Abstract
Hearing loss (HL) is the most common sensory deficit in humans with causative variants in over 140 genes. With few exceptions, however, the population-specific distribution for many of the identified variants/genes is unclear. Until recently, the extensive genetic and clinical heterogeneity of deafness precluded comprehensive genetic analysis. Here, using a custom capture panel (MiamiOtoGenes), we undertook a targeted sequencing of 180 genes in a multi-ethnic cohort of 342 GJB2 mutation-negative deaf probands from South Africa, Nigeria, Tunisia, Turkey, Iran, India, Guatemala and the United States (South Florida). We detected causative DNA variants in 25% of multiplex and 7% of simplex families. The detection rate varied between 0% and 57% based on ethnicity, with Guatemala and Iran at the lower and higher end of the spectrum, respectively. We detected causative variants within 27 genes without predominant recurring pathogenic variants. The most commonly implicated genes include MYO15A, SLC26A4, USH2A, MYO7A, MYO6 and TRIOBP. Overall, our study highlights the importance of family history and generation of databases for multiple ethnically discrete populations to improve our ability to detect and accurately interpret genetic variants for pathogenicity.
Keywords: Deafness, Gene panel, Pathogenic variants, Variants of uncertain significance
Introduction
Hearing loss (HL) is one of the most common sensory impairment in humans. It is estimated that one child in 1000 is born with a prelingual HL that can have significant impact on normal speech and language skills (Yoshinaga-Itano 2000). Approximately 10% of the population is affected with disabling HL by the age of 60 years and ∼50% by the age of 80 years (Davis 1995). HL can be due to environmental factors, genetic factors, or a combination thereof. However, genetic factors are now regarded as the leading cause of childhood HL in developed countries since other causes are generally prevented by vaccines, antibiotics and workplace regulations (Nance 2003). It is estimated that approximately 30% of all genetic HL is syndromic in nature, i.e., (syndromic HL, SHL) (Online Mendelian Inheritance in Man; http://www.ncbi.nlm.nih.gov/omim/), and approximately 70% of genetic HL is non-syndromic (NSHL), wherein hearing impairment is the only feature observed (Gorlin et al. 1995). NSHL generally is due to mutations in single genes. Approximately 80% of NSHL is autosomal-recessive (ARNSHL), 20% is autosomal dominant (ADNSHL), 1% is X-linked, and < 1% is mitochondrial. Most ARNSHL is pre-lingual severe-to-profound, whereas ADNSHL is often post-lingual and progressive (Angeli et al. 2012).
The genetic basis of HL is heterogeneous with numerous loci/genes already identified in humans. Over 140 loci have been described for NSHL (Hereditary Hearing Loss Homepage; http://hereditaryhearingloss.org). Over 700 syndromes may feature HL (Online Mendelian Inheritance in Man; http://www.ncbi.nlm.nih.gov/omim/). The same clinical syndrome can be caused by different genes and different mutations in the same gene may result in SHL and NSHL (Yan and Liu 2008). For some genes, there are both dominant and recessive alleles. Even the same variant in a single gene can be associated with quite variable phenotypes (Hutchin et al. 2003).
Recent technical advances have revealed new molecular mechanisms of HL and provided improved diagnostic methods. Molecular genetic testing for several HL-associated genes is now part of the standard protocol for the etiologic diagnosis of HL (King et al. 2012). An immediate benefit is that the identification of the specific genetic variant responsible for HL can establish or confirm a clinical diagnosis, and allow the implementation of personalized approaches to medical management. The information also facilitates risk assessment for affected families and enables reproductive decision-making.
Decades of experience have proven the diagnostic utility of Mendelian disorders by serial additive Sanger sequencing of candidate genes (Maddalena et al. 2005; Richards et al. 2008). However, this approach is labor intensive and not cost effective for a disorder as heterogeneous as HL. An array-based method has also been developed, but it contains a limited number of genes, is expensive, and only known mutations can be analyzed (Kothiyal et al. 2010). A disorder with high heterogeneity such as HL is often difficult to dissect with these techniques because of the necessity of identifying the candidate genes for testing. Today, the revolutionary targeted capture and next-generation sequencing (NGS) technologies provide a viable alternative because of their massively parallel sequencing capability, which enables the simultaneous screening of multiple HL genes in multiple samples (Shearer et al. 2010; Brownstein et al. 2011; Yan et al. 2013; Tekin et al. 2016). Gene panels are useful when multiple genes are involved in a particular disorder or when there is extensive phenotypic overlap between different disorders. Panels are also more cost effective and results can be obtained more rapidly than a traditional gene by gene approach. In this study, we undertook a targeted sequencing of 180 known and candidate HL causing genes in a multi-ethnic cohort of 342 GJB2-mutation negative probands.
Materials and methods
Subjects
This study was approved by the University of Miami Institutional Review Board (USA), the Madras ENT Research Foundation (P) Ltd (MERF) (India), the University Hospital of Mahdia (Tunisia), the Growth and Development Research Ethics Committee (Iran), the Ethics Committee of University of Ibadan (Nigeria), the Ankara University Medical School Ethics Committee (Turkey), the University Hospital of Sfax Ethics Committee (Tunisia), University of Pretoria School of Medicine Ethics Committee (South Africa), Institute for Research on Genetic and Metabolic Diseases, INVEGEM (Guatemala). A signed informed-consent form was obtained from each participant or, in the case of a minor, from the parents.
We have included in this study a total of 342 GJB2 mutation-negative families of diverse ethnicity. Of these, 185 were simplex and 157 were multiplex with at least 2 affected individuals. Since a 3-generation pedigree was not available in some cases, we did not group multiplex families according to inheritance pattern. The multi-ethnic cohort was comprised of 91 indigenous families from South Africa, 90 from Nigeria, 53 from the USA (South Florida), 38 from Tunisia, 23 from India, 21 from Iran, 19 from Turkey, and 7 from Guatemala. The diagnosis of SNHL was established via standard audiometry in a soundproofed room according to current clinical standards. HL was congenital or prelingual-onset with a severity ranging from mild to profound. Clinical evaluation included a thorough physical examination and otoscopy in all cases. Additional evaluations, including a high-resolution, thin-section computed tomography (CT) and magnetic resonance imaging (MRI) of the temporal bone, were performed when possible. None of the recruited individuals was diagnosed with a syndrome. DNA was extracted from peripheral blood leukocytes of probands according to standard procedures.
Sequencing
Using the Agilent SureDesign online tool (https://earray.chem.agilent.com/suredesign/), a SureSelect custom kit (Agilent, Santa Clara, CA, https://www.agilent.com) was designed to include all exons, 5′ UTRs and 3′ UTRs of 180 known and candidate deafness causing genes (Supplementary Table S1) (Tekin et al. 2016). This custom capture panel (MiamiOtoGenes), with a target size of approximately 1.158 MB encompassing 3494 regions, covers genes associated with both syndromic and non-syndromic forms of HL. The targeted sequencing was processed at the Hussman Institute for Human Genomics (HIHG) Sequencing core, University of Miami. The Agilent’s SureSelect Target Enrichment (Agilent, Santa Clara, CA) of coding exons and flanking intronic sequences in-solution hybridization capture system was used following the manufacturer’s standard protocol. Adapter sequences for the Illumina HiSeq2000 were ligated and the enriched DNA samples were prepared using the standard methods for the HiSeq2000 instrument (Illumina). Through the sample preparation average insert size was 180 bp and paired end reads were used. Regions with lower coverage were not subjected to additional sequencing.
Bioinformatics Analysis
The Illumina CASAVA v1.8 pipeline was used to assemble 99 bp sequence reads. Burrows-Wheeler Aligner (BWA) was applied for alignment of sequence reads to the human reference genome (hg19) (Li and Durbin 2010) and variants were called using FreeBayes (Garrison and Marth 2012). Genesis 2.0 (https://www.genesis-app.com/) was then used for variant filtering based on quality/score read depth and minor allele frequency (MAF thresholds of 0.005 for ARNSHL and 0.0005 for ADNSHL variants) as reported in dbSNP141, the National Heart, Lung, and Blood Institute Exome Sequencing Project Exome Variant Server, Seattle, WA Project (Exome Variant Server, 2012), Exome Aggregation Consortium (ExAC) browser (http://exac.broadinstitute.org/), the 1000 Genome Project Database and our internal database of > 3,000 samples from European, Asian, and American ancestries. Variants meeting these criteria were further annotated based on their presence and pathogenicity information in Human Gene Mutation Database (HGMD; http://www.hgmd.cf.ac.uk), the Deafness Variation Database (DVD) (deafnessvariationdatabase.org), and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/). In the final step, all variants were re-classified based on the American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) guidelines (Richards et al. 2015). These guidelines recommend the use of specific standard terminology for DNA variants in five categories to include pathogenic, likely pathogenic, uncertain significance, likely benign, and benign. They describe criteria using evidence from population data, computational data, functional data and segregation data for variant interpretation. Copy number variations (CNV) calling was performed using an R-based tool (Nord at al. 2011). This method normalizes read-depth data by sample batch and compares median read-depth ratios using a sliding-window approach.
Sanger sequencing was used for the confirmation of variant calls and PCR for the CNVs. Family members, when available, were used for segregation, de novo status and trans configuration of biallelic variants. During the interpretation we also considered phenotypic correlations between the gene variants and their reported phenotypes.
Results
Targeted Capture Sequencing
Targeted capture genome enrichment (TGE) and massively parallel sequencing (MPS) were performed on all probands. An average of 99%, 87%, 60% of the targeted bases were covered at 10×, 50×, and 100×, respectively (Supplementary Fig. S1).
Molecular Findings among Probands in the Multi-Ethnic Cohort
After QC and filtration (read depth >8, Genotype Quality> 35 and QUAL >20), we detected 151 variants in 119 families that we classified as likely pathogenic, pathogenic or variant of uncertain significance based on ACMG guidelines. Of these, 44% (66/151) have been reported in at least one of the following 3 databases: ClinVar, HGMD, DVD (Supplementary Table S2).
HL Causative Genes in the Cohort
When only pathogenic and likely pathogenic variants were taken into consideration, the underlying genetic cause was identified in 53 families, providing an etiologic diagnostic rate of 15% (53/342) in the cohort. The detection rates in different groups were 0% (0/7, Guatemala), 4% (4/91, South Africa), 4% (4/90, Nigeria), 17% (9/53, South Florida), 26% (10/38, Tunisia), 26% (6/23, India), 42% (8/19, Turkey), and 57% (12/21, Iran) (Table 1 and Fig. 1A). Causative variants were detected in 7% (13/185) of the simplex families and 25% (40/157) of the multiplex families (Fig. 1A).
Table 1.
Identified likely pathogenic and pathogenic variants in the solved families.
| ID | M/S | Country | Gene | Transcript | cDNA | Protein | Zygosity | Reference | ACMG1 |
|---|---|---|---|---|---|---|---|---|---|
| F15A | S | India | ILDR1 | NM_001199799.1 | c.58+1G>A | Splice | HM | Novel | P |
| F22A | S | India | OTOF | NM_194248.2 | c.5669G>A | p.W1890* | HM | Novel | P |
| F24A | M | India | TMC1 | NM_138691.2 | c.236+1G>C | Splice | HM | Yang (2013) | P |
| F25A | S | India | MYO7A | NM_000260.3 | c.4485G>A | p.W1495* | HM | Novel | P |
| F27A | M | India | MYO7A | NM_000260.3 | c.3978C>A | p.C1326* | HM | Novel | P |
| F29A | S | India | GIPC3 | NM_133261.2 | c.662C>T | p.T221I | HM | Rehman (2011) | LP |
| 2075 | M | Iran | TMPRSS3 | NM_024022.2 | c.46C>T | p.R16* | HM | Novel | P |
| 2081 | M | Iran | TECTA | NM_005422.2 | c.651dupC | p.N218Qfs*31 | HM | Naz (2003) | P |
| 2082 | M | Iran | MYO15A | NM_016239.3 | c.2280delC | p.S761Lfs*20 | HM | Novel | P |
| 2083 | M | Iran | SLC26A4 | NM_000441.1 | c.235C>T | p.R79* | HM | Wu (2010) | P |
| 2085 | M | Iran | MARVELD2 | NM_001038603.2 | c.1550delA | p.K517Rfs*16 | HM | Babanejad (2012) | P |
| 2088 | M | Iran | MYO15A | NM_016239.3 | c.6273+1G>A | Splice | HM | Novel | P |
| 2094 | M | Iran | ESPN | NM_031475.2 | c.2440C>T | p.Q814* | HM | Sloan-Heggen (2015) | P |
| 2099 | M | Iran | PTPRQ | NM_001145026.1 | c.4009G>T | p.E1337* | HM | Novel | P |
| 2105 | M | Iran | CDH23 | NM_022124.5 | c.2192+1G>C | Splice | HM | Novel | P |
| 2109 | M | Iran | MYO6 | NM_004999.3 | c.392−1G>A | Splice | HM | Novel | P |
| 2111 | S | Iran | DFNB59 | NM_001042702.3 | c.547C>T | p.R183W | HM | Delmaghani (2006) | LP |
| 2113 | S | Iran | USH2A | NM_206933.2 | c.1001G>A | p.R334Q | HM | Baux (2007) | P |
| 1717 | M | Nigeria | MYO7A | NM_000260.3 | c.287C>T | p.T96M | HT | Novel | LP |
| 1717 | M | Nigeria | MYO7A | NM_000260.3 | c.1708C>T | p.R570* | HT | Yoshimura (2014) | P |
| 1746 | M | Nigeria | MYO6 | NM_004999.3 | c.1477_1487delCAAGAACTCTA | p.Q493Sfs*8 | HM | Novel | P |
| 1768 | M | Nigeria | SLC26A4 | NM_000441.1 | c.737delA | p.N246Tfs*43 | HM | Novel | P |
| 1869 | M | Nigeria | SLC26A4 | NM_000441.1 | c.164+1G>C | Splice | HT | Chu (2015) | P |
| 1869 | M | Nigeria | SLC26A4 | NM_000441.1 | c.2171A>T | p.D724V | HT | Novel | LP |
| BS06 6 | S | South Africa | POU3F4 | NM_000307.4 | c.986G>C | p.R329P | HZ | Lee (2009) | LP |
| BS07 4 | S | South Africa | SIX1 | NM_005982.3 | c.373G>A | p.E125K | HT | Mosrati (2011) | LP |
| TS00 5 | M | South Africa | TRIOBP | NM_001039141.2 | c.572delC | p.P191Rfs*50 | HT | Novel | LP |
| TS00 5 | M | South Africa | TRIOBP | NM_001039141.2 | c.3510_3513dupTGCA | p.P1172Cfs*13 | HT | Novel | LP |
| TS05 8 | M | South Africa | MARVEL D2 | NM_001038603.2 | c.1555−1G>A | Splice | HM | Novel | LP |
| FT21 | M | Tunisia | USH2A | NM_206933.2 | c.14586T>G | p.Y4862* | HM | Baux (2014) | P |
| FT22 | M | Tunisia | MYO15A | NM_016239.3 | c.7395+3G>C | Splice | HM | Riahi (2012) | LP |
| FT23 | M | Tunisia | USH2A | NM_206933.2 | c.14586T>G | p.Y4862* | HM | Baux (2014) | P |
| FT24 | M | Tunisia | MYO15A | NM_016239.3 | c.5417T>C | p.L1806P | HM | Riahi (2012) | LP |
| FT25 | M | Tunisia | EPS8 | NM_004447.5 | c.115delA | p.T39Qfs*32 | HM | Novel | P |
| FT28 | M | Tunisia | USH2A | NM_206933.2 | CNV | CNV | HM | Novel | LP |
| FT33 | M | Tunisia | CIB2 | NM_006383.3 | c.247_257delGAGGGGAACCT | p.E83Hfs*30 | HM | Novel | P |
| FT36 | M | Tunisia | PCDH15 | NM_033056.3 | CNV | CNV | HM | Novel | LP |
| FT39 | M | Tunisia | LRTOMT | NM_001145308.4 | c.242G>A | p.R81Q | HM | Ahmed (2008) | LP |
| FT42 | M | Tunisia | LRTOMT | NM_001145308.4 | c.242G>A | p.R81Q | HM | Ahmed (2008) | LP |
| 1384 | M | Turkey | USH1G | NM_173477.2 | c.387dupC | p.K130Qfs*5 | HM | Novel | P |
| 1405 | M | Turkey | MYO15A | NM_016239.3 | c.8309_8311delAGG | p.E2770del | HM | Sloan-Heggen (2015) | P |
| 1580 | M | Turkey | TMIE | NM_147196.2 | c.250C>T | p.R84W | HM | Naz (2002) | LP |
| 1583 | M | Turkey | ILDR1 | NM_001199799.1 | c.583C>T | p.Q195* | HM | Borck (2011) | P |
| 274 | M | Turkey | MYO15A | NM_016239.3 | c.8090T>C | p.V2697A | HT | Schrauwen (2013) | LP |
| 274 | M | Turkey | MYO15A | NM_016239.3 | c.10492-2dupA | Splice | HT | Novel | LP |
| 714 | S | Turkey | OTOF | NM_194248.2 | c.3679C>T | p.R1227* | HM | Novel | P |
| 924 | M | Turkey | SLC26A4 | NM_000441.1 | c.397T>A | p.S133T | HM | Fugazzola (2002) | P |
| 952 | M | Turkey | TRIOBP | NM_001039141.2 | c.2521C>T | p.R841* | HM | Novel | P |
| 1087 | S | USA | MYO7A | NM_000260.3 | c.999T>G | p.Y333* | HM | Weston (1996) | P |
| 1088 | S | USA | MYO15A | NM_016239.3 | c.7226delC | p.P2409Qfs*8 | HM | Novel | P |
| 1255 | S | USA | SLC26A4 | NM_000441.1 | c.2162C>T | p.T721M | HM | Usami (1999) | P |
| 1554 | S | USA | USH2A | NM_206933.2 | c.2299delG | p.E767Sfs*21 | HT | Eudy (1998) | P |
| 1554 | S | USA | USH2A | NM_206933.2 | c.15200delT | p.I5067Tfs*23 | HT | Novel | P |
| NSD F207 | M | USA | DFNB31 | NM_015404.3 | c.1573_1574delAC | p.T525Gfs*43 | HM | Novel | P |
| NSD F253 | M | USA | MYO6 | NM_004999.3 | c.1452dupT | p.N485* | HT | Novel | P |
| NSD F288 | M | USA | TMC1 | NM_138691.2 | c.1939T>C | p.S647P | HM | Brownstein (2011) | LP |
| NSD F362 | M | USA | POU4F3 | NM_002700.2 | c.705delT | p.L236Sfs*6 | HT | Novel | LP |
| NSD F431 | M | USA | TRIOBP | NM_001039141.2 | c.2581C>T | p.R861* | HT | Gu (2015) | P |
| NSD F431 | M | USA | TRIOBP | NM_001039141.2 | c.3089delC | p.P1030Lfs*183 | HT | Novel | P |
S: Simplex, M: Multiplex, P: Pathogenic, LP: Likely Pathogenic, HM: Homozygous, HT: Heterozygous, HZ:Hemizygous;
ACMG: Standards and guidelines for the interpretation of sequence variants (Richards S, 2015).
Fig. 1.
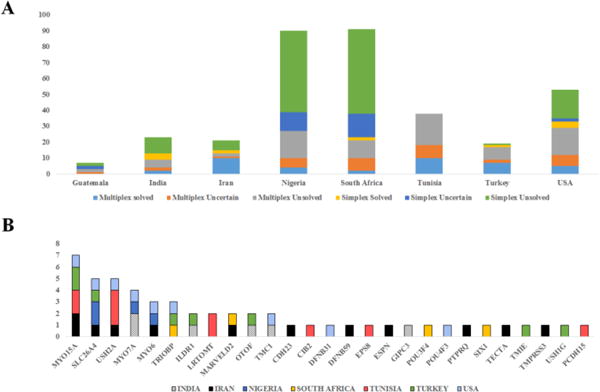
Representation of solved, unsolved and uncertain families, based on ethnicity and simplex/multiplex status (A). Number of solved families for each gene and ethnicity (B).
Of the 119 families, 66 (55%) were classified as uncertain families. Those uncertain families had at least one allele with a variant of unknown significance (VUS) even if they had another allele classified as likely pathogenic or pathogenic. The uncertain family rates in the multiplex families were 22% (6/27) in Nigeria, 38% (8/21) in South Africa, 21% (8/38) in Tunisia, 22% (2/9) in India, 33% (1/3) in Guatemala, 12% (2/17) in Turkey, 8% (1/13) in Iran, and 26% (7/27) in USA (Supplementary Table S3).
In this multi-ethnic cohort, sequence variants were identified in a total of 48 genes (Supplementary Table S2) while 27 different genes had variants in solved families. Genes identified in at least 3 solved families include MYO15A (MIM 602666) (13%; 7/53), SLC26A4 (MIM 605646) (9%; 5/53), USH2A (MIM 608400) (9%; 5/53), MYO7A (MIM 276903) (8%; 4/53), TRIOBP (MIM 609761) (6%; 3/53), and MYO6 (MIM 600970) (6%; 3/53) (Fig. 1B).
Of the 57 unique HL-causing variants identified in solved families, 26 have previously been reported in the literature (Table 1). The remaining 31 novel variations were considered pathogenic or likely pathogenic according to ACMG guidelines (Table 1). Of note in solved families, 81% (43/53) of the 53 probands found to carry causative variants were homozygous for the identified HL-causing variant (autosomal recessive), 11% (6/53) were compound heterozygou (autosomal recessives), 6% (3/53) were heterozygous for a single causative variant (autosomal dominant) and 1 individual was hemizygous for an X-linked variant (Table 1).
Two novel homozygous CNVs were identified in Tunisian families, one consisted of a large deletion of approximately 86.3kb with breakpoints within exons 21 and 22 of USH2A and one deletion of approximately 12.3kb, spanning exons 12 and 13 of the PCDH15 gene (Supplementary Table S4). Deleted exons did not amplify with confirmatory PCR in probands.
While we specifically queried parental consanguinity when obtaining family history, we did not incorporate it into the analysis due to concerns regarding the reliability of self-reported consanguinity in different populations. When we reviewed the variants, we noted that all Indian and Iranian and most Turkish and Tunisian probands were homozygous for pathogenic, likely pathogenic, and VUS, indicating shared ancestry between their parents.
Discussion
In the present study, we have used a panel of 180 genes sequenced by NGS for variant detection in a multi-ethnic group of 342 probands. We identified causative variants in 27 genes without predominant recurring pathogenic variants in the identified genes. The most commonly implicated genes include MYO15A, SLC26A4, USH2A, MYO7A, MYO6, and TRIOBP. As expected, most of the identified variants are autosomal recessive.
Use of the MiamiOtoGene panel established a genetic diagnosis for 28% of all probands from non-sub-Saharan African countries including Guatemala, USA, Tunisia, India, Turkey, and Iran. On the other hand, the etiologic diagnostic rate for families from sub-Saharan Africa (Nigeria, South Africa) is 4%. All the variants detected in the Guatemalan probands were classified as VUS resulting in a “solved” rate of 0% in this ethnic group. Molecular diagnostic rates for Turkish and Iranian probands are very similar to those reported by Shearer et al. (Shearer et al. 2013) using OtoSCOPE and Bademci et al. (Bademci et al. 2015) using whole exome sequencing. It should be noted that a positive family history of deafness is an important indication for a genetic etiology. In our cohort, the distribution of simplex and multiplex cases was remarkably diverse in different ethnicities. Moreover, parental consanguinity is traditionally common in Turkey, Iran, and Tunisia, which increases the chance of having rare autozygous mutations. The current study found solved rates of around 7% for the simplex families compared to 25% for multiplex families. Across a variety of studies utilizing NGS, the diagnostic rate overall averaged 41% and ranged from a low of 10% to 83% (Shearer and Smith 2015). In an analysis of simplex cases, Gu et al. (Gu et al. 2014) found a diagnostic rate of 13%. Direct comparison between studies is difficult because of the fundamental differences in study design. These include prescreening for GJB2 variations, and the number of genes included on a “comprehensive” test, ranging from 34 to 246 different genes (Shearer and Smith 2015). Additionally, the genes selected for each platform vary based on whether only NSHL genes or also SHL genes are included (and which syndromes), and also whether candidate genes identified though animal models or human studies as in the case of our platform, the MiamiOtoGenes panel, are included. Overall, our data highlight the importance of family history and generation of databases with ethnically diverse samples to improve our ability to detect and accurately evaluate genetic variants for pathogenicity. The type of mutations evaluated should also be taken into account, when considering a comprehensive genetic test. While all platforms include analysis of point mutations and small deletions, not all the studies screened for large CNVs (Shearer et al. 2013; Shearer et al. 2014). In the current study, CNVs account for 4% of causative alleles, yet rates as high as 13–19% have been reported.
As NGS technology is becoming more widespread in the diagnostic setting, interpreting the clinical meaning of newly discovered variants will be one of the major challenges of ‘genomic’, or ‘precision’, medicine (Chun-Hui and Liu 2014; Aronson and Rehm 2015). Classifying variants is an important issue. The online prediction programs such as PolyPhen2 and SIFT can provide an indication of whether a variant that changes the amino acid at a certain position could be deleterious; but they are unreliable, can be incorrect and alone should not be used to determine whether a variant is likely to be disease causing (Tchernitchko et al. 2004; Thusberg and Vihinen 2009). HGMD, ClinVar, and DVD are commonly checked to decide about the pathogenicity of a detected variant for HL. However, these databases are not always in agreement for the classification of DNA variants. While recent ACMG-AMP Guidelines provide a solution to this problem, some criteria suggested are subjective that would lead to disagreement between different labs (Richards et al. 2015). Recently nine molecular diagnostic laboratories which involved in the Clinical Sequencing Exploratory Research (CSER) tested ACMG-AMP guidelines for the variant interpretation. Interestingly concordance across laboratories was only 34% and after consensus discussions and detailed review of the ACMG-AMP criteria, authors mentioned that concordance increased to 71% (Amendola et al. 2016).
In our study, the overall diagnostic rate is 15%. 19% of the families were classified as uncertain because the probands in these families had at least one VUS. In order to solve these families, more functional, computational, or literature evidence is needed.
Supplementary Material
Supplementary material fig. S1: Coverage percentages of the targeted genes at 10×, 50× and 100×.
Acknowledgments
This study was supported by R01 DC05575, R01 DC01246, 2P50DC000422-Sub-Project 6432, and R01 DC012115 from the National Institutes of Health/National Institute on Deafness and Other Communication Disorders to Xuezhong Liu and R01DC09645 and R01DC012836 to Mustafa Tekin; University of Pretoria RDP fund to RI Kabahuma.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare no conflict of interest.
References
- Ahmed ZM, Masmoudi S, Kalay E, Belyantseva IA, Mosrati MA, Collin RW, Riazuddin S, Hmani-Aifa M, Venselaar H, Kawar MN, Tlili A, van der Zwaag B, Khan SY, Ayadi L, Riazuddin SA, Morell RJ, Griffith AJ, Charfedine I, Caylan R, Oostrik J, Karaguzel A, Ghorbel A, Riazuddin S, Friedman TB, Ayadi H, Kremer H. Mutations of LRTOMT, a fusion gene with alternative reading frames, cause nonsyndromic deafness in humans. Nat Genet. 2008;40:1335–40. doi: 10.1038/ng.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, Berg JS, Biswas S, Bowling KM, Conlin LK, Cooper GM, Dorschner MO, Dulik MC, Ghazani AA, Ghosh R, Green RC, Hart R, Horton C, Johnston JJ, Lebo MS, Milosavljevic A, Ou J, Pak CM, Patel RY, Punj S, Richards CS, Salama J, Strande NT, Yang Y, Plon SE, Biesecker LG, Rehm HL. Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeli S, Lin X, Liu XZ. Genetics of hearing and deafness. Anat Rec (Hoboken) 2012;295:1812–29. doi: 10.1002/ar.22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson SJ, Rehm HL. Building the foundation for genomics in precision medicine. Nature. 2015;526:336–42. doi: 10.1038/nature15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babanejad M, Fattahi Z, Bazazzadegan N, Nishimura C, Meyer N, Nikzat N, Sohrabi E, Najmabadi A, Jamali P, Habibi F, Smith RJ, Kahrizi K, Najmabadi H. A comprehensive study to determine heterogeneity of autosomal recessive nonsyndromic hearing loss in Iran. Am J Med Genet A. 2012;158A:2485–92. doi: 10.1002/ajmg.a.35572. [DOI] [PubMed] [Google Scholar]
- Bademci G, Foster J, 2nd, Mahdieh N, Bonyadi M, Duman D, Cengiz FB, Menendez I, Diaz-Horta O, Shirkavand A, Zeinali S, Subasioglu A, Tokgoz-Yilmaz S, Huesca-Hernandez F, de la Luz Arenas-Sordo M, Dominguez-Aburto J, Hernandez-Zamora E, Montenegro P, Paredes R, Moreta G, Vinueza R, Villegas F, Mendoza-Benitez S, Guo S, Bozan N, Tos T, Incesulu A, Sennaroglu G, Blanton SH, Ozturkmen-Akay H, Yildirim-Baylan M, Tekin M. Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med. 2016;18:364–71. doi: 10.1038/gim.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baux D, Blanchet C, Hamel C, Meunier I, Larrieu L, Faugere V, Vache C, Castorina P, Puech B, Bonneau D, Malcolm S, Claustres M, Roux AF. Enrichment of LOVD-USHbases with 152 USH2A genotypes defines an extensive mutational spectrum and highlights missense hotspots. Hum Mutat. 2014;35:1179–86. doi: 10.1002/humu.22608. [DOI] [PubMed] [Google Scholar]
- Baux D, Larrieu L, Blanchet C, Hamel C, Ben Salah S, Vielle A, Gilbert-Dussardier B, Holder M, Calvas P, Philip N, Edery P, Bonneau D, Claustres M, Malcolm S, Roux AF. Molecular and in silico analyses of the full-length isoform of usherin identify new pathogenic alleles in Usher type II patients. Hum Mutat. 2007;28:781–9. doi: 10.1002/humu.20513. [DOI] [PubMed] [Google Scholar]
- Borck G, Ur Rehman A, Lee K, Pogoda HM, Kakar N, von Ameln S, Grillet N, Hildebrand MS, Ahmed ZM, Nurnberg G, Ansar M, Basit S, Javed Q, Morell RJ, Nasreen N, Shearer AE, Ahmad A, Kahrizi K, Shaikh RS, Ali RA, Khan SN, Goebel I, Meyer NC, Kimberling WJ, Webster JA, Stephan DA, Schiller MR, Bahlo M, Najmabadi H, Gillespie PG, Nurnberg P, Wollnik B, Riazuddin S, Smith RJ, Ahmad W, Muller U, Hammerschmidt M, Friedman TB, Riazuddin S, Leal SM, Ahmad J, Kubisch C. Loss-of-function mutations of ILDR1 cause autosomal-recessive hearing impairment DFNB42. Am J Hum Genet. 2011;88:127–37. doi: 10.1016/j.ajhg.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein Z, Friedman LM, Shahin H, Oron-Karni V, Kol N, Abu Rayyan A, Parzefall T, Lev D, Shalev S, Frydman M, Davidov B, Shohat M, Rahile M, Lieberman S, Levy-Lahad E, Lee MK, Shomron N, King MC, Walsh T, Kanaan M, Avraham KB. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol. 2011;12:R89. doi: 10.1186/gb-2011-12-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CW, Chen YJ, Lee YH, Jaung SJ, Lee FP, Huang HM. Government-funded universal newborn hearing screening and genetic analyses of deafness predisposing genes in Taiwan. Int J Pediatr Otorhinolaryngol. 2015;79:584–90. doi: 10.1016/j.ijporl.2015.01.033. [DOI] [PubMed] [Google Scholar]
- Davis A. Hearing in Adults: The Prevalence and Distribution of Hearing Impairment and Reported Hearing Disability in the MRC Institute of Hearing Research’s National Study of Hearing. Whurr publisher; London: 1995. [Google Scholar]
- Delmaghani S, del Castillo FJ, Michel V, Leibovici M, Aghaie A, Ron U, Van Laer L, Ben-Tal N, Van Camp G, Weil D, Langa F, Lathrop M, Avan P, Petit C. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat Genet. 2006;38:770–8. doi: 10.1038/ng1829. [DOI] [PubMed] [Google Scholar]
- Eudy JD, Weston MD, Yao S, Hoover DM, Rehm HL, Ma-Edmonds M, Yan D, Ahmad I, Cheng JJ, Ayuso C, Cremers C, Davenport S, Moller C, Talmadge CB, Beisel KW, Tamayo M, Morton CC, Swaroop A, Kimberling WJ, Sumegi J. Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science. 1998;280:1753–7. doi: 10.1126/science.280.5370.1753. [DOI] [PubMed] [Google Scholar]
- Fugazzola L, Cerutti N, Mannavola D, Crino A, Cassio A, Gasparoni P, Vannucchi G, Beck-Peccoz P. Differential diagnosis between Pendred and pseudo-Pendred syndromes: clinical, radiologic, and molecular studies. Pediatr Res. 2002;51:479–84. doi: 10.1203/00006450-200204000-00013. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv. 2012 1207.3907. [Google Scholar]
- Gu X, Guo L, Ji H, Sun S, Chai R, Wang L, Li H. Genetic testing for sporadic hearing loss using targeted massively parallel sequencing identifies 10 novel mutations. Clin Genet. 2015;87:588–93. doi: 10.1111/cge.12431. [DOI] [PubMed] [Google Scholar]
- Gorlin RJ, Toriello HV, Cohen MM. Hereditary hearing loss and its syndromes. Oxford University Press; Oxford: 1995. [Google Scholar]
- Hutchin TP, Parker MJ, Young ID, Davis AC, Pulleyn LJ, Deeble J, Lench NJ, Markham AF, Mueller RF. A novel mutation in the mitochondrial tRNA(Ser(UCN)) gene in a family with non-syndromic sensorineural hearing impairment. J Med Genet. 2000;37:692–4. doi: 10.1136/jmg.37.9.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King PJ, Ouyang X, Du L, Yan D, Angeli SI, Liu XZ. Etiologic diagnosis of nonsyndromic genetic hearing loss in adult vs pediatric populations. Otolaryngol Head Neck Surg. 2012;147:932–6. doi: 10.1177/0194599812453553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothiyal P, Cox S, Ebert J, Husami A, Kenna MA, Greinwald JH, Aronow BJ, Rehm HL. High-throughput detection of mutations responsible for childhood hearing loss using resequencing microarrays. BMC Biotechnol. 2010;10:10. doi: 10.1186/1472-6750-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Song MH, Kang M, Lee JT, Kong KA, Choi SJ, Lee KY, Venselaar H, Vriend G, Lee WS, Park HJ, Kwon TK, Bok J, Kim UK. Clinical and molecular characterizations of novel POU3F4 mutations reveal that DFN3 is due to null function of POU3F4 protein. Physiol Genomics. 2009;39:195–201. doi: 10.1152/physiolgenomics.00100.2009. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalena A, Bale S, Das S, Grody W, Richards S, Committee ALQA. Technical standards and guidelines: molecular genetic testing for ultra-rare disorders. Genet Med. 2005;7:571–83. doi: 10.1097/01.GIM.0000182738.95726.ca. [DOI] [PubMed] [Google Scholar]
- Mosrati MA, Hammami B, Rebeh IB, Ayadi L, Dhouib L, Ben Mahfoudh K, Hakim B, Charfeddine I, Mnif J, Ghorbel A, Masmoudi S. A novel dominant mutation in SIX1, affecting a highly conserved residue, result in only auditory defects in humans. Eur J Med Genet. 2011;54:e484–8. doi: 10.1016/j.ejmg.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Nance WE. The genetics of deafness. Ment Retard Dev Disabil Res Rev. 2003;9:109–19. doi: 10.1002/mrdd.10067. [DOI] [PubMed] [Google Scholar]
- Naz S, Alasti F, Mowjoodi A, Riazuddin S, Sanati MH, Friedman TB, Griffith AJ, Wilcox ER, Riazuddin S. Distinctive audiometric profile associated with DFNB21 alleles of TECTA. J Med Genet. 2003;40:360–3. doi: 10.1136/jmg.40.5.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz S, Giguere CM, Kohrman DC, Mitchem KL, Riazuddin S, Morell RJ, Ramesh A, Srisailpathy S, Deshmukh D, Riazuddin S, Griffith AJ, Friedman TB, Smith RJ, Wilcox ER. Mutations in a novel gene, TMIE, are associated with hearing loss linked to the DFNB6 locus. Am J Hum Genet. 2002;71:632–6. doi: 10.1086/342193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Lee M, King MC, Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics. 2011;12:184. doi: 10.1186/1471-2164-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, Gul K, Morell RJ, Lee K, Ahmed ZM, Riazuddin S, Ali RA, Shahzad M, Jaleel AU, Andrade PB, Khan SN, Khan S, Brewer CC, Ahmad W, Leal SM, Riazuddin S, Friedman TB. Mutations of GIPC3 cause nonsyndromic hearing loss DFNB72 but not DFNB81 that also maps to chromosome 19p. Hum Genet. 2011;130:759–65. doi: 10.1007/s00439-011-1018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riahi Z, Bonnet C, Zainine R, Louha M, Bouyacoub Y, Laroussi N, Chargui M, Kefi R, Jonard L, Dorboz I, Hardelin JP, Salah SB, Levilliers J, Weil D, McElreavey K, Boespflug OT, Besbes G, Abdelhak S, Petit C. Whole exome sequencing identifies new causative mutations in Tunisian families with non-syndromic deafness. PLoS One. 2014;9:e99797. doi: 10.1371/journal.pone.0099797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE, Molecular Subcommittee of the ALQAC ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Sommen M, Corneveaux JJ, Reiman RA, Hackett NJ, Claes C, Claes K, Bitner-Glindzicz M, Coucke P, Van Camp G, Huentelman MJ. A sensitive and specific diagnostic test for hearing loss using a microdroplet PCR-based approach and next generation sequencing. Am J Med Genet A. 2013;161A:145–52. doi: 10.1002/ajmg.a.35737. [DOI] [PubMed] [Google Scholar]
- Shearer AE, Black-Ziegelbein EA, Hildebrand MS, Eppsteiner RW, Ravi H, Joshi S, Guiffre AC, Sloan CM, Happe S, Howard SD, Novak B, Deluca AP, Taylor KR, Scheetz TE, Braun TA, Casavant TL, Kimberling WJ, Leproust EM, Smith RJ. Advancing genetic testing for deafness with genomic technology. J Med Genet. 2013;50:627–34. doi: 10.1136/jmedgenet-2013-101749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J, 2nd, Scherer S, Scheetz TE, Smith RJ. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:21104–9. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Eppsteiner RW, Booth KT, Ephraim SS, Gurrola J, 2nd, Simpson A, Black-Ziegelbein EA, Joshi S, Ravi H, Giuffre AC, Happe S, Hildebrand MS, Azaiez H, Bayazit YA, Erdal ME, Lopez-Escamez JA, Gazquez I, Tamayo ML, Gelvez NY, Leal GL, Jalas C, Ekstein J, Yang T, Usami S, Kahrizi K, Bazazzadegan N, Najmabadi H, Scheetz TE, Braun TA, Casavant TL, LeProust EM, Smith RJ. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am J Hum Genet. 2014;95:445–53. doi: 10.1016/j.ajhg.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Smith RJ. Massively Parallel Sequencing for Genetic Diagnosis of Hearing Loss: The New Standard of Care. Otolaryngol Head Neck Surg. 2015;153:175–82. doi: 10.1177/0194599815591156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM, Babanejad M, Beheshtian M, Simpson AC, Booth KT, Ardalani F, Frees KL, Mohseni M, Mozafari R, Mehrjoo Z, Jamali L, Vaziri S, Akhtarkhavari T, Bazazzadegan N, Nikzat N, Arzhangi S, Sabbagh F, Otukesh H, Seifati SM, Khodaei H, Taghdiri M, Meyer NC, Daneshi A, Farhadi M, Kahrizi K, Smith RJ, Azaiez H, Najmabadi H. Characterising the spectrum of autosomal recessive hereditary hearing loss in Iran. J Med Genet. 2015;52:823–9. doi: 10.1136/jmedgenet-2015-103389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchernitchko D, Goossens M, Wajcman H. In silico prediction of the deleterious effect of a mutation: proceed with caution in clinical genetics. Clin Chem. 2004;50:1974–8. doi: 10.1373/clinchem.2004.036053. [DOI] [PubMed] [Google Scholar]
- Tekin D, Yan D, Bademci G, Feng Y, Guo S, Foster J, 2nd, Blanton S, Tekin M, Liu X. A next-generation sequencing gene panel (MiamiOtoGenes) for comprehensive analysis of deafness genes. Hear Res. 2016;333:179–84. doi: 10.1016/j.heares.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J, Vihinen M. Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Hum Mutat. 2009;30:703–14. doi: 10.1002/humu.20938. [DOI] [PubMed] [Google Scholar]
- Tsai AC, Liu X. Toward Best Practice in Using Molecular Diagnosis to Guide Medical Management, Are We There Yet? N Am J Med Sci (Boston) 2014;7:199–200. [PMC free article] [PubMed] [Google Scholar]
- Usami S, Abe S, Weston MD, Shinkawa H, Van Camp G, Kimberling WJ. Non-syndromic hearing loss associated with enlarged vestibular aqueduct is caused by PDS mutations. Hum Genet. 1999;104:188–92. doi: 10.1007/s004390050933. [DOI] [PubMed] [Google Scholar]
- Weston MD, Kelley PM, Overbeck LD, Wagenaar M, Orten DJ, Hasson T, Chen ZY, Corey D, Mooseker M, Sumegi J, Cremers C, Moller C, Jacobson SG, Gorin MB, Kimberling WJ. Myosin VIIA mutation screening in 189 Usher syndrome type 1 patients. Am J Hum Genet. 1996;59:1074–83. [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Lu YC, Chen PJ, Yeh PL, Su YN, Hwu WL, Hsu CJ. Phenotypic analyses and mutation screening of the SLC26A4 and FOXI1 genes in 101 Taiwanese families with bilateral nonsyndromic enlarged vestibular aqueduct (DFNB4) or Pendred syndrome. Audiol Neurootol. 2010;15:57–66. doi: 10.1159/000231567. [DOI] [PubMed] [Google Scholar]
- Yan D, Liu XZ. Cochlear molecules and hereditary deafness. Front Biosci. 2008;13:4972–83. doi: 10.2741/3056. [DOI] [PubMed] [Google Scholar]
- Yan D, Tekin M, Blanton SH, Liu XZ. Next-generation sequencing in genetic hearing loss. Genet Test Mol Biomarkers. 2013;17:581–7. doi: 10.1089/gtmb.2012.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Wei X, Chai Y, Li L, Wu H. Genetic etiology study of the non-syndromic deafness in Chinese Hans by targeted next-generation sequencing. Orphanet J Rare Dis. 2013;8:85. doi: 10.1186/1750-1172-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura H, Iwasaki S, Nishio SY, Kumakawa K, Tono T, Kobayashi Y, Sato H, Nagai K, Ishikawa K, Ikezono T, Naito Y, Fukushima K, Oshikawa C, Kimitsuki T, Nakanishi H, Usami S. Massively parallel DNA sequencing facilitates diagnosis of patients with Usher syndrome type 1. PLoS One. 2014;9:e90688. doi: 10.1371/journal.pone.0090688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga-Itano C. Successful outcomes for deaf and hard-of-hearing children. Semin Hear. 2000;21:309–326. doi: 10.1055/s-2000-13462. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material fig. S1: Coverage percentages of the targeted genes at 10×, 50× and 100×.


