Abstract
The mammalian cerebral cortex is a complex brain structure integral to our higher cognition. During embryonic cortical development, radial glial progenitors (RGCs) produce neurons and serve as physical structures for migrating neurons. Recent discoveries highlight new roles for RNA localization and local translation in RGCs, both at the cell body and at distal structures called basal endfeet. By implementing technologies from the field of RNA research to brain development, investigators can manipulate RNA-binding proteins as well as visualize single-molecule RNAs, live movement of mRNAs and their binding proteins, and translation. Going forward, these studies establish a framework for investigating how post-transcriptional RNA regulation helps shape RGC function and triggers neurodevelopmental diseases.
Keywords: neurogenesis, RNA localization, translation
The cerebral cortex is a highly organized structure, containing the neurons responsible for our higher cognitive functions. Its development is orchestrated by a vast array of molecular and cellular mechanisms at play in multiple cell types. The adult neocortex is composed of about 80% excitatory neurons and 20% inhibitory neurons, both of which are produced via radial glial cells (RGCs). These radial glial cells are commonly called neural stem cells, for their ability to generate not only neurons but also glia [1,2]. At initial stages of cortical development, the brain is composed of neuroepithelial progenitors which divide symmetrically to expand the precursor pool. As cortical development proceeds, these are replaced by RGCs which sequentially generate subtypes of excitatory neurons (Fig. 1A–C). RGCs produce neurons directly or indirectly by generating transit amplifying progenitors. In mice, the predominant transit progenitors are intermediate progenitors (IPs), whereas in human and nonhuman primates, outer radial glia progenitors are abundant [3–5]. Newly generated excitatory neurons migrate radially toward the pia, forming distinct layers II–VI.
Fig. 1.
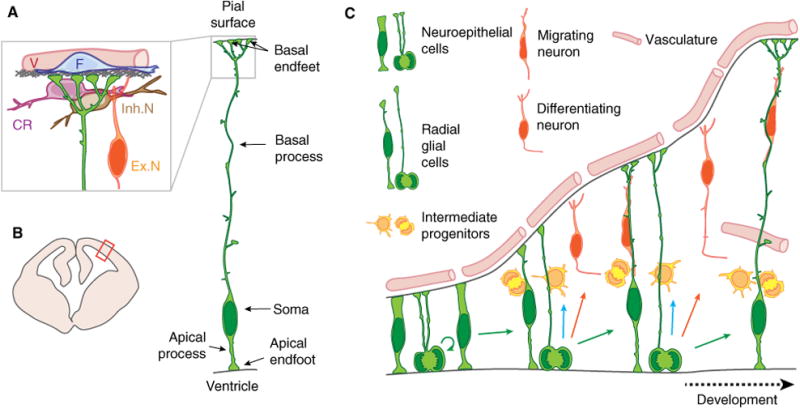
Cartoon of developing brain and neural stem cells with highlighted anatomy of a radial glial progenitor cell. (A) Anatomy of a radial glial progenitor and the endfoot niche, including the basement membrane (gray), inhibitory neurons (Inh.N), excitatory neurons (Ex.N), cajal retzius cells (CR), vasculature (V), and fibroblasts (F). (B) Schematic representation of a coronal section of an embryonic mouse brain during midcorticogenesis. Red box points to the location represented in (A). (C) Cartoon representation of mouse cortical development. This panel shows the different cell types referred to in the present paper. During early corticogenesis, neuroepithelial cells divide symmetrically to expand the precursor pool. As development proceeds, neuroepithelial cells convert into radial glial cells that mainly divide asymmetrically to produce a new RGC and either a neuron or an IPs. IPs divide away from the ventricular border to generate neurons.
The term RGC was first coined based upon the unique morphology of these cells which resemble glial cells with long radial extensions (Fig. 1A). Extensive electron microscopy (EM) studies were invaluable for describing RGC anatomy, revealing a cell body adjacent to the ventricle, and apical endfeet [6,7]. Extending from their cell bodies, RGCs have a basal process that spans the entire thickness of the developing cerebral cortex, forming basal endfeet at the pia (Fig. 1A). In younger brains the basal process is quite short, whereas in older brains it can extend up to several hundred micrometers; and even millimeters in human brains. This structure provides a guidepost for excitatory neurons to migrate from their birthplace in the germinal zones to their final destination in the cortical plate. EM studies together with immunolabeling reveal that organelles are subcellularly localized within RGCs —with Golgi found only within the cell body, and endoplasmic reticulum (ER) distributed throughout the cell body and radial fibers [8]. RGC cell bodies form distinct interconnected clusters linked by Gap junctions, through which calcium signaling can propagate [9,10]. Cell–cell interactions between RGC cell bodies and newborn IPs and neurons can enable signaling such as via the Notch pathway [11].
Following the discovery that RGCs are neuronal and glial precursors, the hypothesis emerged that asymmetric segregation of determinants within apical and basal structures could influence whether RGCs undergo symmetric or asymmetric divisions [12,13]. Local absence of cadherin staining at apical RGC structures (termed cadherin hole) was implicated during asymmetric RGC divisions [14]. Additionally, asymmetric inheritance of cell fate markers, such as Numb, Notch, and EGFR, has also been observed [13,15]. Live imaging of dividing RGCs reveal the basal process itself may influence cell fate, as new cells that inherit the basal process disproportionately retain RGC proliferative behavior [16–18]. This led to the notion that the basal process, in addition to the cell body, could contain asymmetrically segregated fate determinants. Recent studies further indicate that asymmetric RGC fate can be influenced by exogenous signals, such as from the choroid plexus residing within the lateral ventricles [19].
At the pial cortical surface, RGC basal endfeet are tethered to the basal membrane and form a barrier between the cerebral cortex and the overlying meninges. Disruption of this barrier leads to overmigration of cortical neurons into the meninges [20,21]. It is important to note these basal RGC structures reside in a local niche composed of interneurons, excitatory neurons, a basement membrane, and outside the cortex, vasculature, and fibroblasts [22] (Fig. 1A). Notably, signals from meninges as well as upper layer neurons can influence RGC proliferation [23–28]. Moreover, basal process and endfeet structures are not simply passive borders, as live imaging reveals dynamic remodeling within the niche [29]. However, there is a dearth of understanding regarding how information is communicated between RGC basal structures and niche cells.
The morphological and cell biological features of RGCs make them well suited for RNA localization and local translation. Because apical cell bodies and basal endfeet can be separated by hundreds of microns, local control of gene expression in both compartments could mediate RGC functions in cell fate specification, neuronal migration, and signaling. In Saccharomyces cerevisiae, Xenopus, and Drosophila, inheritance of asymmetrically localized mRNAs is associated with cell fate specification [30,31]. In migrating fibroblasts, β-actin mRNA localized to the leading migratory edge allows for local and rapid remodeling of the cytoskeleton [32]. Similarly, in newborn neurons, local translation at the growth cone promotes asymmetric axon growth in response to extracellular cues, and in dendritic spines, local control of gene expresssion modulates spine growth in response to local synaptic activity [30,31]. Thus, in many systems, mRNA localization and local translation promote cell fate, cytoskeletal remodeling, and rapid cellular responses.
In this review we highlight emerging roles for mRNA localization and local translation in mammalian RGCs. We first discuss relevant RNA and developmental technologies for interrogating these processes in RGCs. We then describe RNA-binding proteins (RBPs) and mRNAs implicated in localization and translation within the cell body and basal RGC structures. Finally, we discuss how these new discoveries are beginning to influence and challenge our current understanding of neurogenesis.
Technologies to study RNA localization and translation in mammalian RGCs
RNA technologies
A traditional approach to visualize RNA localization within cells or tissues is fluorescent in situ hybridization (FISH). In contrast to conventional in situ hybridization techniques, recent developments in FISH technology enable higher resolution detection of mRNAs, with the ability to resolve and quantify single mRNA molecules (smFISH) [33]. This approach can be coupled with other fluorescent labeling, such as antibody staining or expression of exogenous fluorescent proteins, to interrogate colocalization of RNA with other molecules and within specific subcellular compartments [34–36].
One of the most highly utilized approaches for visualizing RNA transcripts in live cells is the MS2 system made popular by Robert Singer's group (Fig. 2A). Briefly, MS2 stem loop sequences are included within the localization element of an RNA of interest. This can be achieved by transfecting reagents (DNA plasmids or viruses) or by generating knockin mice [37,38]. Cells must also express the fluorescent fusion protein, EGFP-MCP (MS2 Coat Protein), which has very high affinity for MS2 stem loops and contains a nuclear localization element to minimize background cytoplasmic signal. With this strategy RNAs become decorated with multiple EGFP proteins, enabling the indirect detection of transcripts. A variant of this technique, the PP7 system, also uses RNA stem loop and coat proteins (a forthcoming Review by Kiebler, due to be published in the same issue as this article, addresses this topic) [30]. Combining both MS2 and PP7 systems makes it possible to visualize the dynamics of multiple RNA species simultaneously within a live cell. Use of these RNA imaging approaches have been deployed in many systems including cultured neurons, fibroblasts, zebrafish, and most recently in organotypic embryonic mammalian brain slices [30,34,37–40]. Imaging and labeling technologies continue to be improved, such as with application of CRISPR/Cas9 technology [41].
Fig. 2.
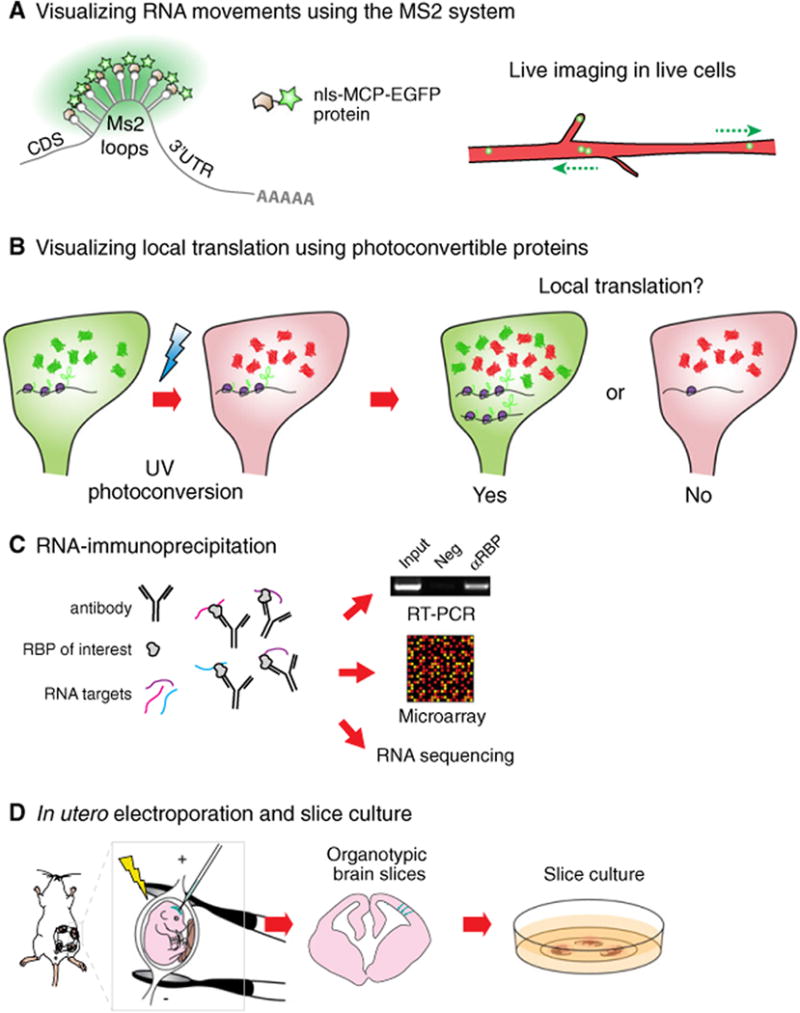
Tools used to study RNA localization and local translation in the developing mouse cortex. (A) In the Ms2 system, DNA constructs expressing an RNA bearing Ms2 loops and a localization sequence are introduced into cells together with Ms2-loop coat protein fused to EGFP (MCP-EGFP). A nuclear localization signal (nls) is added to the MCP-EGFP to reduce background signal of unbound protein in the cytoplasm, and ensure that local events outside the nucleus are actively controlled. RNAs can harbor 24 EGFP molecules, which is sufficient to detect single transcripts. (B) Photoconvertible proteins such as DENDRA2 or KAEDE are valuable to visualize local translation in a distal region of a cell. Reagents expressing RNAs bearing the protein expression cassette together with a localization signal are introduced into cells. After photoconversion, local recovery of the green signal over time indicates local translation (YES), whereas no new green signal suggests the absence of local translation (NO). (C) RNA-immunoprecipitation allows for the identification of RNA targets of RBPs. RNAs pulled down using an antibody targeting a candidate RBP can be analyzed using RT-PCR, microarray or RNA sequencing analyses. This approach has many modifications including crosslinking. (D) In utero electroporation allows for the transfection of RGCs in vivo. Briefly, nucleic acids or proteins are injected into the lateral ventricle of embryonic brains. An electric current is applied to make cell membranes permeable and to direct nucleic acids or proteins into the cells lining the ventricular border. Following electroporation, organotypic brain slices can be cultured for up to several days and subsequently used for live imaging of electroporated cells.
A number of techniques enable visualization of protein translation in cellular compartments. A powerful and flexible approach is the use of photoconvertible proteins coupled with live imaging (Fig. 2B). With this methodology, constructs expressing either Dendra or Kaede coding sequence and containing an RNA localization sequence are introduced into cells [42]. Native Dendra or Kaede proteins fluoresce in the green color spectrum, however, following exposure to UV, are irreversibly converted to red fluorescence. Thus, after conversion, the recovery of a new localized green signal is indicative of de novo protein translation. Given that green photoconvertible proteins can also diffuse from other subcellular locations, it is routine to use a form engineered to associate with the cell membrane to delay diffusion. Alternatively, this experiment can be conducted after physical isolation of the cellular compartment in which local translation is being measured [34,42,43]. Beyond photoconvertible proteins, additional techniques exist for monitoring translation, including fluorescent labeling of ribosomal components, nascent proteins (SINAPS), and use of modified amino acids (BONCAT, FUNCAT) [44–48]. Genome-wide translation analyses can be employed using the latter approaches or ribotag approaches [49–51]. These cutting-edge tools continue to be optimized for use in vivo.
Identification of RNA targets of RBPs within a given cell type or tissue relies upon an approach called RNA-immunoprecipitation (RIP) [52]. This technique utilizes an antibody against an RBP of interest to isolate RNA–RBP complexes. Purified RNAs can then be identified by qPCR, microarray (RIP-CHIP) or RNA sequencing (RIP-seq) (Fig. 2C). Modifications to traditional RIPs employ cross-linking to isolate both low- and high-affinity RNA–protein interactions, since traditional RIPs are thought to primarily detect high-affinity interactions. This includes CLIP, PAR-CLIP, eCLIP, DO-RIP, and HITS-CLIP, the latter two of which can be used in vivo [53–56]. Both traditional and cross-linking-based RIP techniques have been criticized for potentially yielding false positive discoveries due to reassortment of RNA–RBP complexes or nonspecific binding. It is critical to validate findings from genomic studies using additional candidate approaches as described above.
Applications to the brain
The embryonic mammalian cortex is well suited for the application of these RNA technologies. In utero electroporation is invaluable for transfecting nucleic acids and proteins into RGCs in vivo (Fig. 2D) [57]. Using this technique one can modulate gene expression by overexpression or knockdown, as well as label proteins [58]. Of note, as RGCs give rise to neurons, astrocytes and oligodendrocytes in different regions and at distinct developmental stages, it is possible to target specific populations by simply adjusting electroporation parameters. Thus, one can evaluate localization, transport, local translation, and local transcriptomes by electroporating fluorescently tagged molecules or fusion proteins. Such tags can be used for subsequent biochemical use, as for cell-specific RIPs [34].
Another valuable technique for imaging RNA transport and translation is embryonic organotypic brain slices which have the amazing capacity to be cultured ex vivo for several days. Developmental mechanisms, such as RGC proliferation or neuronal migration, are maintained in this ex vivo setting. Coupled with in utero electroporation, this technique reaches its full potential when one performs live imaging with slices generated from electroporated brains (Fig. 2D). Thus, it is possible to assess the behavior of either RNAs or RBPs. As their transport can reach speeds of several microns per second, high temporal and spatial resolution are essential for such assays. This can be achieved using state of the art microscopy approaches such as spinning-disk microscopy with high-resolution objectives.
RNA localization and translation in the RGC cell body
Localization: Staufen2 and its targets
Asymmetric cell division is a hallmark of RGCs during the peak stages of neuron production at midcorticogenesis [17,59–61]. Fundamentally, asymmetric divisions rely upon the unequal segregation of fate determinants in two daughter cells during mitosis [62]. For example, RGCs asymmetrically segregate fate determinants Notch and EGFR, although the functional impact of this is poorly understood [12,15]. Recent studies highlight RNA and RBPs which are subcellularly localized in the cell body to impact asymmetric cell fate.
In back-to-back studies, the labs of Sally Temple and Freda Miller discovered that the RBP STAU-FEN2 (STAU2) controls asymmetric mRNA segregation during RGC mitosis (Fig. 3A) [63,64]. STAU2 is expressed in RGCs throughout corticogenesis, and enriched at the ventricular border within apical RGC endfeet. During prophase and metaphase stages of mitosis, STAU2 accumulates asymmetrically in RGCs. During neurogenic phases, this results in asymmetric distribution to one daughter cell, particularly a newborn IP. Both in vitro and in vivo experiments using shRNA showed Stau2 depletion in RGCs increases differentiation and decreases self-renewal. Importantly, this phenotype is not rescued by expression of an RNA-binding deficient STAU2, indicating STAU2 impacts RGC cell fate by binding RNA. Both studies tested this interesting hypothesis. Vessey et al. [63] drew clues from Drosophila neuroblasts [65], whose asymmetric division relies upon staufen-mediated segregation of prospero mRNA. In the developing mouse, they found that STAU2 binds Prox, the mammalian Prospero ortholog, and that along with STAU2, Prox is asymmetrically segregated in mitotic RGCs during the neurogenic period. Kusek et al. [64] used RIP-CHIP to discover transcriptome-wide STAU2 targets in whole embryonic cortices. Although this tissue contains various cell types, the authors successfully identified about 1500 STAU2-bound mRNAs. Notably, for several targets, Stau2 depletion disrupted their RNA localization in dividing RGCs and subsequent protein distribution in progeny.
Fig. 3.
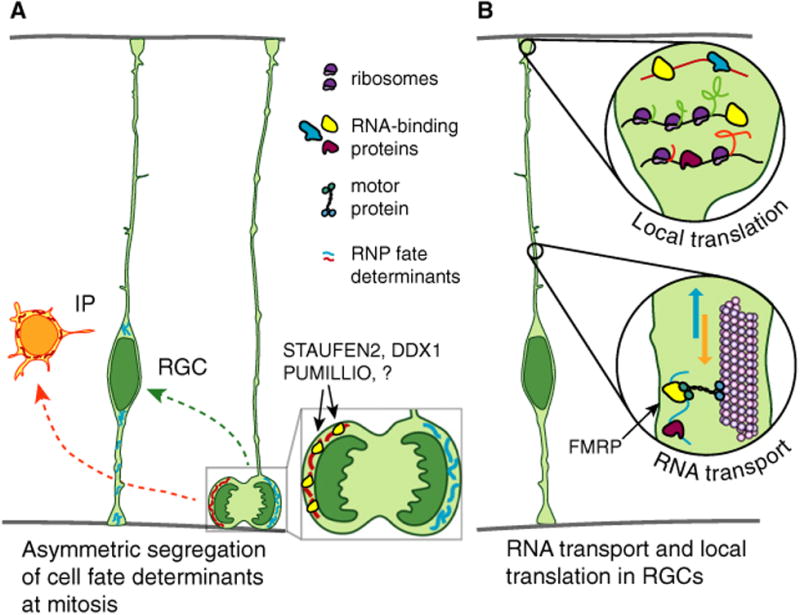
Cartoon representing dynamics and putative roles of RNA localization and regulated translation in the cell body. (A) RGC asymmetric divisions rely on the asymmetric segregation of mRNAs encoding cell fate determinants. This mechanism was shown to rely on the RNA-binding proteins STAU2, DDX1, and PUMILLIO but additional as yet unidentified RNA-binding proteins are also likely important. (B) mRNAs are actively transported in the basal process from the RGC cell body to the basal endfeet. RNA transport is controlled by FMRP, but it is likely additional RBPs also influence RNA transport. In the basal endfeet, these mRNAs can be locally translated.
These findings lay the foundation for investigating how STAU2 controls RNA metabolism to influence cell fate. STAU2 targets were disproportionately enriched for genes involved in cell cycle regulation, including cell cycle exit. Given that cell cycle exit and differentiation are tightly linked in RGCs [66–68], it is plausible that inheritance of cell cycle-related targets promotes differentiation of newborn progeny into IPs or neurons. Without STAU2, both daughter cells would inherit equivalent RNAs, causing differentiation of newborn progeny. Interestingly, STAU2 disproportionately binds RNAs encoding ‘cilia assembly’ proteins, raising the interesting possibility that STAU2 also influences RGC cell fate by controlling RNAs important for the maintenance of the primary cilium. Consistent with a possible role in cilia, STAU2 is most abundant apically in RGCs, where centrosomes reside and from which the primary cilium emanates [69]. It is also interesting to consider how STAU2 controls RNA localization in RGCs. In Drosophila neuroblasts and mammalian neurons, asymmetric RNA accumulation relies upon transport of mRNAs, either passively or actively along the cytoskeleton, as well as anchoring to the cellular cortex [70]. In postmitotic neurons, STAU2 regulates nuclear RNA export and localization to dendrites, where it is required for spine maturation [71,72]. Thus, in RGCs, STAU2 association with the cytoskeleton and centrosomes could also mediate its roles in RNA localization and potentially in local translation.
During mitosis, transcription and translation of mRNA is largely silenced, although some low levels may occur [73]. Thus, it is likely that asymmetrically localized mRNAs are translationally repressed until after mitosis, when they can be rapidly translated to drive the fate and behavior of progeny. Although a potential role for STAU2 in translational regulation within RGCs remains to be explored, it is plausible, given that RGCs contain a large cohort of translation-ally repressed RNAs encoding differentiation factors [74,75].
Beyond STAU2, additional RBPs, including SMAUG2, PUM2, and DDX1, are implicated in translational regulation and asymmetric division [36,63,74,75]. Going forward it will be exciting to consider roles for these proteins in mitotic segregation of mRNAs, translational regulation, as well as how they cooperate with STAU2. Thus, while STAU2 exemplifies how RBPs can influence asymmetric RNA segregation and cell fate in RGCs, these functions are likely broadly applicable to many additional RBPs.
RNA localization and local translation in RGC endfeet
At the distal endfeet of RGCs, mRNAs can also asymmetrically localize. ABBA, also known as Mtss1l, strongly localizes to the putative RGC endfeet [76]. In RGC-like C6-R cells, ABBA depletion perturbs lamellipodial dynamics and extension of cellular processes. These defects could be mediated by ABBA binding to both G-actin and the small GTPase Rac. This suggests a potential role for ABBA in membrane dynamics in RGC endfeet, although local translation or functions for ABBA in endfeet are not yet described. Another mRNA enriched in RGC endfeet is Nestin, best known as a canonical marker of RGCs [77]. Again implications of this subcellular localization or the exact function of NESTIN in RGC endfeet are ill-defined.
A seminal study describing mRNA localization within endfeet demonstrated asymmetric localization of CyclinD2 (Ccnd2) mRNA [78]. This finding was surprising given that Ccnd2 encodes a cell cycle regulator, which is primarily known to act in the nucleus. In addition to discovering that Ccnd2 mRNA and protein are both asymmetrically localized to endfeet, this study made a number of additional discoveries. First, the authors used a series of reporter constructs, introduced by in utero electroporation, to define sequences within the 3′ UTR of Ccnd2 that are sufficient for its endfoot localization. This demonstrated that 3′ UTR elements can confer localization in mammalian RGCs, paralleling findings in other cell types and organisms. The authors then employed fluorescent reporters containing this 3′ UTR along with nuclear localization signals to detect fluorescence within endfeet. From this the authors concluded that RNA is locally translated, however, there are caveats to this conclusion, given the possibility that protein could still diffuse from the cell body. Although this study defined 3′ UTR sequences sufficient for localization, it is not clear whether these sequences are also necessary for local translation. Finally, the authors used clonal cell analysis to argue that Ccnd2 inheritance within new progeny containing a basal process promotes RGC cell fate. This localized pool of Ccnd2 mRNA would be translated following mitosis, and eventually promote entry into another cell cycle.
Many fascinating questions were raised by this study. It remains unclear why the mRNA encoding a cell cycle regulator, CCND2, is localized distally in RGCs. Could endfoot-localized Ccnd2 mRNA travel back to the cell body after mitosis, or does locally synthesized CCND2 protein function within endfeet? Relevant for these possibilities, it is intriguing to consider that CCND2 could function noncanonically in RGC endfeet, independent of the cell cycle. Indeed, there is precedence that other Cyclins, Cdks, and associated regulators can act outside of the cell cycle machinery, including within the nervous system [79–81]. Thus, it will be of interest to determine if other cell cycle-related transcripts also localize and function in end-feet.
Live imaging: RNA and RNA-binding proteins move and function in distinct compartments of RGCs
While previous studies demonstrated that mRNAs can subcellularly localize to radial glia endfeet, a major question was whether they do so actively or passively by diffusion, and whether this is controlled by RBPs. Our group addressed these questions using live imaging and RNA biochemistry [34]. First, we tested the hypothesis that mRNA moves actively in RGCs by employing MS2 technology to investigate mRNA transport. As a proof of principle, 3′ UTR Ccnd2 reporters, discovered by Tsunekawa et al. [78], were introduced into the developing brain via in utero electroporation, followed 16 h later by preparation and imaging of embryonic brain slices. This approach showed that, strikingly, mRNA is competent to move in RGCs at speeds and run-lengths consistent with active, microtubule-based transport (Fig. 3B). RNA movements were distinct between E14.5 and E16.5, in terms of directionality, speed, and run length. Although the nature of these differences remains unclear, it could suggest that RNA transport influences generation of distinct subtypes of excitatory neurons, produced at these different developmental stages. Alternatively, inherent temporal differences in micro-tubule orientation or the presence of distinct RBPs could influence RNA trafficking.
One potential function of RNA localization is local translation (Fig. 3B). In postmitotic neurons, local translation is frequently investigated using axotomy assays or chambers, which enables one to assess local translation within a specific subcellular compartment, without the complication of protein diffusion from the cell body [82]. In the developing cortex this is challenging, given that no current methodologies exist for culturing RGCs to recapitulate their unique and complex anatomy. Therefore, we devised a way to remove the top layer of RGCs in an ‘endfoot preparation.’ This preparation contains RGC endfeet as well as cellular constituents of the pial niche, including vasculature, meninges, basement membrane, and neuronal processes. To monitor de novo protein translation specifically within RGC endfeet, photoconvertible proteins were employed. Photoconvertible reporters introduced via in utero electroporation demonstrated endfeet exhibit local translation, which was blocked by treatment with the translational inhibitor anisomycin. Interestingly, not all endfeet or mRNA reporters exhibited equivalent levels of translation, suggesting that this may be a regulated process. Intrinsically, RBPs, cell cycle stage, or inherent differences in progenitors could influence translation. Local protein production could also be influenced by extrinsic signals generated by the endfoot niche, including meninges and neurons, such as those previously implicated in proliferation control and feedback signaling [23,24].
In order to understand the intrinsic regulation of RNA localization and translation in endfeet, we investigated the RBP, FMRP, with established roles and expression in the embryonic and adult cortex [83,84]. During corticogenesis, FMRP is implicated in neuronal migration and RGC proliferation [85–87]. Both endogenous and exogenous FMRP localized to endfeet and decorated the length of RGC basal processes, which may correspond to FMRP ribonucleoprotein granules being transported to basal endfeet. Live imaging of E16.5 brain slices support this hypothesis, as FMRP moved at similar speeds and behaviors to mRNA reporters at this age [34].
The question of which mRNAs FMRP binds and regulates in endfeet was addressed with traditional RIP approaches, without cross-linking, followed by microarray. Within endfeet, FMRP associated with 115 transcripts significantly enriched for cytoskeletal regulators, signaling molecules, and autism-associated genes, as well as previously identified FMRP targets [83,88,89]. One criticism of RIP-CHIP is the possibility of reassortment following cell dissociation, which could lead to nonspecific hits. However, independent RIPs and FISH assays successfully validated 6/6 candidates tested [34]. FMRP is implicated in both RNA transport and translational inhibition [84,90]. Consistent with the former function, localization of two targets, Kif26a and Dst, but not Apc, was reduced in Fmrp mutant endfeet and Kif26a transport was significantly slower in Fmrp mutant RGCs compared to control. This suggests that FMRP controls RNA transport in RGCs, as in neurons [90]. Based upon this, it will be of interest to observe the cotransport of FMRP with its targets [91], an experiment which is possible but technically challenging due to the requirement of multiple fluorescent proteins imaged simultaneously. In addition to transport, FMRP may also influence protein translation, depending upon the target considered or the developmental stage. Indeed a more comprehensive analysis of how FMRP controls its downstream targets in RGCs is warranted.
This study raises broad questions regarding how RNA is shuttled to endfeet. Beyond FMRP, additional RBPs, STAU2, and PUMILLIO, localize to endfeet [34]. This is interesting especially given the critical role of STAU2 in RGC asymmetric division and RNA localization in the cell body, as described above [63,64]. In other systems FMRP and STAU2 cooperatively bind targets, begging the question of whether this occurs in RGCs [92]. Evaluating the targets of STAU2 and other RBPs in endfeet will help elucidate whether the FMRP-related transcriptome is representative of all transcripts in endfeet. Another RBP of interest is adenomatous polyposis coli (APC), which can also bind microtubules and influence endfoot remodeling [93,94]. Apc mRNA is also localized to endfeet, begging the question of whether it functions locally in endfeet. Beyond RBPs, it is unclear which molecular motors and adaptor proteins influence RNA transport and how RNAs are ultimately tethered at endfoot structures. By defining the cis- and transmachinery controlling RNA localization, one can endeavor to develop potential tools for interrogating function.
How might mRNA localization influence cell fate of RGCs? In the cell body, mitosis itself can control segregation of cell fate components to new progeny. Osumi and colleagues have suggested similar functions may also hold true for localized mRNAs in RGC end-feet [95]. This may be most plausible in early stage RGCs, when transcripts would travel a much shorter distance than in older stage RGCs. While mRNAs can be rapidly transported bidirectionally in late stage RGCs, it remains unknown if individual mRNAs transit from the cell body to the endfeet and then back to the cell body. Defining the lifetime and fate of mRNAs in endfeet will be valuable to address this possibility, although this is technically challenging. It is also conceivable that RGC endfeet are mRNA warehouses, where their local translation (perhaps in response to a cue) results in new protein shuttling back to the nucleus, where it can influence fate. Indeed, the FMRP-bound transcriptome included chromatin factors, and there is precedence for nuclear factors shuttling in a retrograde fashion in neurons [43,82].
The control of local translation in RGCs is also ripe for study. Both Ccnd2 and Kif26a 3′ UTR mRNA reporters were competent to be localized and translated at endfeet. But are all subcellularly localized mRNAs translated to a similar extent? Of note, our study highlights differences in Kif26a and Ccnd2 reporter translation. This could be due to technical or biological differences. Indeed whether a given transcript is translated could be related to a number of factors including mRNA reporter, RGC cell cycle stage, developmental stage, or signals emanating from surrounding niche. Beyond local translation at endfeet, it is interesting to consider that within basal processes, translation ‘hot spots’ may also exist, as suggested by a recent study which showed ER are scattered along the basal process [8].
Locally produced protein in RGC endfeet may enable cell–cell communication between the pia and ventricular compartments (Fig. 4). This is supported by the observation that transcripts encoding signaling molecules of the Rho GTPase and Map Kinase signaling pathways were abundant in the FMRP-bound local transcriptome. Locally produced signaling molecules could transit back to the cell body, and/or communicate with the surrounding niche. Indeed, feedback signaling from upper layer neurons influences subsequent progenitor cell fate, but how this occurs at a molecular level is still largely unknown [24]. Likewise, meninges secrete signals including retinoic acid to influence RGC proliferation [23]. Toward understanding local RGC function, parallels may be drawn between RGCs and astrocytes. RGCs are morphologically similar to astrocytes, generate astrocytes, and astrocytes have endfeet which contact blood vessels in the mature brain. Moreover, there is a strong overlap of transcripts enriched in endfeet and within astrocyte protrusions, suggesting that similar mechanisms may be at play [34].
Fig. 4.
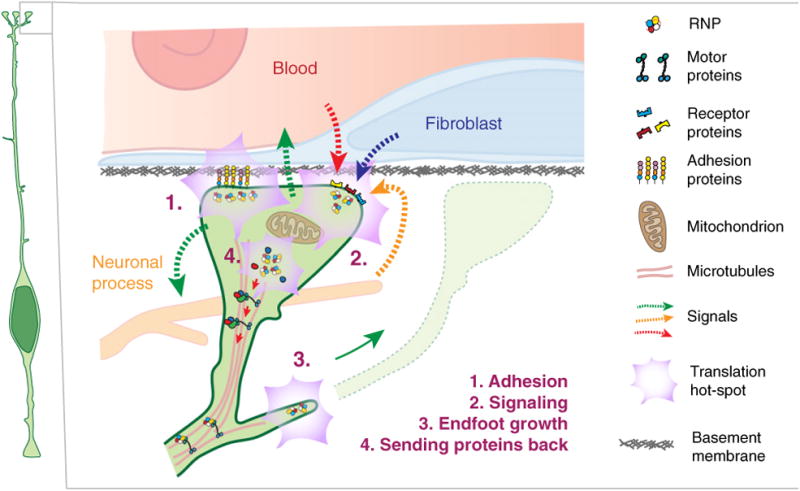
Cartoon representing putative roles of RNA localization and regulated translation in the basal endfeet. Proposed functions, as discussed in this review, are listed and denoted as numbers.
Beyond signaling back to RGC cell bodies, signaling molecules as well as locally produced cytoskeletal proteins may influence RGC structure (Fig. 4). This could include production and maintenance of RGC basal processes and endfeet, as the brain expands radially [96]. Besides ABBA, several transcripts encoding relevant cytoskeletal regulators are enriched in RGC endfeet. Local translation could impact dynamic filopodia-like structures of the basal process, similar to mechanisms within the growth cone of growing axons [29]. The FMRP interactome at basal endfeet also contains some mRNAs encoding membrane and secreted proteins. However, use of both EM and fluorescent proteins marking Golgi components, indicate that RGC basal processes and endfeet lack conventional golgi apparatus, as do neuronal axons [8,97]. This raises the question of how some locally translated proteins are appropriately modified in the absence of Golgi. One possibility is that RGCs employ alternative ER and Golgi pathways which regulate trafficking of appropriate proteins to the membrane, as has been reported in axons [98]. Further studies of endfeet mRNAs encoding membrane-bound and secreted proteins, as well as the presence of such pathways, will be needed to investigate this idea.
Conclusions and perspectives
Dysregulation of RGC proliferation, differentiation, and neuronal migration can cause neurodevelopmental disease. Thus, it is important to consider the potential disease relevance of RNA localization and local translation in RGCs. The observation that FMRP influences RNA transport in RGCs begs the question as to whether its role in distal compartments of RGCs is relevant for Fragile X syndrome. Notably, several FMRP-associated targets in RGC endfeet are themselves implicated in autism, schizophrenia, and primrose syndrome [34]. Likewise, STAU2 associates with cilia-encoding transcripts, suggesting that STAU2-mediated RNA localization could contribute to ciliopathies [64]. Another endfoot-localized transcript, CCND2, is also implicated in the neurodevelopmental disease, megalencephaly, due to stabilization of the CCND2 protein. De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria- polydactyly-hydrocephalus syndrome [99]. It is interesting to consider that diseases dependent upon proper RGC scaffolding and neuronal migration, such as autism, cobblestone microcephaly, and lissencephaly could be affected by local translation in RGCs [100]. This is because the integrity and structure of RGC basal processes is essential for neuronal migration [101]. Indeed local translation could promote maintenance of endfoot attachment to the basal lamina, thus preventing overmigration of neurons. It will be fascinating to consider roles for local translation in the etiology of these and other neurodevelopmental disorders.
RNA localization and local translation have well-established roles in the developing and adult nervous systems including within growing or wounded axons, neuronal synapses, and astrocytes [30,82]. In comparison, in RGCs, RNA localization and translation is a field in its infancy. Nevertheless, as highlighted in this review, RGCs are a cell type ripe for discovery on this front and a new emerging model of RNA localization and translation. Continued investigation promises to yield important insights—not only into cortical development but also into universal principles of RNA localization and translation.
Acknowledgments
We thank members of the Silver lab for helpful discussions including Ashley Lennox and Jeremy Rouanet for reading the manuscript. We apologize to those whose work we could not discuss due to space limitations. We thank funding sources, R01NS083897 to DLS and Duke Regeneration Next Fellowship to LJP.
Abbreviations
- Ccnd2
CyclinD2
- EM
electron microscopy
- ER
endoplasmic reticulum
- FISH
fluorescent in situ hybridization
- IPs
intermediate progenitors
- RBPs
RNA-binding proteins
- RGCs
radial glial cells
- RIP
RNA-immunoprecipitation
Footnotes
Author contributions: LJP and DLS wrote the manuscript.
References
- 1.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 2.Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 3.Dehay C, Kennedy H, Kosik KS. The outer subventricular zone and primate-specific cortical complexification. Neuron. 2015;85:683–694. doi: 10.1016/j.neuron.2014.12.060. [DOI] [PubMed] [Google Scholar]
- 4.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilsch-Bräauninger M, Florio M, Huttner WB. Neocortex expansion in development and evolution – from cell biology to single genes. Curr Opin Neurobiol. 2016;39:122–132. doi: 10.1016/j.conb.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 6.de Aguilar PB, Caviness VS, Jr, Evrard P. Neuron migration within the radial glial fiber system of the developing murine cerebrum: an electron microscopic autoradiographic analysis. Brain Res Dev Brain Res. 1990;52:39–56. doi: 10.1016/0165-3806(90)90220-s. [DOI] [PubMed] [Google Scholar]
- 7.Schmechel DE, Rakic P. A Golgi study of radial glial cells in developing monkey telencephalon: morphogenesis and transformation into astrocytes. Anat Embryol. 1979;156:115–152. doi: 10.1007/BF00300010. [DOI] [PubMed] [Google Scholar]
- 8.Taverna E, Mora-Bermudez F, Strzyz PJ, Florio M, Icha J, Haffner C, Norden C, Wilsch-Bräauninger M, Huttner WB. Non-canonical features of the Golgi apparatus in bipolar epithelial neural stem cells. Sci Rep. 2016;6:21206. doi: 10.1038/srep21206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weissman TA, Riquelme PA, Ivic L, Flint AC, Kriegstein AR. Calcium waves propagate through radial glial cells and modulate proliferation in the developing neocortex. Neuron. 2004;43:647–661. doi: 10.1016/j.neuron.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Rash BG, Ackman JB, Rakic P. Bidirectional radial Ca(2+) activity regulates neurogenesis and migration during early cortical column formation. Sci Adv. 2016;2:e1501733. doi: 10.1126/sciadv.1501733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 12.Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 13.Shen Q, Zhong W, Jan Y, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- 14.Kosodo Y, Röper K, Haubensak W, Marzesco AM, Corbeil D, Huttner WB. Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 2004;23:2314–2324. doi: 10.1038/sj.emboj.7600223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y, Goderie SK, Temple S. Asymmetric distribution of EGFR receptor during mitosis generates diverse CNS progenitor cells. Neuron. 2005;45:873–886. doi: 10.1016/j.neuron.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 16.Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- 17.Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosodo Y, Toida K, Dubreuil V, Alexandre P, Schenk J, Kiyokage E, Attardo A, Mora-Bermudez F, Arii T, Clarke JDW, et al. Cytokinesis of neuroepithelial cells can divide their basal process before anaphase. EMBO J. 2008;27:3151–3163. doi: 10.1038/emboj.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myshrall TD, Moore SA, Ostendorf AP, Satz JS, Kowalczyk T, Nguyen H, Daza RAM, Lau C, Campbell KP, Hevner RF. Dystroglycan on radial glia end feet is required for pial basement membrane integrity and columnar organization of the developing cerebral cortex. J Neuropathol Exp Neurol. 2012;71:1047–1063. doi: 10.1097/NEN.0b013e318274a128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Jin Z, Koirala S, Bu L, Xu L, Hynes RO, Walsh CA, Corfas G, Piao X. GPR56 regulates pial basement membrane integrity and cortical lamination. J Neurosci. 2008;28:5817–5826. doi: 10.1523/JNEUROSCI.0853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjornsson CS, Apostolopoulou M, Tian Y, Temple S. It takes a village: constructing the neurogenic niche. Dev Cell. 2015;32:435–446. doi: 10.1016/j.devcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siegenthaler JA, Ashique AM, Zarbalis K, Patterson KP, Hecht JH, Kane MA, Folias AE, Choe Y, May SR, Kume T, et al. Retinoic acid from the meninges regulates cortical neuron generation. Cell. 2009;139:597–609. doi: 10.1016/j.cell.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave KA, Huylebroeck D, Tarabykin V. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci. 2009;12:1373–1380. doi: 10.1038/nn.2409. [DOI] [PubMed] [Google Scholar]
- 25.Radakovits R, Barros CS, Belvindrah R, Patton B, Müller U. Regulation of radial glial survival by signals from the meninges. J Neurosci. 2009;29:7694–7705. doi: 10.1523/JNEUROSCI.5537-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haubst N, Georges-Labouesse E, De Arcangelis A, Mayer U, Gotz M. Basement membrane attachment is dispensable for radial glial cell fate and for proliferation, but affects positioning of neuronal subtypes. Development. 2006;133:3245–3254. doi: 10.1242/dev.02486. [DOI] [PubMed] [Google Scholar]
- 27.Hartfuss E, Förster E, Bock HH, Hack MA, Leprince P, Luque JM, Herz J, Frotscher M, Gotz M. Reelin signaling directly affects radial glia morphology and biochemical maturation. Development. 2003;130:4597–4609. doi: 10.1242/dev.00654. [DOI] [PubMed] [Google Scholar]
- 28.Griveau A, Borello U, Causeret F, Tissir F, Boggetto N, Karaz S, Pierani A. A novel role for Dbx1-derived Cajal-Retzius cells in early regionalization of the cerebral cortical neuroepithelium. PLoS Biol. 2010;8:e1000440. doi: 10.1371/journal.pbio.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokota Y, Eom TY, Stanco A, Kim WY, Rao S, Snider WD, Anton ES. Cdc42 and Gsk3 modulate the dynamics of radial glial growth, inter-radial glial interactions and polarity in the developing cerebral cortex. Development. 2010;137:4101–4110. doi: 10.1242/dev.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2014;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 33.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilaz LJ, Lennox AL, Rouanet JP, Silver DL. Dynamic mRNA transport and local translation in radial glial progenitors of the developing brain. Curr Biol. 2016;26:3383–3392. doi: 10.1016/j.cub.2016.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Amadei G, Zander MA, Yang G, Dumelie JG, Vessey JP, Lipshitz HD, Smibert CA, Kaplan DR, Miller FD. A Smaug2-based translational repression complex determines the balance between precursor maintenance versus differentiation during mammalian neurogenesis. J Neurosci. 2015;35:15666–15681. doi: 10.1523/JNEUROSCI.2172-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, Lopez-Jones M, Meng X, Singer RH. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science. 2014;343:422–424. doi: 10.1126/science.1239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8:165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buxbaum AR, Wu B, Singer RH. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014;343:419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell PD, Chao JA, Singer RH, Marlow FL. Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Development. 2015;142:1368–1374. doi: 10.1242/dev.118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelles DA, Fang MY, O'Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell. 2016;165:488–496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung KM, van Horck FPG, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cosker KE, Fenstermacher SJ, Pazyra-Murphy MF, Elliott HL, Segal RA. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat Neurosci. 2016;19:690–696. doi: 10.1038/nn.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu B, Eliscovich C, Yoon YJ, Singer RH. Translation dynamics of single mRNAs in live cells and neurons. Science. 2016;352:1430–1435. doi: 10.1126/science.aaf1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katz ZB, English BP, Lionnet T, Yoon YJ, Monnier N, Ovryn B, Bathe M, Singer RH. Mapping translation “hot-spots” in live cells by tracking single molecules of mRNA and ribosomes. eLife. 2016;5:1–36. doi: 10.7554/eLife.10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hinz FI, Dieterich DC, Schuman EM. Teaching old NCATs new tricks: using non-canonical amino acid tagging to study neuronal plasticity. Curr Opin Chem Biol. 2013;17:738–746. doi: 10.1016/j.cbpa.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan X, Hoek TA, Vale RD, Tanenbaum ME. Dynamics of translation of single mRNA molecules in vivo. Cell. 2016;165:976–989. doi: 10.1016/j.cell.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halstead JM, Lionnet T, Wilbertz JH, Wippich F, Ephrussi A, Singer RH, Chao JA. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science. 2015;347:1367–1671. doi: 10.1126/science.aaa3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanz E, Yang L, Su T, Morris D, McKnight G, Amieux P. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci USA. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 53.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. Transcriptome-wide Identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci USA. 2016;113:10322–10327. doi: 10.1073/pnas.1600008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- 58.Mikuni T, Nishiyama J, Sun Y, Kamasawa N, Yasuda R. High-throughput, high-resolution mapping of protein localization in mammalian brain by in vivo genome editing. Cell. 2016;165:1803–1817. doi: 10.1016/j.cell.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 60.Noctor SC, Martìnez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 61.Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- 62.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 63.Vessey JP, Amadei G, Burns SE, Kiebler MA, Kaplan DR, Miller FD. An asymmetrically localized Staufen2-dependent RNA complex regulates maintenance of mammalian neural stem cells. Cell Stem Cell. 2012;11:517–528. doi: 10.1016/j.stem.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 64.Kusek G, Campbell M, Doyle F, Tenenbaum SA, Kiebler MA, Temple S. Asymmetric segregation of the double-stranded RNA binding protein Staufen2 during mammalian neural stem cell divisions promotes lineage progression. Cell Stem Cell. 2012;11:505–516. doi: 10.1016/j.stem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Broadus J, Fuerstenberg S. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- 66.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 67.Pilaz LJ, Patti D, Marcy G, Ollier E, Pfister S, Douglas RJ, Betizeau M, Gautier E, Cortay V, Doerflinger N, et al. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci USA. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pilaz LJ, McMahon JJ, Miller EE, Lennox AL, Suzuki A, Salmon E, Silver DL. Prolonged mitosis of neural progenitors alters cell fate in the developing brain. Neuron. 2016;89:83–99. doi: 10.1016/j.neuron.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilsch-Bräuninger M, Peters J, Paridaen JTML, Huttner WB. Basolateral rather than apical primary cilia on neuroepithelial cells committed to delamination. Development. 2012;139:95–105. doi: 10.1242/dev.069294. [DOI] [PubMed] [Google Scholar]
- 70.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 71.Kiebler MA, Hemraj I, Verkade P, Köhrmann M, Fortes P, Marión RM, Ortín J, Dotti CG. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goetze B, Tuebing F, Xie Y, Dorostkar MM, Thomas S, Pehl U, Boehm S, Macchi P, Kiebler MA. The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J Cell Biol. 2006;172:221–231. doi: 10.1083/jcb.200509035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D. The translational landscape of the mammalian cell cycle. Mol Cell. 2013;52:574–582. doi: 10.1016/j.molcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kraushar ML, Thompson K, Wijeratne HRS, Viljetic B, Sakers K, Marson JW, Kontoyiannis DL, Buyske S, Hart RP, Rasin MRR. Temporally defined neocortical translation and polysome assembly are determined by the RNA-binding protein Hu antigen R. Proc Natl Acad Sci USA. 2014;111:E3815–3824. doi: 10.1073/pnas.1408305111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang G, Smibert CA, Kaplan DR, Miller FD. An eIF4E1/4E-T complex determines the genesis of neurons from precursors by translationally repressing a proneurogenic transcription program. Neuron. 2014;84:723–739. doi: 10.1016/j.neuron.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 76.Saarikangas J, Hakanen J, Mattila PK, Grumet M, Salminen M, Lappalainen P. ABBA regulates plasma-membrane and actin dynamics to promote radial glia extension. J Cell Sci. 2008;121:1444–1454. doi: 10.1242/jcs.027466. [DOI] [PubMed] [Google Scholar]
- 77.Dahlstrand J, Lardelli M, Lendahl U. Nestin mRNA expression correlates with the central nervous system progenitor cell state in many, but not all, regions of developing central nervous system. Brain Res Dev Brain Res. 1995;84:109–129. doi: 10.1016/0165-3806(94)00162-s. [DOI] [PubMed] [Google Scholar]
- 78.Tsunekawa Y, Britto JM, Takahashi M, Polleux F, Tan SS, Osumi N. Cyclin D2 in the basal process of neural progenitors is linked to non-equivalent cell fates. EMBO J. 2012;31:1879–1892. doi: 10.1038/emboj.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 80.Hydbring P, Malumbres M, Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol. 2016;17:280–292. doi: 10.1038/nrm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmetsdorf S, Gärtner U, Arendt T. Constitutive expression of functionally active cyclindependent kinases and their binding partners suggests noncanonical functions of cell cycle regulators in differentiated neurons. Cereb Cortex. 2007;17:1821–1829. doi: 10.1093/cercor/bhl091. [DOI] [PubMed] [Google Scholar]
- 82.Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 84.Richter JD, Bassell GJ, Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci. 2015;16:595–605. doi: 10.1038/nrn4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saffary R, Xie Z. FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J Neurosci. 2011;31:1427–1439. doi: 10.1523/JNEUROSCI.4854-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Castrén ML, Tervonen T, Kärkkäinen V, Heinonen S, Castrén E, Larsson K, Bakker CE, Oostra BA, Akerman K. Altered differentiation of neural stem cells in fragile X syndrome. Proc Natl Acad Sci USA. 2005;102:17834–17839. doi: 10.1073/pnas.0508995102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.La Fata G, Gärtner A, Domınguez-Iturza N, Dresselaers T, Dawitz J, Poorthuis RB, Averna M, Himmelreich U, Meredith RM, Achsel T, et al. FMRP regulates multipolar to bipolar transition affecting neuronal migration and cortical circuitry. Nat Neurosci. 2014;17:1693–1700. doi: 10.1038/nn.3870. [DOI] [PubMed] [Google Scholar]
- 88.Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 90.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ehses J, Kiebler MA, Fernández-Moya SM. RNA transport: from head to toe in radial glial cells. Curr Biol. 2016;26:R1285–R1287. doi: 10.1016/j.cub.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 92.Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yokota Y, Kim WY, Chen Y, Wang X, Stanco A, Komuro Y, Snider W, Anton ES. The adenomatous polyposis coli protein is an essential regulator of radial glial polarity and construction of the cerebral cortex. Neuron. 2009;61:42–56. doi: 10.1016/j.neuron.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Preitner N, Quan J, Nowakowski DW, Hancock ML, Shi J, Tcherkezian J, Young-Pearse TL, Flanagan JG. APC is an RNA-binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell. 2014;158:368–382. doi: 10.1016/j.cell.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tsunekawa Y, Kikkawa T, Osumi N. Asymmetric inheritance of Cyclin D2 maintains proliferative neural stem/progenitor cells: a critical event in brain development and evolution. Dev Growth Differ. 2014;56:349–357. doi: 10.1111/dgd.12135. [DOI] [PubMed] [Google Scholar]
- 96.Lu X, Duan M, Song L, Zhang W, Hu X, Zhao S, Chen S. Morphological changes of radial glial cells during mouse embryonic development. Brain Res. 2015;1599:57–66. doi: 10.1016/j.brainres.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 97.Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Merianda TT, Lin AC, Lam JSY, Vuppalanchi D, Willis DE, Karin N, Holt CE, Twiss JL. A functional equivalent of endoplasmic reticulum and Golgi in axons for secretion of locally synthesized proteins. Mol Cell Neurosci. 2009;40:128–142. doi: 10.1016/j.mcn.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mirzaa GM, Parry DA, Fry AE, Giamanco KA, Schwartzentruber J, Vanstone M, Logan CV, Roberts N, Johnson CA, Singh S, et al. De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome. Nat Genetics. 46:510–515. doi: 10.1038/ng.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wegiel J, Kuchna I, Nowicki K, Imaki H, Wegiel J, Marchi E, Ma SY, Chauhan A, Chauhan V, Bobrowicz TW, et al. The neuropathology of autism: defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 2010;119:755–770. doi: 10.1007/s00401-010-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ernst C. Proliferation and differentiation deficits are a major convergence point for neurodevelopmental disorders. Trends Neurosci. 2016;39:290–299. doi: 10.1016/j.tins.2016.03.001. [DOI] [PubMed] [Google Scholar]


