Abstract
Over the past twenty years, exciting developments in optical and molecular imaging approaches have allowed researchers to examine with unprecedented resolution the spatial organization of transcription sites in the nucleus. An attractive model that has developed from these studies is that active genes cluster to preformed transcription factories that contain multiple active RNA polymerases and transcription factor proteins required for efficient mRNA biogenesis. However, this model has been extensively debated in part due to the fact transcription factories and their features have only been documented in fixed cells. In this review, we will focus on recent live-cell imaging studies that are changing our understanding of transcription factories.
Introduction
The genetic information encoded in the genomes of organisms contains all the information necessary for life. How this information is read and regulated has been area of intense study. It’s clear that a large battery of proteins associate with active genes and participate in transcription and cotranscriptional RNA processing, and the exquisite regulation of these processes [1]; therefore, ‘factory’ may be an appropriate descriptor. However, the nature of this factory remains in dispute. Is the factory a ‘brick and mortar’ (stable) structure containing transcription machinery to which genes are delivered, or is the factory transient and built on genes as needed by assembling the transcription machinery? Classical biochemical studies, supported by more recent live-cell imaging analyses, show that transcription factor proteins and RNA Polymerase II are recruited to genes to activate their expression. In contrast, cell biology studies in fixed cells suggest that genes move to stationary preformed factories that contain RNA Polymerase II (Pol II) and other transcription factors to initiate gene expression. In this review, we will highlight recent studies that begin to resolve the features of transcription factories in living cells.
RNA Polymerase II transcription factories -– “seeing is believing”
In the mid-1990s, Cook and colleagues, using confocal microscopy, made the pioneering observation that bromo-uridine triphosphate (BrUTP) labeled nascent transcripts in mammalians cells did not occur diffusely in the nucleoplasm, but at discrete foci [2]. They coined these sites transcription factories. Later studies confirmed this finding using BrUTP and biotin-cytidine triphosphate (biotin-CTP) nucleotide analog labeled RNAs visualized with light microscopy and high resolution electron microscopy. Notably, electron microscopy imaging also revealed that immune-gold labeled nascent transcripts and RNA Polymerase II molecules colocalize in transcription factories [3,4]. This finding was further supported by light microscopy studies showing the spatial colocalization of active Pol II and gene transcripts, using immunostaining to detect Pol II and RNA fluorescence in situ hybridization to detect the transcripts (immuno-RNA FISH). Collectively, these studies provide strong evidence for the spatial organization of transcription in cells [5–7].
Nonetheless, one of the shortcomings of the previously described findings is that they rely on cell fixation, which some argue could create aggregation artifacts. To address this, groups have attempted to image GFP-tagged Pol II in living mammalian cells using confocal microscopy [8–10]. These studies failed to see Pol II clustering, but instead noted diffuse GFP signal in the nucleus. This may be a consequence of inadequate resolution to distinguish between multiple transcription factories and low signal, as it is estimated transcription factories contain a small number of Pol IIs (approximately 8) [8]. To overcome these potential issues, a recent study from Cisse et al. (2013) examined the distribution of Pol II tagged with the photo-convertible protein Dendra2 in living mammalian cells using super-high resolution photoactivation light microscopy (PALM) [11]. This form of microscopy uses iterative cycles of photoconversion (converting fluorescence from green to red for a small fraction of labeled proteins), localization and bleaching (permanently turning off red fluorescence of proteins) to assemble an image one polymerase at a time with a resolution of 20–50 nm [12,13]. Using this approach Cisse et al. identified Pol II clustering, indicating the presence of transcription factories in living cells. Notably, the estimated cluster size was below the diffraction limit of light, around 100 nm, which is consistent with estimates derived from electron microscopy studies [4] and may explain why previous live-cell imaging did not observe them.
Applying a different strategy, Ghamari et al. (2013) used confocal microscopy to visualize the localization of CDK9 fused to the fluorescent protein mCherry, as a proxy for Pol II, in living mouse embryonic stem cells and cells of primary tissues (mouse embryonic fibroblasts, erythroid fetal liver cells, neuronal cells and adult liver cells) [14]. Cdk9 and its partner CyclinT form P-TEFb, a CTD-kinase that binds and phosphorylates the CTD of Paused Pol II and pausing factors at the 5′ end of genes, thereby allowing productive Pol II transcriptional elongation [1,15]. Therefore, the authors of this paper reasoned that Cdk9 could be used as a representative protein of Pol II transcription factories. Interestingly, they observed CDK9-mcherry foci in all cell lines, especially the very flat MEFs (may provide for higher resolution imaging). Moreover, these foci overlapped with transcripts of active genes as measured by immuno-RNA FISH. This finding, as well the work of Cisse and colleagues [11] provide the first evidence for the existence of Pol II transcription clusters in live cells.
Transcription factories –formation before or after function?
One of the most debated features of the transcription factories is whether they are stable pre-assembled structures to which genes are recruited or are they active genes that have recruited the transcription machinery. If the former model were correct, one might expect that factories are stable in the absence of active transcription. Consistent with this, immunostaining experiments in mammalian cells revealed that Pol II remains associated with foci after global inhibition of gene transcription using heat shock or inhibition of elongation using 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside (DRB) [16]. A caveat with this study is that DRB inhibits elongation, but not initiation, which results in an accumulation of promoter proximal paused Pol II at most genes [15,17]. Thus, this particular drug does not completely remove Pol II from genes. Nonetheless, further support for the preassembly model also comes from a study in which activation of the immediate early (IE) proto-oncogenes c-Myc and Fos in mouse B cells correlates with their relocation to transcription factories, as measured by DNA-FISH and antibody staining [18]. Displacement of genes to pre-existing transcription factories has also been reported by other groups [19,20]. These findings provide the evidence that factories are stable structures that form before transcription and recruit genes upon their activation (Figure 1, a–b).
Figure 1. Models for the origin of transcription factories.
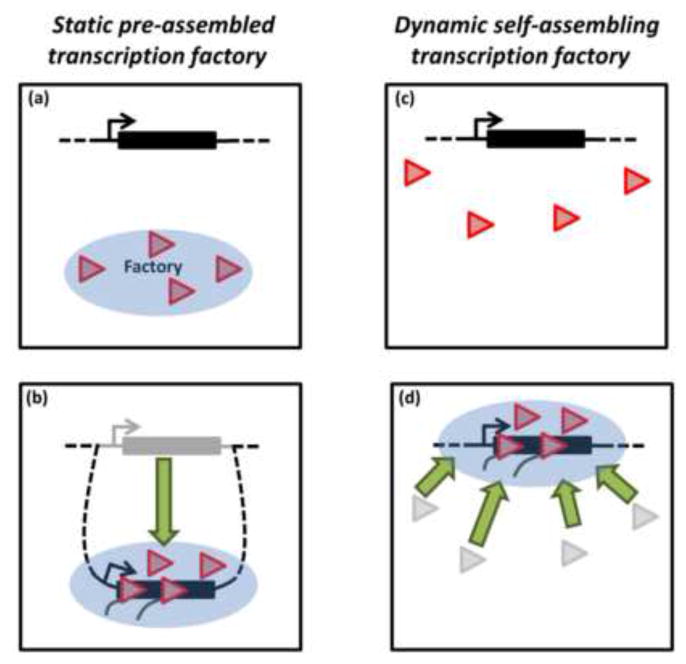
(a) Schematic cartoon of inactive gene. The blue circle indicates a static preassembled transcription factory containing Pol II and other transcription factors. For simplicity, only Pol II is shown (red triangle). (b) Upon activation, the gene moves (or loops out of its chromosome) to the preformed factory for expression. (c) Cartoon of inactive gene surrounded by nucleoplasmic pool of Pol IIs. (d) Gene activation leads to the dynamic self-assembly of Pol II transcription factory onto the gene for expression.
Evidence for the preassembly model has been largely driven by studies in fixed cells. In contrast, live-cell imaging paints a more dynamic picture of transcription, in which factories assemble and disassemble on active genes. Several imaging studies have examined the localization of fluorescent protein-tagged Pol II and other transcription factors to inducible tandem gene arrays. These spatially clustered genes provide a boost in signal to noise and ensure that a majority of fluorescence signal observed at the array is due to interacting Pol II molecules [21]. Using this system in mammalian cells, it has been found that transcription induction leads to GFP-Pol II localization to artificial gene arrays [22,23]. Similarly, in Drosophila, heat shock induction results in the de novo recruitment of GFP-Pol II to the native Hsp70 loci on polytene chromosomes (a natural gene array) [24–26]. Moreover, multicolor-time lapse imaging of fluorescently tagged transcription factors has revealed that they are sequentially recruited. Heat shock factor activator is recruited first to the induced Hsp70 loci (10 sec after HS), Pol II and P-TEFb (120 sec after HS) next, then Spt6 elongation factor (130 sec after HS) and Topoisomerase I (140 sec after HS). Interestingly, Fluorescence recovery after photobleaching (FRAP) experiments indicated that Pol II and other transcription factors dynamically associate with active gene loci [26]. In addition, none of the above studies observed movements of genes upon transcriptional activation.
One of the limitations of the previous live-cell imaging studies is that they make use of artificial tandem gene arrays or polytene arrays that may not accurately reflect the biology of normal genes on chromatids of diploid cells. This was circumvented by Cisse et al. (2013) who examined the assembly dynamics of Dendra2-Pol II clusters in living mammalian diploid cells using a time-lapse version of PALM [11]. Strikingly, their findings revealed that Pol II molecules transiently interact in clusters, with an average life-time of around 5 seconds. This finding is consistent with measured dynamics of GFP-Pol II on mammalian artificial arrays [22]. In addition, global changes in transcription, triggered by serum, alter dramatically the assembly and disassembly of Pol II clusters. This also leads to Pol II foci colocalizing with induction responsive genes, as measured by Immuno-RNA FISH. These findings indicate that the dynamic clustering of Pol II at active genes is a regulated process. Collectively, live-cell imaging studies indicate that factories are dynamic structures that form de novo on genes during transcription (Figure 1, c–d).
Transcription is a physiological process that takes place in an intact living cell. Therefore, live-cell imaging studies present the most biologically compelling analyses of transcription factory biology. Based on these findings we favor a model where transcription factories assemble onto genes with subsequent transient interactions particularly between coregulated and adjacent genes. Self-assembled structures could lead to the formation of high concentrations of factors shared among neighboring genes. Notably, in Drosophila, FRAP studies have shown that during the time course of heat shock Pol II and transcription factors are progressively retained at the Hsp70 loci and are thought to be recycled for subsequent rounds of transcription [26]. One could envision that these factors are recycling between neighboring genes (Figure 2), and there may be cooperative interactions of factors. This could account for chromatin interactions seen by Hi-C that have revealed that genes closer together on chromosomes are more likely to interact [27]. This is further supported by a recent ChIA-PET study, which measured chromatin interactions bound by Pol II and showed clustering of neighboring active genes on chromosomes [28].
Figure 2. Model for local recycling of polymerase and transcription factors among neighboring genes.
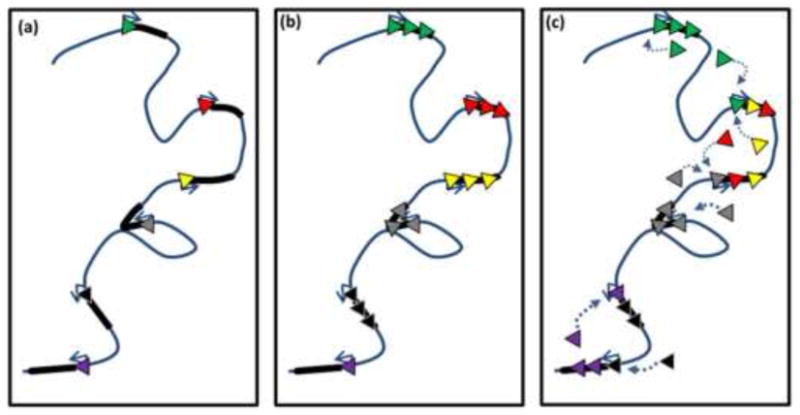
(a) Schematic cartoon of portion of chromosome with six tandem inactive genes containing paused Pol II (multicolor triangles indicate Pol II associated with respective genes). (b) Upon activation of these genes, nucleoplasmic pools of Pol II as well as other transcription factors (for simplicity transcription factors excluded from image) are recruited to genes. (c) As transcription proceeds, the local concentration of polymerases that have completed a round of RNA synthesis builds up around genes. These polymerases can then be shared among neighboring genes to reinitate transcription.
Relating molecular and optical studies of Pol II distributions
Cell biology studies have shown that that the number of Pol II foci in fixed cells varies based on cell type and imaging technique [29]. For example, it has been estimated that Hela cells contain anywhere from 300–8,000 Pol II foci [2,4,30]. These studies used a battery of different imaging approaches that differ in both in their resolution and sensitivity: wide-field microscopy, confocal microscopy and electron microscopy. Therefore, it is possible that these measurements could be in agreement, but are limited by the assays used. The highest resolution images of transcription factories (and thus possibly the best measurement) were obtained by the Cook lab using electron microscopy of cryosectioned human cells, in which they detected approximately 8,000 Pol II foci [30]. Notably, at this time there are no estimates of the total number of Pol II foci using super resolution imaging in living cells.
Another method to visualize nascent transcription complexes is to use molecular approaches such as nuclear run-on assays coupled with massively parallel sequencing to evaluate with high precision genome-wide distribution of transcriptionally engaged polymerases on genes. Such analyses in human cells revealed that 68% of the Pol II transcribed genes (16,882 genes) are expressed in a single cell type [31]. Notably, it would appear then that the number of active genes exceeds the number of Pol II foci found in humans cells. On the surface, these findings would indicate that genes can come together with some frequency in factories. In support this model, there is considerable evidence for spatial clustering of active genes [32]. However, an alternative hypothesis is that factories might be simply resolvable units of Pol II transcription along a largely linear chromosome. In principle, high resolution microscopy techniques such as EM and PALM should be able to resolve Pol II signal at single loci. It would therefore be interesting to correlate genome-wide Pol II signal peaks at comparable sensitivities seen in direct cell observations. Such measurements would also be aided by single cell studies that take into account the fact that transcription does not occur continuously, but as bursts [33] and that not all active genes are expressed at any given moment in every cell. As technologies develop to address these quantitative issues it will be exciting to see how the distribution of Pol II in cells compares molecularly and optically.
Conclusions and future directions
Studies over the past twenty years have provided compelling evidence that Pol II and transcribing genes colocalize at specific foci in cells. However, what has been less understood is how these structures originate. Recent advances in live-cell imaging approaches have allowed the examination of the dynamics of transcription in living cells. These studies are beginning to paint a picture of transcription factories that self-assemble on genes during activation. Given the paucity of live-cell imaging data, additional experiments are required to test this model. For example, it will be interesting to test how general the dynamic nature of Pol II factories are in different species and cell types using a variety of super imaging technologies. In addition, it is still not absolutely clear if the factories move to genes or genes move to factories. This could be tested by examining the dynamics of fluorescently tagged genes (using CRISPR or TALE technology [34,35]) and Pol II in the same cells. We look forward to what will be revealed by such studies and studies yet to be imagined.
Acknowledgments
This work was supported by NIH grant GM25232 to J.T.L.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories’ in human nuclei. J Cell Sci. 1996;109(Pt 6):1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 5.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nature Genetics. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 6.Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morey C, Kress C, Bickmore WA. Lack of bystander activation shows that localization exterior to chromosome territories is not sufficient to up-regulate gene expression. Genome Res. 2009;19:1184–1194. doi: 10.1101/gr.089045.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hieda M, Winstanley H, Maini P, Iborra FJ, Cook PR. Different populations of RNA polymerase II in living mammalian cells. Chromosome Res. 2005;13:135–144. doi: 10.1007/s10577-005-7720-1. [DOI] [PubMed] [Google Scholar]
- 9.Sugaya K, Vigneron M, Cook PR. Mammalian cell lines expressing functional RNA polymerase II tagged with the green fluorescent protein. J Cell Sci. 2000;113(Pt 15):2679–2683. doi: 10.1242/jcs.113.15.2679. [DOI] [PubMed] [Google Scholar]
- 10.Kimura H, Sugaya K, Cook PR. The transcription cycle of RNA polymerase II in living cells. J Cell Biol. 2002;159:777–782. doi: 10.1083/jcb.200206019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341:664–667. doi: 10.1126/science.1239053. The authors report the identification of Pol II foci in living human cells using super resolution microscopy. In addition, this study finds that Pol II molecules transiently interact in foci. This observation refutes the model that Pol II foci are static pre-assembled structures. [DOI] [PubMed] [Google Scholar]
- 12.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 13.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Ghamari A, van de Corput MP, Thongjuea S, van Cappellen WA, van Ijcken W, van Haren J, Soler E, Eick D, Lenhard B, Grosveld FG. In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes Dev. 2013;27:767–777. doi: 10.1101/gad.216200.113. This is the first study to provide evidence that transcription factories are present in living mammalian cells. Specifically, the authors show Cdk9 (a Pol II associated protein) foci in various mouse cell-lines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol. 2008;28:1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou GL, Xin L, Song W, Di LJ, Liu G, Wu XS, Liu DP, Liang CC. Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered housekeeping genes. Mol Cell Biol. 2006;26:5096–5105. doi: 10.1128/MCB.02454-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, et al. Imaging transcription in living cells. Annu Rev Biophys. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker M, Baumann C, John S, Walker DA, Vigneron M, McNally JG, Hager GL. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 2002;3:1188–1194. doi: 10.1093/embo-reports/kvf244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- 25.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 26••.Zobeck KL, Buckley MS, Zipfel WR, Lis JT. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell. 2010;40:965–975. doi: 10.1016/j.molcel.2010.11.022. This study shows that that transcription factors and Pol II are sequentially recruited to the activated Hsp70 loci in living cells. Moreover, they see no movement of genes upon induction. These finding indicate that transcription factors and Pol II move to genes, not vice versa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. The authors provide a genome-wide map of chromatin interactions associated with Pol II using ChIA-PET in five different human cells lines. Their analyses reveals common and cell specific promoter-promoter interactions between neighboring and distant genes on chromosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rieder D, Trajanoski Z, McNally JG. Transcription factories. Front Genet. 2012;3:221. doi: 10.3389/fgene.2012.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pombo A, Jackson DA, Hollinshead M, Wang Z, Roeder RG, Cook PR. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Raj A, van Oudenaarden A. Single-molecule approaches to stochastic gene expression. Annu Rev Biophys. 2009;38:255–270. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 35.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]


