Abstract
The mammalian molecular clock is comprised of a complex network of transcriptional programs that integrate environmental signals with several physiological pathways in a tissue-specific manner. Basic clock features have been elucidated, and emerging technologies are starting to uncover the underlying molecular mechanisms, setting the stage for a ‘systems’ view of the molecular clock. Here we consolidate recent results from genome-wide studies of genetic and epigenomic factors with our understanding of the classic clock mechanism. In addition to its importance in human physiology and disease, the clock mechanism serves as a robust model to uncover or test general principles of dynamic in vivo transcription regulation.
INTRODUCTION
The old adage “you are what you eat” highlights the interconnected relationship between humans and their environment. In addition to the molecules we ingest, we are physically connected to the natural rhythm of the sun, which provides light and heat with a period of 24 hours. This rhythm has shaped everything from human behavior, such as periodic sleeping and eating, to the molecular clocks that exist in most cells of our body. In mammals, peripheral organs including the liver contain molecular clocks that are entrained by fasting-feeding cycles as well as inputs from the brain, which has it own clock controlled by light-dark cycles through the retina-suprachiasmatic nucleus tract situated in the hypothalamus. Molecular clocks coordinate gene expression programs in several physiological systems, allowing cells to preemptively adjust to rhythmic environmental changes. For decades, epidemiological and molecular research have stressed the importance of synchronicity between the 24 hour day and the endogenous molecular clocks, to explain how light pollution and aberrant eating schedules may contribute to disease in industrialized societies1.
The existence of a transcriptionally based eukaryotic circadian clock was discovered in Drosophila as was much of its mechanistic underpinnings2–4. Mammalian clocks, using many of these same principles but far more complicated, have been investigated primarily in mice5,6. In all mammalian tissues, the engine of the molecular clock consists of a transcription-translation feedback loop that is initiated by the basic helix loop helix (bHLH) domain containing transcription factor (TF) BMAL1 (the protein product of Arntl). BMAL1 dimerizes with either Circadian Locomoter Output Cycles Kaput (CLOCK) or Neuronal PAS Domain Protein 2 (NPAS2) bHLH-containing proteins and binds to E-boxes near Period (Per) and Cryptochrome (Cry) during the activation phase. PER/CRY proteins, assembled into a large complexes7, feed back by suppressing BMAL1/CLOCK activity in the repression phase. These proteins ultimately degrade and new BMAL1 complexes re-initiate the activation phase roughly every 24 hours8–10. Additional components of the core clock stimulated by BMAL1/CLOCK include RAR-related Orphan Receptor (ROR) and Reverse of c-erbAα (Rev-erbα), which bind ROR DNA elements (RORE) elements to stimulate or repress transcription, respectively. BMAL1/CLOCK also stimulate the expression of D Site of Albumin Promoter (DBP) and E4 Promoter Binding Protein (E4BP4), which bind to D-box elements in the genome and contribute to the circadian output of the molecular clock.
Recent genome-wide studies have started to unveil the molecular milieu of clock TF binding8,11. Clock TF binding is sequence-specific, and in this context the TFs are direct readers of the genome. However, different regions of the genome are more easily accessible than others due to chromatin and the epigenome (i.e. heterochromatin versus euchromatin), providing additional nodes of transcriptional regulation. Moreover, the ensemble of binding sites at a promoter and associated enhancers underpin the binding energetics of a myriad of tissue-specific TFs that cooperatively bind to regulate transcription. In addition to directly binding DNA, clock factors recruit chromatin-modifying enzymes that alter the local epigenome. Therefore, clock factors, just like TFs, play a fundamental role in bridging the genome with the epigenome.
Clock TF binding motifs
A combination of 3 binding elements -- E-boxes, RORE, and D-boxes -- found near transcriptional starts sites (TSS) and enhancers coordinate core clock TF binding and gene transcription. Additional factors that contribute to entraining the clock to external inputs (light/feeding) include, but are not limited to, humoral factors such as insulin, glucagon and glucocorticoids as wells as TFs such as cAMP response element-binding protein (CREB)12,13. The exact molecular nature of how these external pathways intersect with the core clock mechanism are under active investigation14,15.
Clock factor binding motifs are found throughout the mammalian genome, yet an infinitesimally small proportion is bound by clock TFs underscoring the importance of additional genetic and epigenetic components in the core clock mechanism. Moreover, TFs bind to thousands of sites in the genome, but it is unlikely that each binding event is functional16. Thus, apart from identifying binding sites, additional sources of information are required to define biologically functional cistromes. Functional sites are often enriched with cooperatively binding TFs as well as contain unique epigenetic signatures11,17, highlighting the combinatorial nature of mammalian transcription regulation. Whether epigenetic or genetic mechanisms dominate in this context is very much under intense investigation. It is important to note that biological specificity of TF binding and function is largely dictated by Gibbs free energy, whereby small gains in binding energy through cooperative interactions results in logarithmic increases in affinity. Thus, we favor the view that differential expression of TFs and cofactors underlies much of the tissue specificity of clock factors, and epigenomic elements coordinately contribute to the energetics of cooperative binding.
Genome-wide examination of circadian enhancer RNA (eRNA) expression, which mark active enhancers, by global run-on sequencing (GRO-seq) has identified functional TF cistromes in the mouse liver11. This study showed that enrichment of specific TF motifs in eRNA demarcate different circadian periods. Specifically, the most enriched site at Zeitgeber time 6~9 (ZT; where ZT0 is ‘lights on’ and ZT12 is ‘lights off’), ZT 9~15, ZT 18~24, and ZT 0~3 were found to be E-boxes, D-boxes, RORE/Rev-DR2, and ETS motifs, respectively. This unbiased analysis verified the essential role of TF binding to their motifs in the control of circadian gene transcription. In this section, we will focus on circadian motifs and emerging models of clock TF binding modes.
E-box motif
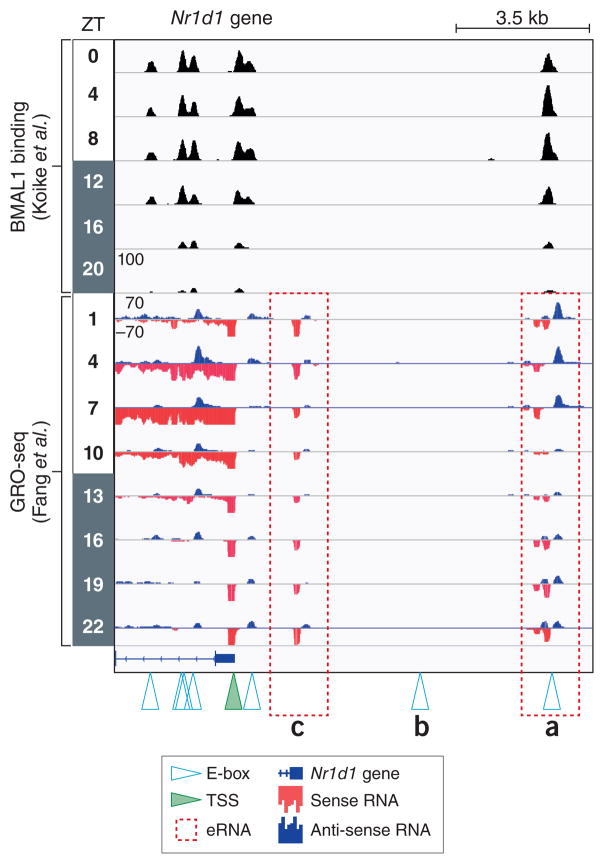
E-box motifs contain a core CANNTG sequence that is recognized by bHLH domain containing TFs. The flanking sequences of E-boxes confer additional levels of specificity to the dozens of bHLH-containing proteins in the mammalian proteome by extending the motif and altering DNA shape18. Its importance to the circadian clock was first appreciated when it was discovered in the promoter of Per in Drosophila19, and later in mammals20. Notably, besides binding to single E-boxes, BMAL1/CLOCK also bind to tandem E-boxes spaced 6 or 7 nucleotides apart with much higher affinity due to cooperative effects, leading to increased recruitment of the transcriptional machinery21. Comparison of the BMAL1 binding and eRNA expression at the Nr1d1 (Rev-erbα) locus shows that not every consensus E-box is bound by BMAL1 (Figure 1a and 1b), indicating its binding is also dictated by the epigenome or interactions with other TFs. Furthermore, BMAL1 target genes may have active enhancers that are not directly bound by BMAL1 (Figure 1c). This, together with the fact that E-box controlled clock targets have variable expression phases8,22 suggest additional higher order levels of clock gene regulation by unknown circadian TFs or epigenomic mechanisms.
Figure 1. The Nr1d1 gene locus.
ChIP-seq data from Koike et al.8 juxtaposed to Global Run-On Sequencing (GRO-Seq) data from Fang et al.11 performed in mouse livers from circadian time points (ZT0 is light on, ZT 12 is lights off). Canonical CACGTG E-box motifs (denoted by open triangles) are bound by BMAL1 with the exception of (b). GRO-seq measures nascent transcription levels of genic and intergenic RNA, including short-lived enhancer RNA (eRNA). Two rhythmically expressed eRNAs (a) and (c) are present near the transcriptional start site (TSS, green triangle) of Nr1d1, but only one (a) is directly bound by BMAL1. Whether these eRNA are linked topologically or whether the eRNA in (c) is independent of BMAL1 activity is not known.
Despite minor differences in E-box preferences among bHLH proteins, several can impinge on BMAL1 binding sites, particularly when their expression levels are altered due to disease23–25. Oncogenic Myc, a bHLH containing protein, directly activates expression of multiple repressors of the clock including Rev-erbα and Rev-erbβ through binding E-boxes at their promoters24. This leads to disruption of circadian BMAL1 oscillation as well as circadian glucose metabolism24. USF1, a bHLH protein, binds to E-box motifs to regulate transcription26 and ChIP analysis has shown that it can bind to enhancers of Dbp, Per1 and Per2. Furthermore, upregulation of USF1 in mice, caused by a genetic variation in the Usf1 promoter sequence, rescues circadian phenotypes in CLOCK mutants25, supporting an activating role for USF1 in E-box mediated transcription.
RORE/RevDR2 motif
The RORE motif is composed of an AT-rich sequence preceding a core motif of PuGGTCA27. It was first discovered as the binding sites for RORs with high specificity27,28, where they function to activate gene transcription29. Later, it was shown that two other members of the nuclear receptor superfamily, Rev-erbα and Rev-erbβ, bind RORE30–32. Rev-erbα not only binds to the RORE as a monomer, but could also function as a homodimer to repress transcription of genes containing a novel sequence of two tandem RORE sites separated by 2 bp (Rev-DR2)33. It has been shown that total body as well as the tissue specific knockout of RORs29,34,35 or Rev-erbs36–39 leads to disruption or phase shift of the molecular clock in various tissues. Indeed, both RORs and Rev-erbs bind to RORE/Rev-DR2 motif in a circadian manner and they work coordinately to maintain the robust circadian expression of core clock genes, such as BMAL1, NPAS2, CRY1, E4BP439.
D-box motif
D-boxes are variants of bZIP motifs and are 9- to 10-bp palindromes composed of two GTAAY half-site sequences. This motif is bound by the proline and acidic amino acid-rich basic leucine zipper (PAR-bZIP) TF family in a phase-specific manner, including DBP, E4BP4, HLF, TEF. All of these TFs are expressed in a circadian manner in several tissues controlled by BMAL1/CLOCK through E-box motifs in their promoters21. DBP, HLF and TEF are transcription activators and are very similar in their primary amino acid sequences, with evidence of redundancy in their function40. Intriguingly, DBP/TEF/HLF triple total body knockout do not affect circadian behaviors and clock gene expression40, suggesting that DBP/HLF/TEF are more likely to control clock output genes instead of the core clock mechanism.
On the other hand, E4BP4 is a transcriptional repressor whose expression is controlled by Rev-erbα through its RevDR2 motif, resulting in expression phase opposite to those of DBP/HLF/TEF. Indeed, the genes and enhancers repressed by Rev-erbα are shared targets of E4BP4 through the D-box motif11. Thus, E4BP4 is thought to control a distinct arm of the clock output that coordinates with DBP/HLF/TEF to switch between the on-off transcriptional states of the target genes in a D-box dependent manner. This regulatory role of E4BP4, however, has yet to be confirmed by genetic loss-of-function or genome-wide ChIP-seq studies.
Models of TF crosstalk at circadian motifs
Competitive binding
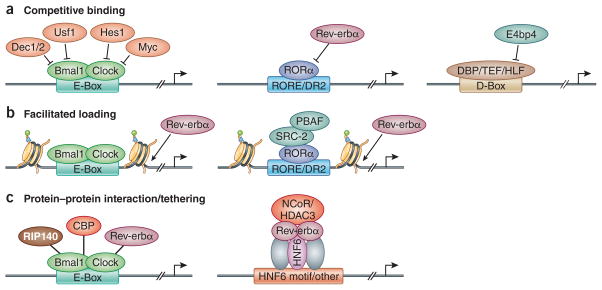
Changes in circadian TF concentrations in the nucleus are major determinants of their binding and transcriptional activity. Moreover, several TFs (activators and repressors) can bind to similar genomic sequences and compete for binding sites near genes they regulate (Figure 2a). As an example, ROR and Rev-erb both bind strongly to RORE sites32. Indeed, although RORα protein levels are nearly constant in the liver29, RORα binding to RORE is very rhythmic and in opposite phase to Rev-erb binding, supporting a competition model dominated by diurnally expressed Rev-erb proteins39. Moreover, knockouts of the ROR co-activator SRC2 or the Rev-erb co-repressors NCOR and HDAC3 lead to much milder changes in BMAL1 expression compared to those of ROR and Rev-erb KO39,41, highlighting that competition between ROR/Rev-erb binding constitutes the major mode of transcriptional regulation, apart from the cofactors they recruit to modify chromatin.
Figure 2. Mechanisms regulating genomic binding of circadian TFs.
(a) Competitive binding among different TFs is observed at E-box, RORE, and D-box motifs. TF binding to the E-box motif, including DEC1/2, USF1, HES1, MYC can inhibit BMAL1 binding. Circadian expressed Rev-erbα competes with RORα, which results in oscillation of RORα binding. E4BP4 competes with other PAR-domain basic leucine zipper TFs DBP/TEF/HLF, which also leads to a robust diurnal switch on/off of their target gene transcription. (b) BMAL1 and CLOCK can function as pioneer TFs to promote the chromatin de-condensation during activation phase. Similarly, RORα could also recruit epigenetic modification machinery to remodel the chromatin structure to facilitate the binding of Rev-erbα. (c) PXDLS peptide motif, which is conserved in CBP, RIP140 and Rev-erbα, can interact with BMAL1/CLOCK and regulate circadian transcription. On the other hand, Rev-erbα can also be tethered to lineage determining TFs, such as HNF6 in the liver, to regulate different metabolic output processes in various tissues.
Competition phenomena have been observed with other TFs. Transcriptional repressors DEC1/2 and activator USF1 can compete for E-boxes with BMAL1/CLOCK25,42, while DBP/HLF/TEF compete with E4BP4 on D-box motifs43. Notably, DNA binding motifs may have different sets of competitors based on slight variations in the motif sequences. For example, at canonical E-boxes, DEC1/2 compete with BMAL1/CLOCK, while at EL-box motifs (an E-box-like motif that contains a N-box motif) HES1 competes with and suppresses BMAL1/CLOCK activity44. Interestingly, the competition model also predicts that other TFs with non-circadian functions can affect clock gene expression, as was shown between c-MYC and BMAL1/CLOCK24. Indeed, clock dysfunction is common to many diseases including cancer and metabolic disorders45, and it remains to be seen whether transcriptional disequilibrium of clock genes is a general marker of disease.
Facilitated Loading
Besides competition, some clock TFs may obtain access to DNA in a cooperative manner that has been termed facilitated loading (Figure 2b). It has been shown that BMAL1 and CLOCK have histone modification activity and can function like pioneer TFs to promote the rhythmic removal of nucleosomes at its binding sites46. This rhythmicity of chromatin opening can potentially lead to (or facilitate) the rhythmic binding of nearby transcription TFs and nuclear receptors, including HNF4, HNF6, CEBPA, STAT5, and Rev-erbs46. Indeed, BMAL1 and RORα can make chromatin more accessible by recruiting PBAF members of the SWI/SNF complex during the activation phase to facilitate the binding of Rev-erbα47. This model, which underscores the importance of the additional TF binding, might also explain the disconnect between BMAL1/CLOCK binding and different transcription initiation rates of their targets.
Protein-Protein Interaction and Tethering
It is well known that circadian TFs recruit chromatin-modifying enzymes through protein-protein interactions. Additionally, clock TFs may also indirectly bind to DNA through protein-protein interactions (Figure 2c), producing functionally distinct outcomes compared with their direct DNA binding activities. For example, Rev-erbα can control the core clock via its DNA binding domain (DBD), but it can also be tethered by lineage determining TFs, such as HNF6, HNF4 and CEBP, to regulate the metabolic output of the core clock in a tissue-specific manner39. In addition, besides CRY and PER, BMAL1 also interacts with proteins containing a short PXDLS peptide motif, which is found in Rev-erbα, RIP140 and CBP48. Indeed, about one third of BMAL1 binding sites are common to the Rev-erbα cistrome, and are found near genes involved in energy homeostasis37, supporting a potential tethering role for BMAL1 at these Rev-erbα targets. There is also potential evidence that BMAL1 may be tethered to chromatin at non-E-box sites8 but this notion has not been directly tested.
Epigenomics of circadian transcription
Mammalian clock models tend to be oversimplified – depicting TFs binding to naked DNA elements upstream of genes. However, in reality, these DNA elements are weaved around nucleosomes and packaged into chromatin. Distinct chromatin states are marked by unique histone post-translational modifications (PTMs). For example, histone H3 lysine 4 methylation (H3K4me) is found in euchromatin and methylation of H3K9, just 5 amino acids away, occurs in heterochromatin. Adding to the complexity, monomethylated H3K4 (H3K4me1) is associated with enhancer elements whereas its trimethylated form (H3K4me3) occurs near transcriptionally active TSS49. Furthermore, the histone variant H2A.Z can be deposited near TSS and enhancers to loosen DNA/nucleosome binding49. Advances in this field have led to an abundance of work highlighting the combinatorial aspect of epigenetic gene regulation, which is also relevant at circadian loci50. Importantly, the histone PTMs discussed in this review and generally studied by researchers constitute a very small proportion of known histone PTMs49, many of which remain under-characterized.
Histone PTMs are thought to function in part by recruiting specific effector complexes that can bind or ‘read’ combinations of PTMs51, akin to TF binding to cognate DNA sequences. Like TFs, multiple histone readers cooperatively bind chromatin through multivalent interactions with histone PTM51. Increasing experimental evidence link the epigenome with the molecular clock. For example, Rev-erbα genomic binding sites vary in different tissues39, which contain the same genomic sequence but vastly different epigenomes and TF expression. Furthermore, several studies have shown that BMAL1/CLOCK binding per se is insufficient to induce transcription, and additional TFs and epigenetic mechanisms are involved22,52–56. In this section we will highlight findings at the intersection of the molecular clock and epigenomics.
Circadian histone acetylation
Histone acetylation is highly rhythmic at clock gene promoters and enhancers. Specifically, H3K27Ac, a marker of active enhancers, and H3K9Ac have been shown to be rhythmic and positively correlated with clock gene expression17,53,57. In fact, it was shown that histone acetylation is more indicative of active gene transcription of Per1/2 and Cry1 genes, which are phase-delayed about 6 to 9 hours, compared to BMAL1/CLOCK binding22. Rhythmic histone acetylation at clock loci is largely mediated by p300 and CBP histone acetyltransferases (HATs)52,53,58. In addition to these HATs, it has been proposed that CLOCK may have intrinsic HAT activity that acetylates histones59. CLOCK, CBP and p300 share many of the same histone targets (e.g. H3K9, H3K14 and histone H4)60, thus their activities may be redundant or, more intriguingly, we speculate they may function at different phases of the clock. For example, CLOCK HAT activity may be important for its pioneer-like activity in acetylating and evicting histones early in the activation phase46,59. Later in the activation phase, p300 or CBP can bind the BMAL1/CLOCK dimer, and acetylate flanking nucleosomes to further loosen chromatin and stimulate transcription52,58,60. Thus, we think that the combined HAT activities of CLOCK, p300 and CBP contribute to robust histone acetylation of chromatin surrounding E-boxes and as yet unknown mechanisms dictate the target specificity and temporal activity of these TFs.
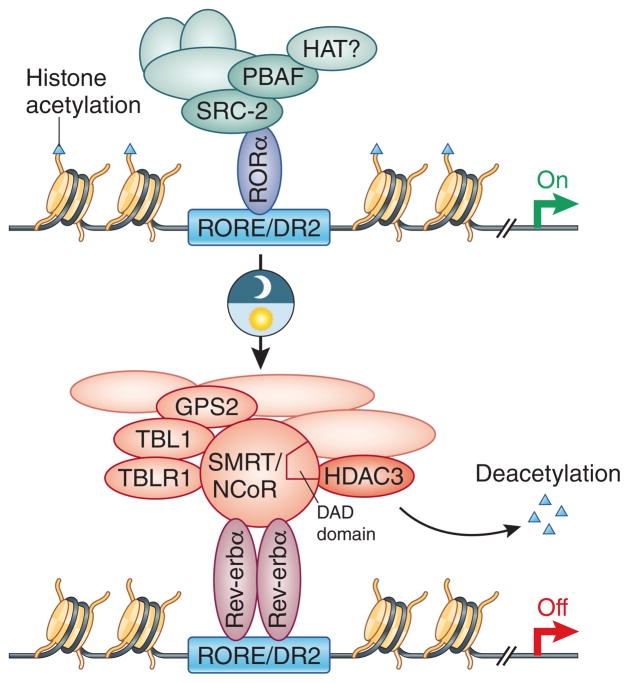
Levels of histone acetylation can be regulated by HATs as well as histone deacetylases (HDAC), which catalytically remove the acetyl modification from lysine residues. Several HDACs have been shown to be important for controlling circadian histone acetylation and pan-specific inhibitors such as trichostatin (TSA) or sodium butyrate increase histone acetylation levels near Per/Cry and alter clock gene expression54,61. HDACs cannot directly bind to DNA and thus they are recruited to chromatin as part of effector complexes (cofactors) that directly interact with transcription factors and other epigenomic factors. For example, Rev-erbα represses transcription in part by recruiting the corepressor complexes N-CoR and/or SMRT to RORE57. A stable member of the N-CoR/SMRT complex is HDAC362, which is required to deacetylate histones near RORE in a circadian manner and affect the epigenomic regulation of clock gene transcription57 (Figure 3).
Figure 3. Rev-erbα and RORα coordinate rhythmic gene expression at RORE/DR2 elements.
Rev-erb represses transcription by two known major mechanisms: 1) Competes with and jettisons RORα and associated co-activators from chromatin; and 2) Recruits N-CoR/SMRT corepressor complexes harboring HDAC3 to deacetylate nearby chromatin and heterochromatin. The deacetylase activation domain (DAD) of N-CoR and SMRT is required to stimulate HDAC3 activity.
One of the major mechanisms of transcriptional repression mediated by CRY and PER is the direct recruitment of the Sin3 complex, which contains HDAC1 and HDAC261,63. Another corepressor complex called NuRD, also contains HDAC1 and HDAC2, binds PER/CRY to deacetylate nearby histones and repress clock genes64. Between the Sin3 and NuRD complexes, there are a myriad protein factors with binding modules that recognize specific histone and DNA modifications including Rbap48 (WD40 repeats), CHD4 (chromodomain and PHD finger), and MBD (methylated DNA reader), but whether they function to ‘read’ the clock epigenome has not been elucidated.
Sirtuins are another class of HDACs involved in the core clock mechanism and levels of their cofactor, nicotinamide adenine dinucleotide, are under tight circadian control in many physiological systems65. Two independent groups found that SIRT1 associates with the BMAL/CLOCK heterodimer to regulate the circadian activity of SIRT1, although they came to opposite conclusions regarding the impact of of SIRT1 on the amplitudes and magnitudes of circadian gene expression66,67. It will be important for additional studies to resolve this controversy.
Thus, HDACs are prolific in the core clock mechanism (Figure 5). Interestingly, although many HDACs seem to target the same histone residues, there appears to be some specificity in their activity since knock down experiments of single HDACs have yielded increases in histone acetylation57,63,67. Therefore, it is tempting to speculate that these HDACs act in concert on different nucleosomes and their net activity contributes to cumulative deacetylation and robust heterochromatin formation of circadian loci. Alternatively, we envisage that HDACs may be part of distinct clock complexes occurring at different genomic sites and that they may also have distinct specificities for acetylated chromatin associated proteins beyond histones.
Circadian histone methylation
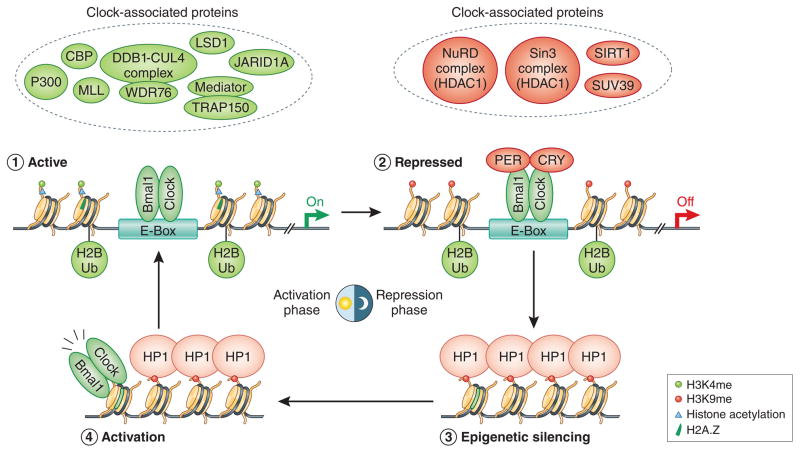
In addition to acetylation, lysine side chains can be methylated by methyltransferases, and often their deacetylation precedes this favoring an acetylation-methylation switch49. In the case of histone H3K9, while acetylation is a strong indicator of euchromatin and active transcription, its methylation promotes heterochromatin formation and transcriptional repression. Rhythmic H3K9 methylation levels near circadian E-boxes are mediated by SUV39 methyltransferase68 and are anti-phase to H3K9 acetylation patterns. Following H3K9 methylation, heterochromatin protein 1 alpha (HP1α) binds H3K9me2 through its chromodomain near E-boxes and mediates rhythmic heterochromatin formation during the repressive phase of the clock53. Intriguingly, clock genes remain repressed well after the degradation of CRY/PER8–10, suggesting that H3K9me/HP1 may serve as epigenetic silencers of clock gene expression until BMAL1/CLOCK re-initiate the cycle53,68 (Figure 5). This mechanism also hints at active circadian H3K9 demethylation upon or before BMAL1/CLOCK binding, but the underlying mechanism is entirely unclear, and identification of a responsible H3K9 demethylase is needed to resolve the molecular events at this critical juncture of the clock. Aside from demethylases, nucleosome-remodeling complexes can also remove modified histones and replace them with unmodified histones or functional variants. Indeed, histone H2A.Z is deposited concurrently with BMAL1/CLOCK binding46 (Figure 5), but whether other histone variants are replaced/deposited is not known and identification of circadian histone chaperones/remodelers that bind BMAL1/CLOCK or even precede their activity is needed.
H3K4 methylation often co-occurs with acetylation of nearby lysines residues within the same nucleosome or histone near transcriptionally active genes and enhancers69, and the clock mechanism is no exception. H3K4me3 levels are dynamically regulated near TSSs of clock genes10,53,70. The MLL family of methyltransferases are responsible for the circadian deposition of H3K4me371–73, with various MLL isoforms having mutually exclusive mechanisms of clock regulation71. In addition to serving as coactivators for BMAL1/CLOCK, MLLs can also act as coactivators for ROR73.
Several putative histone demethylases have been shown to modulate clock gene expression74–77. Curiously, the catalytic activity of these enzymes have been dispensable for their clock dependent activity with the exception of JMJD576, and accordingly none have been shown to target H3K4 methylation. Intriguingly, it was shown that LSD1, which can demethylate H3K4me and H3K9me, is a catalytically-independent coactivator for BMAL1/CLOCK in the clock mechanism77. Thus, H3K4me and H3K9me specific demethylases have yet to be identified for clock genes. Perhaps clues will come from model organisms such as Arabidopsis and Neurospora, where functional clock studies have identified several putative epigenetic modifiers78 including conserved demethylases79.
Circadian histone ubiquitination
Histone H2A monoubiquitination is associated with gene repression and is part of the well-studied Polycomb EZH2-H3K27me3-PRC1 pathway that control heterochromatin formation during cell differentiation and cancer. Interestingly, EZH2 is constitutively bound to BMAL1/CLOCK, but its affect on H3K27me3 around E-boxes is minimal, and whether H3K27me3 levels are circadian near clock genes is unclear80. In contrast, H2B monoubiquitination (H2BUb) levels are rhythmic near circadian E-boxes81 (Figure 4). Ddb1-Cul4 mediated H2BUb occurs late in the activation phase, and is thought to usher in the repression phase of the clock by facilitating the recruitment of the PER/CRY complex to BMAL1/CLOCK bound E-boxes81. Although a different class of E3 ligases was implicated, H2BUb was also shown to occur in plants and found to be important for the repression phase of clock gene transcription82. There are several proteins that can putatively bind or ‘read’ monoubiquitinated H2B83 but none to date have been identified in the PER/CRY repressive complex81. Moreover, the highly conserved SAGA complex has been shown to deubiquitinate H2B from yeast to humans, but whether it has a role in the clock has yet to be determined.
Figure 4. Temporal view of chromatin and clock factors surrounding rhythmic E-boxes.
Recent work has started to unravel the temporal coordination of the molecular clock, including rhythmic changes to histone modifications, histone deposition and chromatin structure. The classical clock proteins (BMAL1/CLOCK/CRY/PER) recruit several chromatin-modifying proteins and complexes that control the epigenome.
Circadian DNA methylation
DNA methylation (DNAme), catalyzed by DNA methyltransferases (DNMT), is a well-studied epigenetic modification with complex roles in gene regulation. In mammals, it predominately occurs on cytosines in cytosine-guanine (CpG) dinucleotides throughout the genome. DNAme patterns are tissue-specific and it is thought that CpG methylation near genes plays a fundamental role in establishing and maintaining cell identity during differentiation. Massive changes in DNAme occur during development and disease, which can take days and weeks to transpire. Coupled with the fact that mechanisms of DNA de-methylation are complex and multifactorial, it is thought that this epigenetic mark is stable and that changes on the order of 24h cycles are unlikely. Studies on circadian DNA methylation in mouse livers and brain cells found no major rhythmic changes, but these analyses focused on broad regions of the genome and rhythmic methylation at single CpG was not interrogated17,84. Nevertheless, there is accumulating evidence for crosstalk between DNA methylation and circadian rhythms, particularly in the brain84,85. DNMT levels are expressed rhythmically in the mouse brain and liver, with some evidence of rhythmic DNA methylation occurring in Line-1 repeat elements86,87. MECP2, a reader of DNA methylation and an important player in Rett syndrome, binds to the N-CoR complex in the brain88. Intriguingly, the canonical E-box motif, CTCGAG, contains a central CpG moiety that can become methylated and we hypothesize influence BMAL1/CLOCK binding. Indeed, CpG methylation alters binding of some bHLH proteins, but this has not be directly tested for BMAL1/CLOCK89. Additionally, whether differential CpG methylation of E-boxes in different tissues and disease contexts regulate BMAL1/CLOCK genomic binding has yet to be determined.
Dynamic chromatin architecture
Recent advances in genome-wide chromatin mapping technologies have stimulated a new appreciation for the 3D architecture of chromatin, and its critical contributions to long-distance cis-acting mechanisms of gene regulation. For example, they have provided genome-wide and mechanistic proof that distal elements such as enhancers physically “loop” near the TSSs they regulate. Additionally, these methods have uncovered the presence of subnuclear chromatin compartments or zones where gene expression is uniformly regulated - e.g. the nuclear periphery is largely repressive for transcription and genes recruited there during differentiation are epigenetically silenced90. Studies in cultured cells have shown that these modes of regulation are also apparent in circadian systems91–94. The Dbp gene loops to over 200 long-range sites in the genome in a BMAL1-dependent manner91. Several looping factors, such as members of the Mediator complex, form interactions with core clock TFs9,95. Deletion of one of these factors important for looping, Smc3, causes major disruptions to the clock94. Interestingly, the clock machinery also interacts with the nuclear envelope in a circadian manner suggesting that translocation of clock genes to the nuclear periphery may constitute a general silencing mechanism for clock controlled gene. Support for this model comes from other studies showing that chromatin gains H3K9me2 levels as it translocates from the center of the nucleus to the periphery96, and this mechanism may underlie the circadian H3K9me2 levels detected in clock loci53. Thus, there is strong indication in vitro that chromatin structure changes in a circadian manner.
Concluding remarks
Over the past few years an explosion of new genome-wide technologies have provided unprecedented access to the intracellular molecular clock. The basic model of the clock has withstood much experimental scrutiny, with recently added molecular and mechanistic details that have enriched our understanding of the physiological clock. Studies in Neurospora and cyanobacteria have also bolstered the non- and post-transcriptional regulation of the molecular clock, including rhythmic peroxiredoxin cycling97. Still, there is a great need to integrate and balance ever-growing genome-wide datasets with biochemical, molecular and genetic approaches. Additionally, most research has focused on the liver and brain clocks, but with increasing sensitivity of sequencing technologies including single-cell capabilities we anticipate a more thorough understanding of cellular clocks throughout the body in the coming years. Given the robustness and synchronicity of the molecular clock, we are confident that promising avenues of research including CRISPR technology, which is already being used to manipulate the clock98, and chromatin capture techniques will be amenable in in vivo settings. Beyond an academic issue, these techniques will no doubt provide therapeutic insights to the expanding list of circadian disorders and circadian GWAS studies in human populations99,100.
The study of clock TFs transcends many scientific fields, providing meaningful opportunities in vivo to address several fundamental questions in epigenomic and transcription regulation. For example, the mechanisms that confer functionality at a subset of TF binding sites are largely unknown. The discovery of molecular rules that dictate why BMAL1/CLOCK are essential to the clock but play ancillary roles at thousands of tissue-specific sites may provide exciting clues to this end. Moreover, the study of BMAL1/CLOCK activity during the early activation phase may elucidate how pioneer TFs, which can theoretically access hundreds of thousands of binding sites in the mammalian genome, home in on specific genomic regions. Indeed, regulation of chromatin structure may inform many of these open-ended questions, thus the study of epigenomic factors near circadian DNA motifs may shed light on the key determinants of transcription regulation.
Acknowledgments
We thank members of the Lazar lab for helpful discussions. Work on circadian rhythms in the Lazar lab is supported by NIH R01 DK45586 and the JBP foundation, and RP is supported by NIH F32 DK108555.
References
- 1.Bedrosian Ta, Fonken LK, Nelson RJ. Endocrine Effects of Circadian Disruption. Annu Rev Physiol. 2015;78:150724172241001. doi: 10.1146/annurev-physiol-021115-105102. [DOI] [PubMed] [Google Scholar]
- 2.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–6. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price JL, et al. double-time is a novel Drosophila clock gene that regulates period protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 4.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 5.King DP, et al. Positional Cloning of the Mouse Circadian Clock Gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 7.Kim JY, Kwak PB, Gebert M, Duong HA, Weitz CJ. Purification and analysis of PERIOD protein complexes of the mammalian circadian clock. Methods Enzymol. 2015;551:197–210. doi: 10.1016/bs.mie.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–54. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lande-Diner L, Boyault C, Kim JY, Weitz CJ. A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc Natl Acad Sci U S A. 2013;110:16021–6. doi: 10.1073/pnas.1305980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Martelot G, et al. Genome-Wide RNA Polymerase II Profiles and RNA Accumulation Reveal Kinetics of Transcription and Associated Epigenetic Changes During Diurnal Cycles. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang B, et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159:1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 13.Yagita K, Okamura H. Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett. 2000;465:79–82. doi: 10.1016/s0014-5793(99)01724-x. [DOI] [PubMed] [Google Scholar]
- 14.Brown SA. Circadian Metabolism: From Mechanisms to Metabolomics and Medicine. Trends in Endocrinology and Metabolism. 2016 doi: 10.1016/j.tem.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 15.O’Neill JS, Reddy AB. The essential role of cAMP/Ca2+ signalling in mammalian circadian timekeeping. Biochem Soc Trans. 2012;40:44–50. doi: 10.1042/BST20110691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitfield TW, et al. Functional analysis of transcription factor binding sites in human promoters. Genome Biol. 2012;13:R50–R50. doi: 10.1186/gb-2012-13-9-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vollmers C, et al. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gord??n R, et al. Genomic Regions Flanking E-Box Binding Sites Influence DNA Binding Specificity of bHLH Transcription Factors through DNA Shape. Cell Rep. 2013;3:1093–1104. doi: 10.1016/j.celrep.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao H, Allen DL, Hardin PE. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17:3687–93. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998;95:5474–9. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C, Etchegaray JP, Cagampang FRA, Loudon ASI, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 23.Xiong W, Li J, Zhang E, Huang H. BMAL1 regulates transcription initiation and activates circadian clock gene expression in mammals. Biochem Biophys Res Commun. 2016;473:1019–25. doi: 10.1016/j.bbrc.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Altman BJ, et al. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metab. 2015;22:1009–1019. doi: 10.1016/j.cmet.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimomura K, et al. Usf1, a suppressor of the circadian Clock mutant, reveals the nature of the DNA-binding of the CLOCK:BMAL1 complex in mice. Elife. 2013;2013 doi: 10.7554/eLife.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferré-D’Amaré AR PP, Roeder RGBS. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giguère V, et al. Isoform-specific amino-terminal domains dictate DNA-binding properties of RORα a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 28.Giguère V, McBroom LD, Flock G. Determinants of target gene specificity for ROR alpha 1: monomeric DNA binding by an orphan nuclear receptor. Mol Cell Biol. 1995;15:2517–26. doi: 10.1128/mcb.15.5.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeda Y, Jothi R, Birault V, Jetten AM. ROR?? directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res. 2012;40:8519–8535. doi: 10.1093/nar/gks630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dumas B, et al. A new orphan member of the nuclear hormone receptor superfamily closely related to Rev-Erb. Mol Endocrinol. 1994;8:996–1005. doi: 10.1210/mend.8.8.7997240. [DOI] [PubMed] [Google Scholar]
- 31.Harding HP, Lazar MA. The orphan receptor Rev-ErbA alpha activates transcription via a novel response element. Mol Cell Biol. 1993;13:3113–21. doi: 10.1128/mcb.13.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forman BM, et al. Cross-talk among ROR alpha 1 and the Rev-erb family of orphan nuclear receptors. Mol Endocrinol. 1994;8:1253–61. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- 33.Harding HP, Lazar MA. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol Cell Biol. 1995;15:4791–802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 35.Sato TK, et al. A functional genomics strategy reveals rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 36.Woldt E, et al. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19:1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bugge A, et al. Rev-erb alpha and Rev-erb beta coordinately protect the circadian clock and normal metabolic function. GENES Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science. 2015;348:1488–92. doi: 10.1126/science.aab3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gachon F, et al. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004;18:1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stashi E, et al. SRC-2 Is an Essential Coactivator for Orchestrating Metabolism and Circadian Rhythm. Cell Rep. 2014;6:633–645. doi: 10.1016/j.celrep.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honma S, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 43.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueshima T, et al. Identification of a new clock-related element EL-box involved in circadian regulation by BMAL1/CLOCK and HES1. Gene. 2012;510:118–125. doi: 10.1016/j.gene.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 45.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 46.Menet JS, Pescatore S, Rosbash M. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev. 2014;28:8–13. doi: 10.1101/gad.228536.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu B, et al. Coactivator-Dependent Oscillation of Chromatin Accessibility Dictates Circadian Gene Amplitude via REV-ERB Loading. Mol Cell. 2015;60:769–783. doi: 10.1016/j.molcel.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shalev M, et al. The PXDLS linear motif regulates circadian rhythmicity through protein-protein interactions. Nucleic Acids Res. 2014;42:11879–11890. doi: 10.1093/nar/gku873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soshnev AA, Josefowicz SZ, Allis CD. Greater Than the Sum of Parts: Complexity of the Dynamic Epigenome. Mol Cell. 2016;62:681–694. doi: 10.1016/j.molcel.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown Sa. A New Histone Code for Clocks? Science (80- ) 2011;333:1833–1834. doi: 10.1126/science.1212842. [DOI] [PubMed] [Google Scholar]
- 51.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct & Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Etchegaray J, Lee C, Wade Pa, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 53.Ripperger Ja, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 54.Stratmann M, Suter DM, Molina N, Naef F, Schibler U. Circadian Dbp Transcription Relies on Highly Dynamic BMAL1-CLOCK Interaction with E Boxes and Requires the Proteasome. Mol Cell. 2012;48:277–287. doi: 10.1016/j.molcel.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 55.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012;2012 doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ukai-Tadenuma M, et al. Delay in feedback repression by cryptochrome 1 Is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 57.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee Y, et al. Coactivation of the CLOCK-BMAL1 complex by CBP mediates resetting of the circadian clock. J Cell Sci. 2010;123:3547–3557. doi: 10.1242/jcs.070300. [DOI] [PubMed] [Google Scholar]
- 59.Doi M, Hirayama J, Sassone-Corsi P. Circadian Regulator CLOCK Is a Histone Acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 60.Curtis AM, et al. Histone Acetyltransferase-dependent Chromatin Remodeling and the Vascular Clock. J Biol Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- 61.Naruse Y, et al. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol Cell Biol. 2004;24:6278–87. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oberoi J, et al. Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nat Struct Mol Biol. 2011;18:177–184. doi: 10.1038/nsmb.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duong Ha, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011;332:1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim JY, Kwak PB, Weitz CJ. Specificity in Circadian Clock Feedback from Targeted Reconstitution of the NuRD Corepressor. Mol Cell. 2014;56:738–748. doi: 10.1016/j.molcel.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 65.Asher G, Sassone-Corsi P. Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161:84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 66.Asher G, et al. SIRT1 Regulates Circadian Clock Gene Expression through PER2 Deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 67.Nakahata Y, et al. The NAD+-Dependent Deacetylase SIRT1 Modulates CLOCK-Mediated Chromatin Remodeling and Circadian Control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duong HA, Weitz CJ. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol. 2014;21:126–132. doi: 10.1038/nsmb.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Young NL, et al. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malapeira J, Khaitova LC, Mas P. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc Natl Acad Sci U S A. 2012;109:21540–5. doi: 10.1073/pnas.1217022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valekunja UK, et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci U S A. 2013;110:1554–9. doi: 10.1073/pnas.1214168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17:1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim DH, et al. Crucial roles of mixed-lineage leukemia 3 and 4 as epigenetic switches of the hepatic circadian clock controlling bile acid homeostasis in mice. Hepatology. 2015;61:1012–1023. doi: 10.1002/hep.27578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DiTacchio L, et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–5. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reischl S, Kramer A. Fbxl11 Is a Novel Negative Element of the Mammalian Circadian Clock. J Biol Rhythms. 2015;XX:1–11. doi: 10.1177/0748730415587407. [DOI] [PubMed] [Google Scholar]
- 76.Jones Ma, et al. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci U S A. 2010;107:21623–21628. doi: 10.1073/pnas.1014204108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nam H, et al. Phosphorylation of LSD1 by PKCα Is Crucial for Circadian Rhythmicity and Phase Resetting. Mol Cell. 2014;53:791–805. doi: 10.1016/j.molcel.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 78.Wang B, Kettenbach AN, Gerber SA, Loros JJ, Dunlap JC. Neurospora WC-1 Recruits SWI/SNF to Remodel frequency and Initiate a Circadian Cycle. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee HG, Lee K, Jang K, Seo PJ. Circadian expression profiles of chromatin remodeling factor genes in Arabidopsis. J Plant Res. 2014;128:187–199. doi: 10.1007/s10265-014-0665-8. [DOI] [PubMed] [Google Scholar]
- 80.Etchegaray JP, et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 81.Tamayo AG, Duong HA, Robles MS, Mann M, Weitz CJ. Histone monoubiquitination by Clock–Bmal1 complex marks Per1 and Per2 genes for circadian feedback. Nat Struct Mol Biol. 2015;22:759–66. doi: 10.1038/nsmb.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Himanen K, et al. Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J. 2012;72:249–260. doi: 10.1111/j.1365-313X.2012.05071.x. [DOI] [PubMed] [Google Scholar]
- 83.Fuchs G, Oren M. Writing and reading H2B monoubiquitylation. Biochimica et Biophysica Acta - Gene Regulatory Mechanisms. 2014;1839:694–701. doi: 10.1016/j.bbagrm.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 84.Azzi A, et al. Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat Neurosci. 2014;17:377–382. doi: 10.1038/nn.3651. [DOI] [PubMed] [Google Scholar]
- 85.Lim ASP, et al. 24-Hour Rhythms of DNA Methylation and Their Relation with Rhythms of RNA Expression in the Human Dorsolateral Prefrontal Cortex. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004792. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Xia L, et al. Daily variation in global and local DNA methylation in mouse livers. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maekawa F, et al. Diurnal expression of Dnmt3b mRNA in mouse liver is regulated by feeding and hepatic clockwork. Epigenetics. 2012;7:1046–1056. doi: 10.4161/epi.21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lyst MJ, et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci. 2013;16:898–902. doi: 10.1038/nn.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu S, et al. DNA methylation presents distinct binding sites for human transcription factors. Elife. 2013;2013 doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bickmore WA, Van Steensel B. Genome architecture: Domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 91.Aguilar-Arnal L, et al. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat Struct Mol Biol. 2013;20:1206–13. doi: 10.1038/nsmb.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin ST, et al. Nuclear envelope protein MAN1 regulates clock through BMAL1. Elife. 2014;3:e02981. doi: 10.7554/eLife.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao H, et al. PARP1- and CTCF-Mediated Interactions between Active and Repressed Chromatin at the Lamina Promote Oscillating Transcription. Mol Cell. 2015;59:984–997. doi: 10.1016/j.molcel.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 94.Xu Y, et al. Long-Range Chromosome Interactions Mediated by Cohesin Shape Circadian Gene Expression. PLoS Genet. 2016;12:e1005992. doi: 10.1371/journal.pgen.1005992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lehmann R, et al. Assembly of a comprehensive regulatory network for the mammalian circadian clock: A bioinformatics approach. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Towbin BD, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 97.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Korge S, Grudziecki A, Kramer A. Highly Efficient Genome Editing via CRISPR/Cas9 to Create Clock Gene Knockout Cells. J Biol Rhythms. 2015;XX:1–7. doi: 10.1177/0748730415597519. [DOI] [PubMed] [Google Scholar]
- 99.Hu Y, et al. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat Commun. 2016;7:10448. doi: 10.1038/ncomms10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qian J, Scheer FAJL. Circadian System and Glucose Metabolism: Implications for Physiology and Disease. Trends in Endocrinology and Metabolism. 2016;27:282–293. doi: 10.1016/j.tem.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]