and pulling on each others (Figure S3A, B). This polarity reversal was also quantified by measuring the centrosome X coordinate along the nucleus-nucleus axis, hereafter referred to as the cell polarity index toward intercellular junction. The coordinate sign change attests to the centrosome repositioning from the nucleus side oriented toward the intercellular junction to the side oriented toward extra-cellular matrix adhesions (Figure 2E). Interestingly, in MCF10A treated with TGF-β, this polarity reversal was due to both centrosomes moving away from the junction to the cell center and nuclei moving away from cell-matrix adhesions toward intercellular junction (Figure 2E, Figure S3C). By contrast, in MDCK, nuclei positioning was not affected by HGF treatment, and centrosome repositioning alone contributed to polarity reversal (Figure 2E).
Matrix stiffness promotes polarity reversal
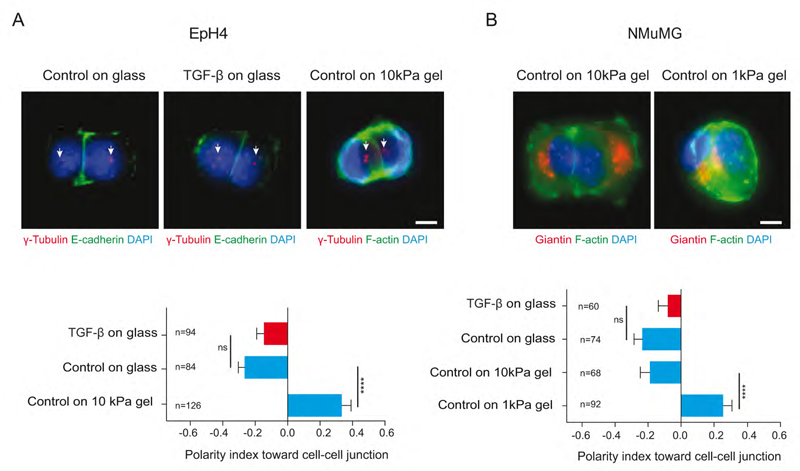
We further investigated centrosome repositioning in other classical models of EMT. NMuMG and EpH4 are luminal mammary cells that are known to be highly sensitive to EMT induction (Lamouille and Derynck, 2007; Montesano et al., 2007) (Figure S2H, I). To our surprise, when plated on micropatterns, both cell types were polarized toward cell matrix adhesion (mesenchyme-like polarity) and not toward inter-cellular junction. Since TGF-β was absent from the growth factor-defined serum-free culture medium, we reasoned that EMT might have been induced by the cell culture substrate. Indeed, matrix stiffness is a potent EMT inducer (Markowski et al., 2012; Wei et al., 2015). Cells were thus plated on micropatterned poly-acrylamide gels of controlled stiffness (Vignaud et al., 2014). On such soft substrates, cell doublets spread to a lesser extent and adopted a more compact geometry. When cultured on 10kPa gels, EpH4 displayed a typical epithelial polarity with the nucleus-centrosome axis oriented toward intercellular junctions, opposite to their polarization on glass (Figure 3A). Stiffness had to be further reduced to 1 kPa for NMuMG to recover a typical epithelial-like polarity (Figure 3B). These results show that matrix stiffness is sufficient to induce polarity reversal in the absence of TGF-β and thereby predispose epithelial cells to a mesenchyme transition. They also revealed that polarity reversal is quite reactive to EMT factors and that it can be easily induced in sensitive epithelial cells in response to mechanical and/or biochemical stimulations.
Figure 3. Matrix stiffness promotes polarity reversal.
(A) Images of control and TGF-β treated EpH4 cell doublets on H-shaped glass micropattern stained for E-cadherin (green), centrosome (red and white arrows) and DNA (blue). Cell doublets on Polyacrylamide gel were labeled for F-actin (green).
(B) Control NMuMG cells on glass and polyacrylamide gels are stained for Giantin (Golgi apparatus marker) (red), F-actin(green) and DNA (blue). Horizontal histograms show quantification of polarity index toward cell-cell junction. Scale bars represent 5 μm. Errors bar indicate SEM. Two tailed-non-parametric Mann-Whitney tests were used. ****: p<0.0001. Error bars indicate SEM.
Polarity reversal is an early feature of EMT
Cell sensitivity to EMT inducers prompted us to evaluate the timing of centrosome repositioning during the EMT process. TGF-β treatment is known to take several days to induce a full EMT but the very first changes appear a few hours after TGF-β addition (D’Souza et al., 2014). Cells were first plated on micropatterns and then treated with TGF-β for increasing periods of time. In MCF10A, centrosome repositioned in less than 24 hours (Figure 2F). In MDCK, TGF-β effects were detectable after 4 hours of treatment and polarity was reversed after 8 hours only. HGF effects on MDCK were even faster; centrosome repositioned almost immediately and inversion was completed within 4 hours (Figure 2F). These data imply that centrosome repositioning is an early sign of EMT concomitant with the first changes in protein expression following TGF-β addition (D’Souza et al., 2014). All further experiments were performed by 5 day treatment of TGF-β to MCF10A and 3 day treatment of HGF to MDCK unless specifically mentioned.
Microtubule network remodeling accompany centrosome re-centering
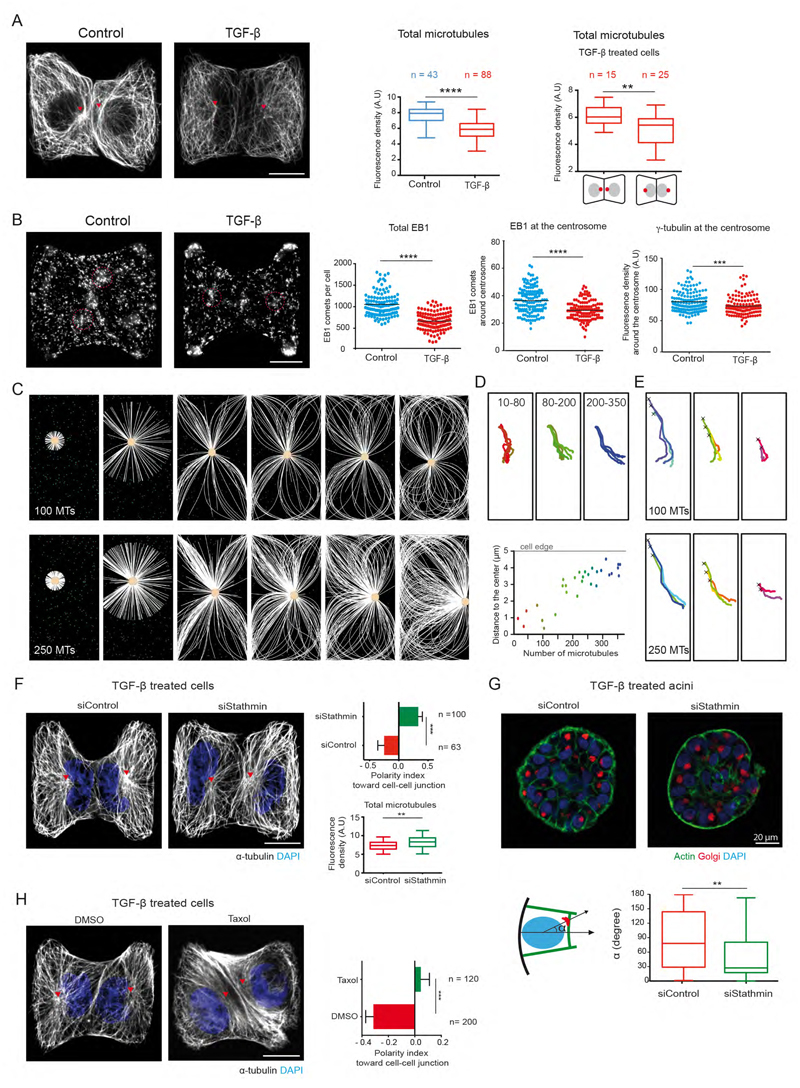
We further investigated the mechanism by which centrosome leaves its off-centered position next intercellular junctions to position at the cell center during EMT. Since centrosome positioning mostly depends on the microtubule network (Mimori-Kiyosue, 2011; Tang and Marshall, 2012), we compared microtubule network architectures before and after induction of EMT to gain further insight in the mechanism supporting centrosome repositioning. TGF-β treatment induced a drop in the total amount of microtubules (Figure 4A). Interestingly, the portion of TGF-β treated cells, which retained their centrosome position toward intercellular junction, like the control cells, harbored more microtubules (Figure 4A). These changes could be quantified by the diminution of polymerized α–tubulin intensity (Figure 4A) as well as the reduction in the number of EB1 comets (Figure 4B). Those differences could be explained by a reduction of centrosomal microtubules, as suggested by the reduction in γ-tubulin intensity (Figure 4B) and the number of EB1 comets at the centrosome (Figure 4B). These observations suggest that the high amount of microtubules may be responsible for centrosome off-centering in epithelial cells and its decrease would promote its recentring in mesenchymal cells. We tested this new hypothesis by performing numerical simulations with Cytosim (Nedelec and Foethke, 2007) in which the number of microtubules was modulated. Asters were constrained to grow in a confined space similar to the cell shape obtained on square micropatterns. Cytoplasmic dyneins were scattered throughout the cytoplasm so that their minus-end directed motion could promote aster centering by exerting pulling forces on microtubules (Wu et al., 2011). Microtubules were allowed to glide along and to push on cell edges as they grow. Interestingly, at low microtubule numbers (10-100), asters moved and stabilized at the cell center whereas with higher numbers of microtubules (200-350), the pushing forces exceeded the centering force and moved the centrosome off-center, toward the cell edge (Figure 4C, movie S2). Measuring centrosome final position in relation to the number of microtubules confirmed this observation (Figure 4D). Importantly, this behavior was quite robust and did not depend on centrosome initial position (Figure 4E). We further experimentally tested whether increasing microtubule number in TGF-β treated cells could restore centrosome off-centering close to the intercellular junction. To do so, we knocked down microtubule destabilizing protein Op18/stathmin in TGF-β treated cells (Belmont and Mitchison, 1996). Forty-eight hour post stathmin siRNA treatment; we observed clear restoration of centrosome position of EMT induced cells along with increase in the microtubule number (Figure 4F). The position of nucleus-golgi axis was also restored in the acini transfected with stathmin siRNA (Figure 4G). In addition, 5 hour of taxol treatment to TGF-β treated cells increased the microtubule bundles and also restored the off-centered centrosome position like that of non-treated cells (Figure 4H). These results suggest that the amount of polymerized tubulin was responsible for the transition from an off-centered microtubule network in epithelial cells to a centered conformation in mesenchymal cells.
Figure 4. Microtubule network remodeling accompanies centrosome re-centering during EMT.
(A) Images of control and TGF-β treated MCF10A cell-doublets on H-shaped micropattern stained for a-tubulin. Box plot shows quantification of total microtubule density on left. Microtubule density in two populations TGF-β treated cells with different nucleus-centrosome axis polarity on right. (AU is arbitrary fluorescence units).
(B) Images of MCF10A cell-doublets stained for EB1. Scatter plots show quantification of total EB1 comets, EB1 comets at the centrosome and γ-tubulin at the centrosome. A circular region of interest of 1.5 μm radius (red dotted circle) was used to count EB1 comets and γ-tubulin intensity at the centrosome.
(C) Numerical simulation showing microtubules (white) and centrosome (yellow) motion in response to varying microtubules number in a rectangular cell. Green dots correspond to cytoplasmic dynein (green).
(D) Effect of varying microtubule number on centrosome trajectory to final position. Different colors represent different microtubule number. Graph represents relationship between number of microtubules and the final position of the centrosome relative to cell center.
(E) Centrosome trajectories when starting from various initial positions (marked by cross) for either 100 (top) or 250 (bottom) microtubules.
(F) Images of TGF-β treated and stathmin knocked down cells with microtubules (white), nuclei (blue). Total Microtubule density (Arbitrary units) of stathmin knocked down cells. Scale bar (A-F) is 10 μm
(G) Images of TGF-β treated acini transfected with siRNA. Nucleus-golgi axis orientation (α) with respect to normal to the base of acini is quantified.
(H) Images of TGF-β treated cells with 5 hours of Taxol treatment. Polarity index is quantified on right. Scale bar is 10 μm.
Two tailed-non-parametric Mann-Whitney tests were used. ***: p<0.001, ****: p<0.0001.
Par3 regulates centrosome repositioning during EMT
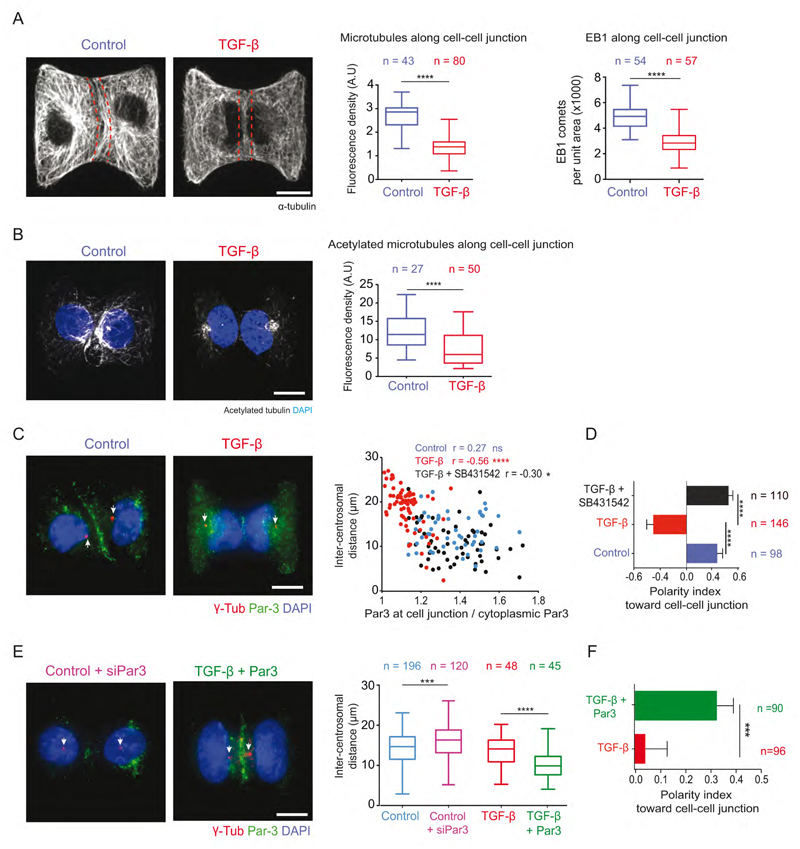
In our numerical simulations, no external bias was added to direct centrosome off-centering toward a specific cell edge. Partitioning defective proteins (PAR) polarity complexes Par3/Par6/aPKC, β-catenin and dyneins along intercellular junctions could actively bias this process by stabilizing and pulling on microtubules (Harris and Peifer, 2007; Ligon et al., 2001; Schmoranzer et al., 2009). To test whether such a mechanism was active in our conditions, we first quantified microtubule abundance along intercellular junction. Both tubulin intensity and EB1 comets were reduced along the junction of TGF-β treated cells (Figure 5A). Furthermore population of stabilized microtubules labeled by acetylated tubulin was significantly reduced at the cell-cell junction (Figure 5B). These results argued in favor of a mechanism of centrosome off-centering toward intercellular junctions in epithelial cells by local microtubule stabilization.
Figure 5. Par3 regulates centrosome position during EMT.
(A) Microtubule density and EB1 comets at the cell-cell junction are measured within the area indicated by red dotted lines of MCF10A cells on H-micropattern. AU is arbitrary units.
(B) Density of acetylated microtubules was measured at the cell-cell junction in the area marked by red dotted lines in panel (A).
(C) Par3 (green), γ-tubulin (red) and DNA (blue) staining of MCF10A cells. Central graph shows the relationship between Par3 enrichment at cell-cell junction and inter-centrosome distance. Pearson's correlation test r, ****: p<0.001, *: p<0.1, ns>0.1.
(D) Vertical histograms show measurement of polarity index toward cell-cell junction.
(E) Image of Par3 siRNA treated control cells and TGF-β treated cells with Par3 overexpression, labeled for Par3 (green), centrosome (red) and DNA (blue). Box plots show quantification of inter-centrosome distance.
(F) Horizontal histogram shows quantification of cell polarity index toward cell-cell junction.
Arrows point at centrosomes. Scale bars represent 10 μm. Errors bar indicate SEM. Two tailed-non-parametric Mann-Whitney test were used ***:p<0.001, ****:p<0.0001.
Intercellular junctions remained but were altered in TGF-β treated cells (Figure S3). Disappearance of Par3 from intercellular junctions could be involved in centrosome release since they it has been shown to regulate centrosome positioning in a wide range of cell types (Feldman and Priess, 2012; Oliaro et al., 2010; Schmoranzer et al., 2009; Solecki et al., 2009; St Johnston and Sanson, 2011). Par3 is notably involved in centrosome translocation from central to apical positioning during mesenchyme-to-epithelium transitions during neurulation (Hong et al., 2010). As intuited, Par3 localization along intercellular junction was drastically reduced after 5-day treatment of TGF-β (Figure 5C). Interestingly the junction density of Par3 along intercellular junctions appeared correlated to centrosome positioning: the higher the concentration of Par3 the closer the centrosome (Figure 5C). Inhibition of signaling downstream of TGF-β type I receptor by SB431542 (Inman et al., 2002) restored Par3 levels and centrosome off-centering toward intercellular junction (Figure 5C, D). Furthermore down-regulation of Par3 levels by Pard3 siRNA in untreated MCF10A cells increased the inter-centrosome distance (Figure 5E), while Par3b overexpression in TGF-β (3 day) treated cells restored centrosome position toward intercellular junction (Figure 5E-F), further confirming Par3 implication in the regulation of centrosome repositioning during EMT.
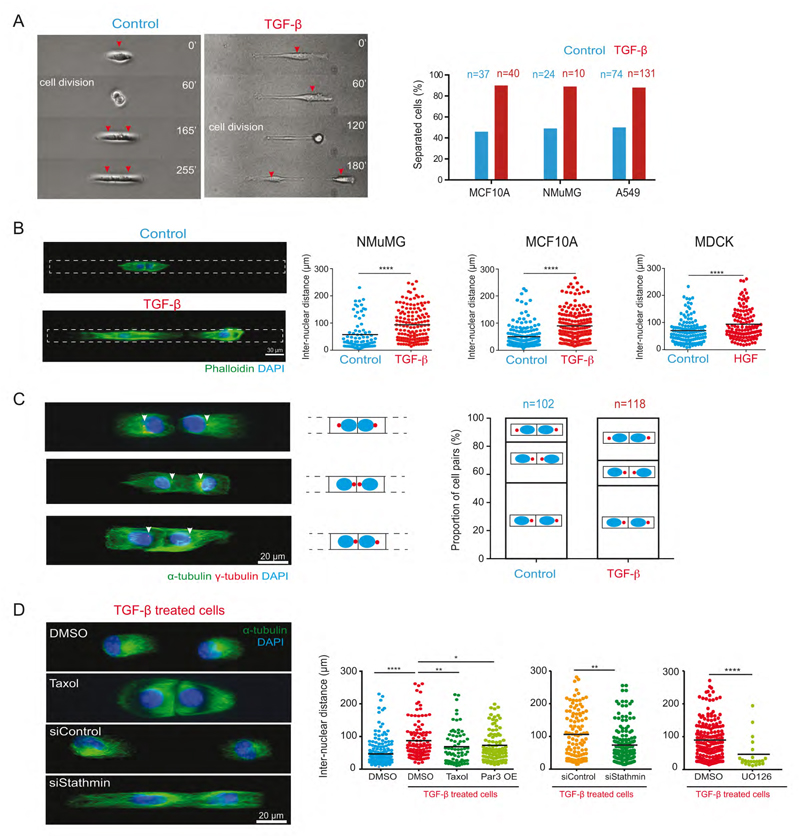
Centrosome repositioning promotes cell scattering
Centrosome repositioning in immobilized cells on H-shaped micropattern showed that it was not a consequence of cell motion. Nucleus-centrosome reorientation toward cell-matrix adhesion even suggested cells were predisposed to separate. But whether centrosome repositioning could actually trigger cell scattering remained to be tested. We first used micropatterned tracks on which cells were free to separate from each other. In three distinct epithelial cell lines, untreated cells stayed in contact whereas the majority of TGF-β treated cells separated after division (Figure 6A, Movie S3). Similarly, single end point assay based on inter-nuclear distance in cell doublets revealed higher cell scattering in TGF-β or HGF treated cells (Figure 6B). Importantly, TGF-β or HGF-treated-cells that were still in contact showed twice more conformations with reversed centrosome position compared to control cells, suggesting that centrosome repositioning preceded cell scattering (Figure 6C). We tested the positive effect of microtubule disassembly in cell scattering upon EMT induction by adding Taxol to TGF-β treated cells. Five hours of taxol treatment did reduce cell scattering (Figure 6D). Par3 overexpression also had a modest reduction effect on cell scattering (Figure 6D). Interestingly, and consistent with our previous observations on centrosome repositioning, high stathmin levels have been shown to stimulate cell migration during EMT and cancer metastasis (Li et al., 2011; Lu et al., 2014). Here, we observed 30% drop in cell scattering upon stathmin knockdown (Figure 6D). As stathmin is activated by a MEK/ERK-dependent phosphorylation (Filbert et al., 2012), we inhibited the MEK pathway in EMT induced cells with UO126 and also observed a strong reduction of cell scattering (Figure 6D). These effects were likely due to centrosome repositioning rather than increased microtubule stability that has been shown to promote rather than impair individual cell migration (Zhang et al., 2011). Taken together these observations support the view that EMT-induced cell separation is intimately coupled to centrosome re-centering via stathmin-dependent microtubule disassembly.
Figure 6. Centrosome repositioning promotes cells scattering during EMT.
(A) Time-lapse sequence NMuMG cells (red arrows) on micro-patterned lines. Quantification of the proportion of cell separation in control and TGF-β treated cells for various cell types. n indicates number events measured for cell separation.
(B) Images of fixed NMuMG cells labeled for F-actin (green) and nucleus (blue) on micropatterned tracks of 300 μm. Histograms show the quantification of inter-nuclear distance between cell pairs of different cell lines.
(C) Images of TGF-β treated MCF10A cell-pairs on micropatterned tracks in different configurations of nucleus-centrosome axis orientation. Arrows indicate centrosomes. The proportion of each configuration is quantified on right.
(D) Images of TGF-β treated MCF10A cell-pairs in the presence of DMSO or Taxol (5 hours of treatment) and cells transfected with control or stathmin siRNA. On the right inter-nuclear distances between the cell-pairs are quantified.
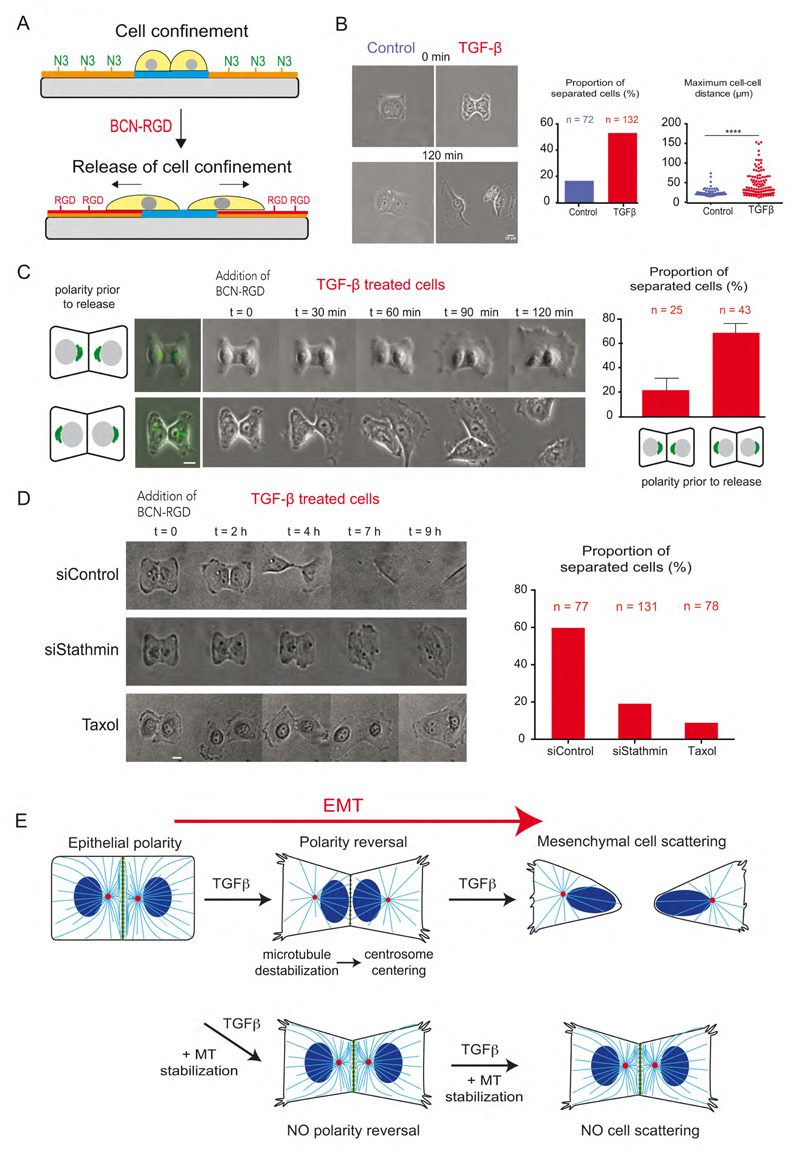
To directly test whether centrosome repositioning was actually causing cell scattering we looked for a way to induce it, or not, before cells had the opportunity to move away from each other. We decided to use dynamic micropatterning to release confined cells with pre-established epithelial or inverted polarities.
Dynamic micropatterning relies on the use of click chemistry to graft RGD peptides to the PEG chains preventing cell adhesion around micropatterns (van Dongen et al., 2013). The mild conditions of the azide-alkyne cycloaddition offer the possibility to graft a RGD-alkyne compound to a PEG-azide chain in the presence of living cells (Figure 7A). Thus, micropatterned cells can start migrating out of the micropattern upon addition and grafting of RGD groups to the PEG chains.
Figure 7. Polarity reversal is necessary for cell scattering.
(A) Schematic depicting principle of dynamic micropatterning with azide-PLL-PEG (orange) and cell motion on BCN-RGD modified substrate (red).
(B) Images of cells on H-shaped micropattern before and after modification of BCN-RGD modified substrate. Histograms show measurement of the proportion of cell separation and maximum inter-nuclear distance between the MCF10A cells two hours after addition of BCN-RGD.
(C) Time-lapse sequence images of TGF-β treated MCF10A cells expressing Golgi apparatus markers (visualized in green at t=0) in response to the addition of BCN-RGD. Histogram shows measurement of the proportion of cell separation depending on their initial polarity orientation. Scale bar is 10 μm.
(D) Time-lapse image sequence of TGF-β treated MCF10A cells with stathmin knockdown or Taxol treatment. Their cell separation post 20 hours of BCN-RGD addition was quantified on right. Scale bar is 10 μm.
(E) Schematic description of microtubule reorganization and centrosome repositioning that causes polarity reversal and finally cell separation during the course of EMT.
We first confirmed the expected outcomes of EMT induction, i.e. that most MCF10A epithelial cells remain in contact with each other while TGF-β treated cells tend to separate within 4 hour after cell release by addition of RGD (Figure 7B). We further took advantage that not all TGF-β treated cells did revert their polarity (Figure 2D, E) to compare the scattering of TGF-β treated cells depending on their pre-established polarity. For this, we used the larger Golgi apparatus rather than small centrosome markers to facilitate their detection by live cell microscopy. We saw a clear difference in scattering behavior of TGF-β treated depending upon the initial orientation of their polarity axis: cells with the polarity axis pointing toward inter-cellular junctions (similar to non-treated epithelial cells) had less propensity (24%, n=25) to separate from each other compared to cells with a polarity axis pointing toward Cell-matrix adhesion (69%, n=43) (Figure 7C and movie S4).
We further attempted to specifically interfere with centrosome repositioning in TGF-b treated cells by reducing microtubule disassembly prior to cell release. Strikingly, cell scattering was reduced by 66% (n=131) in stathmin knock-down cells and by 85% (n=78) in taxol-treated cells compared to the control cells (n=77) (Figure 7D and Movie S5). Noteworthy all cells (Control siRNA, stathmin siRNA and Taxol treatment) formed an intercellular junction when confined, but only cells with stabilized microtubules retained it after release from confinement. These experiments conclusively proved that, upon TGF-β induced EMT, cells that maintained their polarity toward intercellular junction behaved like epithelial cells with less scattering potential while cells with inverted polarity were primed for cell scattering. Hence, we establish that EMT induction involves polarity reversal by centrosome repositioning away from inter-cellular junctions to promote cell separation.
Discussion
These results have revealed the existence of a global intra-cellular rearrangement occurring in the few hours following the addition of TGF-β, which therefore appeared as one of the earliest morphological sign of EMT that precedes the loss of inter-cellular connections. Microtubule network geometry and centrosome position rapidly adapt to early modifications of inter-cellular junction composition and notably to the reduction of Par3 concentration. Decrease in microtubule nucleation and polymerization in addition to the decrease of selective microtubule stabilization along the junction lead to centrosome displacement from the junction to the cell center (Figure 7E). Thereby the centrosome relocates on the opposite side of the nucleus. These intra-cellular rearrangements result into an effective reversal of internal polarity axis and are coordinated with inversion of cortical polarity component localization such as Par3 and Podocalyxin (Figure 5C, Figure S2G). Centrosome repositioning further promotes cell scattering (Figure 7E). As such, the centrosome-microtubule network appears to act as an extensive and sensitive spatial integrator of cell adhesion cues allowing the orientation of cell internal polarity to adapt to changes in cell’s microenvironment.
How the centrosome adopts an off-center position toward inter-cellular junctions in epithelial cells is not well understood. Centrosome decentering is generally considered to rely on the production of pulling forces from a defined part of cell periphery as it has been described during immune synapse formation (Yi et al., 2013) and spindle orientation (Théry et al., 2007). Par3 is present along epithelial cells junctions. It is capable to recruit dynein that pulls on microtubules (Ligon et al., 2001; Schmoranzer et al., 2009) and thereby can direct centrosome position like during planar cell polarity establishment (Jiang et al., 2015; Sipe et al., 2013), intestinal cell polarization (Feldman and Priess, 2012) and neurulation (Buckley et al., 2012; Hong et al., 2010). The correlation we observed between Par3 levels and centrosome position in our conditions (Figure 5C-F) are consistent with this off-centering mechanism.
However, a mechanism based on local pulling only would not be sensitive and efficient as it would have to overcome the centering forces applied on all the other microtubules (Letort et al., 2016). Numerical simulations suggested that some specific reorganizations of microtubule network, including the elongation and increased number of microtubules, could circumvent this limitation by developing decentering pushing forces in the entire network (Letort et al., 2016). Our experimental and numerical experiments are consistent with this view. In epithelial cells, microtubules were long and numerous and the centrosome was off-centered. Whereas in mesenchymal cells microtubules were shorter and less numerous and the centrosome localized at the cell center. Microtubule length increase in mesenchymal cells by down-regulation of stathmin expression level restored centrosome position to an off-centered, epithelial-like, conformation. These observations were also consistent with the previous descriptions of decentered centrosomes in lipid vesicles (Pinot et al., 2009) or microfabricated chambers (Faivre-Moskalenko and Dogterom, 2002) when microtubules were longer than the diameter of the space they were confined in.
The mechanism by which knock down of stathmin prevented microtubule disassembly, centrosome repositioning to the cell center and cell scattering in response to TGF-β is unclear. Stathmin modulates microtubule dynamics via its interaction with tubulin dimers (Belmont and Mitchison, 1996) but, interestingly, also affects microtubule nucleation at the centrosome (Ringhoff and Cassimeris, 2009), which is consistent with our observation of reduced level of γ-tubulin and EB1 at the centrosome upon addition of TGF-β. In parallel, stathmin (also named oncoprotein 18) is known to be overexpressed in several form of human malignancies and has already been suggested to contribute to oncogenic EMT (Li et al., 2011; Lu et al., 2014; Melhems et al., 1991), in which polarity reversal would be worth being investigated.
Importantly, our results show that centrosome-microtubule network geometry adapts to but also actively feedbacks to the adhesion-actin network configuration. TGF-β treated cells that did not undergo polarity reversal could not separate upon the release of spatial constraint suggesting that the centrosome stabilizes the inter-cellular junction as long as it stays close to it and promotes the migration machinery as it comes closer to cell-matrix adhesions at the cell front. Indeed microtubules interact and feedback with both types of adhesions (Akhmanova et al., 2009). The centrosome and the junction exchange materials: some centrosomal proteins transit from centrosome to the junction, allowing local microtubule anchoring and stabilization (Gavilan et al., 2015; Lechler and Fuchs, 2007; Moss et al., 2007). In return, microtubules stabilize the junction (Meng et al., 2008), notably by the dynein-dependent recruitment of intercellular junction components like occludin (Glotfelty et al., 2014). Some translocated centrosomal proteins even promote junction reinforcement and epithelial cell acquisition of a columnar shape (Gavilan et al., 2015). Similarly, when internal polarity axis is directed toward cell adhesion to ECM, microtubule density increases can promote actin polymerization and adhesion turnover and thereby foster cell migration (Etienne-Manneville, 2013). Interestingly, during epithelial scattering occludin and Par3 have been shown to relocalize from inter-cellular junction to the cell front where they promote leading edge protrusion and cell migration (Du et al., 2010). Par3 relocalization was also observed in our working conditions on micropatterns. These considerations account for the capacity of centrosome relocalization from intercellular junction toward ECM adhesions to actively weaken inter-cellular interaction and promote the cell migration machinery even before effective cell displacement and thereby primes cell scattering.
Thus our results provide new evidences for the key role of the centrosome-microtubule network interplay with the adhesion-actin system in the regulation of complex tissue remodeling.
Experimental Procedures
The description of the details of mouse embryo and mammary gland manipulation, 3D acini formation, cell culture and transfection, DNA constructs, drug treatments, antibodies, micropatterning, microscopy can be found in Supplemental Experimental Procedures.
Cell culture and EMT induction
MCF10A cells were cultured in Lonza MEGM medium (Lonza #CC3150) as described by ATCC protocol. MDCK, NMuMG and EpH4 cells were cultured as described in Supplemental experimental procedures. EMT was induced by addition of 5 ng/ml TGF-β1 to MCF10A for 5 days, 2 ng/ml to NMuMG & EpH4 for 3 days and 10 ng/ml of HGF for MDCK for 3 days. MCF10A 3D cultures were prepared by cultivating cells in Matrigel as described previously (Debnath et al., 2003).
Cell micropatterning
Micropatterns were fabricated by coating poly-ethylene-glycol on glass and exposing this coating to deep UV light through a chromium photomask (Azioune et al., 2010). Soft substrates micropatterning was achieved by polymerizing a mix of acrylamide and bis-acrylamide onto a micropatterned glass slide to transfer proteins from the glass onto the hydrogel (Vignaud et al., 2014). Traction forces were calculated with an Image J plugin to measure gel relaxation field after cell detachment (Martiel et al., 2015). Dynamic micropatterning was performed using click chemistry to bind BCN-RGD onto azide groups grafted on PLL-PEG (van Dongen et al., 2013).
Cell Scattering on line micropatterns and video-microscopy
150,000 cells per were plated onto micropatterned lines on a 20x20 mm glass coverslip. Non attached cells were rinsed away by medium exchange once few cells were attached to the micropattern to avoid obtaining several cells per line. Coverslips were then mounted on video chambers (Chamlide, CM-s20-1). Cells motion was then video-recorded in transmitted light with a Nikon Eclipse Ti-E with 10x phase contrast objective. Time interval was set to 15 min. Cell separation after division was then counted by visual inspection.
Quantification of centrosome position
Image analysis for centrosome positioning was performed using a series of Macros in Image J. Briefly; z-projection of each color channel was obtained using 'maximum z-projection' plugin followed by merging the 4 channels to obtain a composite image. Using Pattern Alignment plugin (https://sites.google.com/site/qingzongtseng/template-matching-ij-plugin) images were aligned with the reference micropattern image. Nuclei in DAPI channel were detected using image thresh-holding and object size criteria. Centrosomes were detected with similar thresh-holding and by using nuclei Region Of Interest (ROI) as spatial reference. Finally nucleus-centrosome vector was computed by subtracting co-ordinates of centrosome from nucleus co-ordinates and was normalized by the nucleus radius as indicated in Figure 2.
Quantification of EB1 comets
Z-projected images of EB1 comets were obtained as described above. Background subtraction was performed using a rolling ball radius of 50. For analyzing nucleation capacity of the centrosome, ROI of 1.5µm radius was drawn around the centrosome labeled by Ninein. EB1 comets were detected in the selected ROI by 'Find Maxima' Process.
Quantification of microtubule intensity
Images of microtubules were acquired using CSUX1-A1N Nikon Spinning Disk microscope (Yokogawa) with 100X, 1.3 NA objective and Evolve 512 EMCCD camera (Photometrics). Z-projected images 'Maximum z intensity' of alpha-tubulin staining were obtained as described above. A ROI 46x46 µm was selected containing the cell pair on <H> micropatterns. Total intensity of the image was measured in Image J for each image to quantify total microtubule density in each cell-pair. To quantify microtubule density at the cell-cell junction, a ROI of 3.2 µm thickness was drawn along the cell-cell junction and intensity of alpha-tubulin staining was measured.
Quantification of acini polarization
Images of acini were acquired on a Nikon Spinning Disk microscope at 60x oil objective with a z-step of 500 nm. To determine angle alpha formed between normal to cell periphery and the nucleus-centrosome vector (Figure 1), 4-5 central planes of acini images were selected to obtain single layers of cells at the center of the acini. A spatial reference line was drawn along the basal surface of cells, which was marked by phalloidin staining. Using the 'angle' measurement in Image J, the value of alpha (0-180°) was determined providing the angle formed by vectors in the same plane. Only the plane in the middle of acini was considered for these measurements.
Numerical Simulations using Cytosim Software
Simulations were performed using the Cytosim software (www.cytosim.org). Microtubules are considered as elastic fibers surrounded by a viscous fluid following Langevin dynamics (Nedelec and Foethke, 2007). We simulated only microtubules nucleated from and anchored to a centrosomal complex. New nucleation of microtubules, microtubules unbinding from centrosome or steric interactions between microtubules are not taken into account. Microtubules can grow at a force-dependent speed, can undergo catastrophe events and shrink, and can undergo rescue events. Their bending elasticity is modelled following Euler's buckling description. Microtubules are initially uniformly distributed around the centrosome and can freely rotate around it. They are constrained into a rectangular space with an Hookean rappel force and will push against this border but can glide freely along it. Cytoplasmic dyneins are modeled as immobile objects spread into the cellular space that can bind/unbind microtubules and move toward the microtubule minus end when bound, thus generating a pulling force on the centrosome.
Statistical Analysis
Mann-Whitney non-parametric test was used to compare differences between the samples. Error bars indicate standard error mean (SEM). N indicates numbers of experiments while n indicates sample size.
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures, three figures, and five movies.
Acknowledgments
We thank Priscilla Soulie for providing EpH4 cells, James Sillibourne for providing human-Ninein antibody, Benoit Ladoux for providing anti-GM135 antibody, Matthieu Piel and his team for providing APP & BCN-RGD compounds and advices for dynamic patterning and Benoit Vianay for his help with microscopy. We are grateful to Jean-Paul Thierry for interesting discussions, Laurent Blanchoin for his useful comments throughout the project and Maxence Nachury for his thoughtful advices about the potential role of stathmin. This work was supported by grants from the ERC (SpiCy, StG-310472) and the BPI France (ETICS, I1107018W). M.B. received a fellowship from the ANRT (CIFRE 2011/1730) and M.P. a fellowship from the Laboratory of Excellence (Grenoble Alliance for Integrated Structural Cell Biology). GB and SL are supported by fellowships from the Wellcome Trust. OFC is supported by La Ligue Nationale et Regionale contre le Cancer and Espoir Foundation.
Footnotes
Authors contribution
M.B. performed most experimental work and data analyses. M.P. performed most experiments with MDCK. S.T. performed experiments on mouse mammary glands. G.B. performed experiments on mouse embryos. Q.T. performed preliminary experiments and contributed to software developments for image analysis. G.L performed numerical simulations. G.B. and S.L. developed image analysis tools. O.F.C. designed experiments on 3D cell culture and provided key information about EMT to the consortium. J.Y. provided critical comments about the work and edited the manuscript. M.T. directed the project, analyzed the data and wrote the manuscript.
References
- Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A, Stehbens SJ, Yap AS. Touch, grasp, deliver and control: functional cross-talk between microtubules and cell adhesions. Traffic. 2009;10:268–274. doi: 10.1111/j.1600-0854.2008.00869.x. [DOI] [PubMed] [Google Scholar]
- Akhtar N, Streuli CH. An integrin-ILK-microtubule network orients cell polarity and lumen formation in glandular epithelium. Nat Cell Biol. 2013;15:17–27. doi: 10.1038/ncb2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstrom JA, Raff RA. Sea urchin primary mesenchyme cells: Relation of cell polarity to the epithelial-mesenchymal transformation. Dev Biol. 1988;130:57–66. doi: 10.1016/0012-1606(88)90413-7. [DOI] [PubMed] [Google Scholar]
- Azioune A, Carpi N, Tseng Q, Théry M, Piel M. Protein micropatterns: A direct printing protocol using deep UVs. Methods Cell Biol. 2010;97:133–146. doi: 10.1016/S0091-679X(10)97008-8. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Bornens M. Organelle positioning and cell polarity. Nat Rev Mol Cell Biol. 2008;9:874–886. doi: 10.1038/nrm2524. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Roignot J, Datta A, Overeem AW, Kim M, Yu W, Peng X, Eastburn DJ, Ewald AJ, Werb Z, et al. A Molecular Switch for the Orientation of Epithelial Cell Polarization. Dev Cell. 2014;31:171–187. doi: 10.1016/j.devcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley CE, Ren X, Ward LC, Girdler GC, Araya C, Green MJ, Clark BS, Link Ba, Clarke JDW. Mirror-symmetric microtubule assembly and cell interactions drive lumen formation in the zebrafish neural rod. EMBO J. 2012;32:30–44. doi: 10.1038/emboj.2012.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burute M, Thery M. Spatial segregation between cell-cell and cell-matrix adhesions. Curr Opin Cell Biol. 2012;24:628–636. doi: 10.1016/j.ceb.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Carney PR, Couve E. Cell polarity changes and migration during early development of the avian peripheral auditory system. Anat Rec. 1989;225:156–164. doi: 10.1002/ar.1092250211. [DOI] [PubMed] [Google Scholar]
- D’Souza R, Knittle aM, Nagaraj N, van Dinther M, Choudhary C, ten Dijke P, Mann M, Sharma K. Time-resolved dissection of early phosphoproteome and ensuing proteome changes in response to TGF- Sci Signal. 2014;7:rs5–rs5. doi: 10.1126/scisignal.2004856. [DOI] [PubMed] [Google Scholar]
- Das RM, Storey KG. Apical abscission alters cell polarity and dismantles the primary cilium during neurogenesis. Science (80-. ) 2014;343:200–204. doi: 10.1126/science.1247521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Desai RA, Gao L, Raghavan S, Liu WF, Chen CS. Cell polarity triggered by cell-cell adhesion via E-cadherin. J Cell Sci. 2009 doi: 10.1242/jcs.028183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen SFM, Maiuri P, Marie E, Tribet C, Piel M. Triggering cell adhesion, migration or shape change with a dynamic surface coating. Adv Mater. 2013;25:1687–1691. doi: 10.1002/adma.201204474. [DOI] [PubMed] [Google Scholar]
- Du D, Xu F, Yu L, Zhang C, Lu X, Yuan H, Huang Q, Zhang F, Bao H, Jia L, et al. The tight junction protein, occludin, regulates the directional migration of epithelial cells. Dev Cell. 2010;18:52–63. doi: 10.1016/j.devcel.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Microtubules in cell migration. Annu Rev Cell Dev Biol. 2013;29:471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective Epithelial Migration and Cell Rearrangements Drive Mammary Branching Morphogenesis. Dev Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre-Moskalenko C, Dogterom M. Dynamics of microtubule asters in microfabricated chambers: the role of catastrophes. Proc Natl Acad Sci U S A. 2002;99:16788–16793. doi: 10.1073/pnas.252407099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Priess JR. A role for the centrosome and PAR-3 in the hand-off of MTOC function during epithelial polarization. Curr Biol. 2012;22:575–582. doi: 10.1016/j.cub.2012.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbert EL, Le Borgne M, Lin J, Heuser JE, Shaw AS. Stathmin regulates microtubule dynamics and microtubule organizing center polarization in activated T cells. J Immunol. 2012;188:5421–5427. doi: 10.4049/jimmunol.1200242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavilan MP, Arjona M, Zurbano A, Formstecher E, Martinez-Morales JR, Bornens M, Rios RM. Alpha-catenin-Dependent Recruitment of the Centrosomal Protein CAP350 to Adherens Junctions Allows Epithelial Cells to Acquire a Columnar Shape. PLOS Biol. 2015;13:e1002087. doi: 10.1371/journal.pbio.1002087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotfelty La, Zahs A, Iancu C, Shen L, Hecht Ga. Microtubules Are Required for Efficient Epithelial Tight Junction Homeostasis and Restoration. Am J Physiol Cell Physiol. 2014;307:245–254. doi: 10.1152/ajpcell.00336.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godde NJ, Galea RC, Elsum Ia, Humbert PO. Cell polarity in motion: redefining mammary tissue organization through EMT and cell polarity transitions. J Mammary Gland Biol Neoplasia. 2010;15:149–168. doi: 10.1007/s10911-010-9180-2. [DOI] [PubMed] [Google Scholar]
- Harris TJC, Peifer M. aPKC controls microtubule organization to balance adherens junction symmetry and planar polarity during development. Dev Cell. 2007;12:727–738. doi: 10.1016/j.devcel.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AM, Duboff B, Casaletto JB, Gladden AB, McClatchey AI. Merlin/ERM proteins establish cortical asymmetry and centrosome position. Genes Dev. 2012;26:2709–2723. doi: 10.1101/gad.194027.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30:276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Hinck L, Silberstein GB. Key stages in mammary gland development: the mammary end bud as a motile organ. Breast Cancer Res. 2005;7:245–251. doi: 10.1186/bcr1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E, Jayachandran P, Brewster R. The polarity protein Pard3 is required for centrosome positioning during neurulation. Dev Biol. 2010;341:335–345. doi: 10.1016/j.ydbio.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RY-J, Guilford P, Thiery JP. Early events in cell adhesion and polarity during epithelial-mesenchymal transition. J Cell Sci. 2012:4417–4422. doi: 10.1242/jcs.099697. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolás FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Jiang T, McKinley RFA, McGill MA, Angers S, Harris TJC. A Par-1-Par-3-Centrosome Cell Polarity Pathway and Its Tuning for Isotropic Cell Adhesion. Curr Biol. 2015;25:2701–2708. doi: 10.1016/j.cub.2015.08.063. [DOI] [PubMed] [Google Scholar]
- Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J Cell Biol. 2007;176:147–154. doi: 10.1083/jcb.200609109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letort G, Nedelec F, Blanchoin L, Théry M. Centrosome centering and decentering by microtubule network rearrangement. Mol Biol Cell. 2016;27:2833–2843. doi: 10.1091/mbc.E16-06-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Jiang P, Du W, Wu Z, Li C, Qiao M, Yang X, Wu M. Siva1 suppresses epithelial-mesenchymal transition and metastasis of tumor cells by inhibiting stathmin and stabilizing microtubules. Proc Natl Acad Sci U S A. 2011;108:12851–12856. doi: 10.1073/pnas.1017372108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon LA, Karki S, Tokito M, Holzbaur ELF. Dynein binds to β -catenin and may tether microtubules at adherens junctions. Nat Cell Biol. 2001;3:913–917. doi: 10.1038/ncb1001-913. [DOI] [PubMed] [Google Scholar]
- Lu Y, Liu C, Xu YF, Cheng H, Shi S, Wu CT, Yu XJ. Stathmin destabilizing microtubule dynamics promotes malignant potential in cancer cells by epithelial-mesenchymal transition. Hepatobiliary Pancreat Dis Int. 2014;13:386–394. doi: 10.1016/s1499-3872(14)60038-2. [DOI] [PubMed] [Google Scholar]
- Luxton GWG, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr Opin Cell Biol. 2011;23:579–588. doi: 10.1016/j.ceb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowski MC, Brown AC, Barker TH. Directing epithelial to mesenchymal transition through engineered microenvironments displaying orthogonal adhesive and mechanical cues. J Biomed Mater Res - Part A. 2012;100 A:2119–2127. doi: 10.1002/jbm.a.34068. [DOI] [PubMed] [Google Scholar]
- Martiel J-L, Leal A, Kurzawa L, Balland M, Wang I, Vignaud T, Tseng Q, Théry M. Measurement of cell traction forces with ImageJ. Methods Cell Biol. 2015;125:269–287. doi: 10.1016/bs.mcb.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Melhems RF, Zhu X, Hailat N, Strahler JR, Hanash SM. Characterization of the Gene for a Proliferation-related. 1991;266:17747–17753. [PubMed] [Google Scholar]
- Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135:948–959. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y. Shaping microtubules into diverse patterns: Molecular connections for setting up both ends. Cytoskeleton. 2011;68:603–618. doi: 10.1002/cm.20540. [DOI] [PubMed] [Google Scholar]
- Montesano R, Carrozzino F, Soulié P. Low concentrations of transforming growth factor-beta-1 induce tubulogenesis in cultured mammary epithelial cells. BMC Dev Biol. 2007;7:7. doi: 10.1186/1471-213X-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss DK, Bellett G, Carter JM, Liovic M, Keynton J, Prescott AR, Lane EB, Mogensen MM. Ninein is released from the centrosome and moves bi-directionally along microtubules. J Cell Sci. 2007;120:3064–3074. doi: 10.1242/jcs.010322. [DOI] [PubMed] [Google Scholar]
- Nedelec F, Foethke D. Collective Langevin dynamics of flexible cytoskeletal fibers. New J Phys. 2007;9:427–427. [Google Scholar]
- Nelson WJ. Remodeling epithelial cell organization: transitions between front-rear and apical-basal polarity. Cold Spring Harb Perspect Biol. 2009;1:a000513. doi: 10.1101/cshperspect.a000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch L, Wollman SH. Ultrastructure of intermediate stages in polarity reversal of thyroid epithelium in follicles in suspension culture. J Cell Biol. 1980;86:875–880. doi: 10.1083/jcb.86.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojakian KG, Schwimmer R. Regulation of epithelial cell surface polarity reversal by beta 1 integrins. J Cell Sci. 1994;107:561–576. [PubMed] [Google Scholar]
- Oliaro J, Van Ham V, Sacirbegovic F, Pasam A, Bomzon Z, Pham K, Ludford-Menting MJ, Waterhouse NJ, Bots M, Hawkins ED, et al. Asymmetric Cell Division of T Cells upon Antigen Presentation Uses Multiple Conserved Mechanisms. J Immunol. 2010;185:367–375. doi: 10.4049/jimmunol.0903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinot M, Chesnel F, Kubiak JZ, Arnal I, Nedelec FJ, Gueroui Z. Effects of confinement on the self-organization of microtubules and motors. Curr Biol. 2009;19:954–960. doi: 10.1016/j.cub.2009.04.027. [DOI] [PubMed] [Google Scholar]
- Pollack AL, Runyan RB, Mostov KE. Morphogenetic Mechanisms of Epithelial Tubulogenesis : MDCK Cell Polarity Is Transiently Rearranged without Loss of Cell – Cell Contact during Scatter Factor / Hepatocyte Growth. Dev Biol. 1998;79:64–79. doi: 10.1006/dbio.1998.9091. [DOI] [PubMed] [Google Scholar]
- Pouthas F, Girard P, Lecaudey V, Ly TBN, Gilmour D, Boulin C, Pepperkok R, Reynaud EG. In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum. J Cell Sci. 2008;121:2406–2414. doi: 10.1242/jcs.026849. [DOI] [PubMed] [Google Scholar]
- Revenu C, Streichan S, Donà E, Lecaudey V, Hufnagel L, Gilmour D. Quantitative cell polarity imaging defines leader-to-follower transitions during collective migration and the key role of microtubule-dependent adherens junction formation. Development. 2014;141:1282–1291. doi: 10.1242/dev.101675. [DOI] [PubMed] [Google Scholar]
- Ringhoff DN, Cassimeris L. Abl tyrosine kinase phosphorylates nonmuscle Myosin light chain kinase to regulate endothelial barrier function. Mol Biol Cell. 2009;20:3451–3458. doi: 10.1091/mbc.E09-10-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Macara IG. Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Biol. 2014;15:225–242. doi: 10.1038/nrm3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli AE, Auzan M, Alonso Ma, Bornens M, Martin-Belmonte F. Cell confinement controls centrosome positioning and lumen initiation during epithelial morphogenesis. J Cell Biol. 2012;198:1011–1023. doi: 10.1083/jcb.201203075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa E, Szabó A, Bibonne A, Theveneau E, Parsons M, Mayor R. Cadherin Switch during EMT in Neural Crest Cells Leads to Contact Inhibition of Locomotion via Repolarization of Forces. Dev Cell. 2015;34:421–434. doi: 10.1016/j.devcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoranzer J, Fawcett JP, Segura M, Tan S, Vallee RB, Pawson T, Gundersen GG. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol. 2009;19:1065–1074. doi: 10.1016/j.cub.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seton-Rogers SE, Lu Y, Hines LM, Koundinya M, LaBaer J, Muthuswamy SK, Brugge JS. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc Natl Acad Sci U S A. 2004;101:1257–1262. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe CW, Liu L, Lee J, Grimsley-Myers C, Lu X. Lis1 mediates planar polarity of auditory hair cells through regulation of microtubule organization. Development. 2013;140:1785–1795. doi: 10.1242/dev.089763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki DJ, Trivedi N, Govek E-E, Kerekes Ra, Gleason SS, Hatten ME. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Sanson B. Epithelial polarity and morphogenesis. Curr Opin Cell Biol. 2011;23:540–546. doi: 10.1016/j.ceb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Takesono A, Heasman SJ, Wojciak-Stothard B, Garg R, Ridley AJ. Microtubules regulate migratory polarity through Rho/ ROCK signaling in T cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP, Behringer RR. Mouse gastrulation: the formation of a mammalian body plan. Mech Dev. 1997;68:3–25. doi: 10.1016/s0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- Tang N, Marshall WF. Centrosome positioning in vertebrate development. J Cell Sci. 2012;125:4951–4961. doi: 10.1242/jcs.038083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M. Micropatterning as a tool to decipher cell morphogenesis and functions. J Cell Sci. 2010;123:4201–4213. doi: 10.1242/jcs.075150. [DOI] [PubMed] [Google Scholar]
- Théry M, Jiménez-Dalmaroni A, Racine V, Bornens M, Jülicher F. Experimental and theoretical study of mitotic spindle orientation. Nature. 2007;447:493–496. doi: 10.1038/nature05786. [DOI] [PubMed] [Google Scholar]
- Tseng Q, Duchemin-Pelletier E, Deshiere A, Balland M, Guillou H, Filhol O, Théry M. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci U S A. 2012;109:1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignaud T, Ennomani H, Théry M. Polyacrylamide hydrogel micropatterning. Methods Cell Biol. 2014;120:93–116. doi: 10.1016/B978-0-12-417136-7.00006-9. [DOI] [PubMed] [Google Scholar]
- Wang H, Lacoche S, Huang L, Xue B, Muthuswamy SK. Rotational motion during three-dimensional morphogenesis of mammary epithelial acini relates to laminin matrix assembly. Proc Natl Acad Sci. 2013;110:163–168. doi: 10.1073/pnas.1201141110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZA, Ojakian KG, Nelson WJ. Steps in the morphogenesis of a polarized epithelium II. Disassembly and assembly of plasma membrane domains during reversal of epithelial cell polarity in multicellular epithelial (MDCK) cysts. J Cell Sci. 1990;95:153–165. doi: 10.1242/jcs.95.1.153. [DOI] [PubMed] [Google Scholar]
- Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, Chen AC, Sah RL, Taylor SS, Engler AJ, et al. Matrix stiffness drives epithelial–mesenchymal transition and tumour metastasis through a TWIST1–G3BP2 mechanotransduction pathway. Nat Cell Biol. 2015;17:678–688. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Misra G, Russell RJ, Ladd AJC, Lele TP, Dickinson RB. Effects of dynein on microtubule mechanics and centrosome positioning. Mol Biol Cell. 2011;22:4834–4841. doi: 10.1091/mbc.E11-07-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Wu X, Chung AH, Chen JK, Kapoor TM, Hammer Ja. Centrosome repositioning in T cells is biphasic and driven by microtubule end-on capture-shrinkage. J Cell Biol. 2013;202:779–792. doi: 10.1083/jcb.201301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Lam P-y, Eichelberg MR, Zasadil L, Bement WM, Huttenlocher A. The role of microtubules in neutrophil polarity and migration in live zebrafish. J Cell Sci. 2012;125:5702–5710. doi: 10.1242/jcs.108324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, O’Brien LE, Wang F, Bourne H, Mostov KE, Zegers MMP. Hepatocyte growth factor switches orientation of polarity and mode of movement during morphogenesis of multicellular epithelial structures. Mol Biol Cell. 2003;14:748–763. doi: 10.1091/mbc.E02-06-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Datta A, Leroy P, O’Brien LE, Mak G, Jou T-S, Matlin KS, Mostov KE, Zegers MMP. Beta1-integrin orients epithelial polarity via Rac1 and laminin. Mol Biol Cell. 2005;16:433–445. doi: 10.1091/mbc.E04-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvon AC, Walker JW, Danowski B, Fagerstrom C, Khodjakov A, Wadsworth P. Centrosome Reorientation in Wound-Edge Cells Is Cell Type Specific. Mol Biol Cell. 2002;13:1871–1880. doi: 10.1091/mbc.01-11-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Grode KD, Stewman SF, Diaz-Valencia JD, Liebling E, Rath U, Riera T, Currie JD, Buster DW, Asenjo AB, et al. Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat Cell Biol. 2011;13:361–370. doi: 10.1038/ncb2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tian X-J, Zhang H, Teng Y, Li R, Bai F, Elankumaran S, Xing J. TGF-β-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci Signal. 2014;7:ra91. doi: 10.1126/scisignal.2005304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.