Abstract
Triple-negative breast cancers (TNBCs) are among the most aggressive cancers characterized by a high propensity to invade, metastasize and relapse. We previously reported that the TNBC-specific inhibitor, AMPI-109, significantly impairs the ability of TNBC cells to migrate and invade by reducing levels of the metastasis-promoting phosphatase, PRL-3. Here, we examined the mechanisms by which AMPI-109 and loss of PRL-3 impede cell migration and invasion. AMPI-109 treatment or knock down of PRL-3 expression were associated with deactivation of Src and ERK signaling and concomitant downregulation of RhoA and Rac1/2/3 GTPase protein levels. These cellular changes led to rearranged filamentous actin networks necessary for cell migration and invasion. Conversely, overexpression of PRL-3 promoted TNBC cell invasion by upregulating matrix metalloproteinase 10, which resulted in increased TNBC cell adherence to, and degradation of, the major basement membrane component laminin. Our data demonstrate that PRL-3 engages the focal adhesion pathway in TNBC cells as a key mechanism for promoting TNBC cell migration and invasion. Collectively, these data suggest that blocking PRL-3 activity may be an effective method for reducing the metastatic potential of TNBC cells.
Keywords: Metastasis, laminin, MMP, triple-negative breast cancer, PRL-3
1. Introduction
Breast cancer is the principle cause of cancer-related mortality in women worldwide [1]. Gene expression profiling has clustered the disease into five major subtypes based on estrogen receptor (ER) expression, progesterone receptor (PR) expression and human epidermal growth factor receptor 2 (HER2) amplification [2]. Anti-hormonal therapies are available for breast cancer patients with tumors expressing ER or PR, while targeted therapy with the monoclonal antibodies trastuzumab and pertuzumab, are indicated for patients with tumors exhibiting HER2 amplification. Unfortunately, triple-negative breast cancers (TNBCs), which comprise 15–20% of all newly diagnosed cases of breast cancer, and are frequently diagnosed as high grade invasive tumors, lack expression of these three molecular markers [3]. As a result, cytotoxic chemotherapy is most frequently utilized as the standard of care for women diagnosed with metastatic TNBC and there are currently no approved targeted agents for these patients. Metastatic TNBC tumors also have a higher risk of distant recurrence and death compared to other breast cancers [4]. Paradoxically, patients with non-metastatic TNBC treated with neoadjuvant chemotherapy have better response rates than other breast cancer subtypes [5], suggesting a relative driver of poor outcome in TNBC is the development of metastatic disease. It is therefore critical to identify new therapeutic agents that can either slow or prevent the metastatic dissemination of TNBC cells.
Our laboratory previously examined the effects of the novel TNBC-specific small molecule inhibitor AMPI-109 on TNBC cell migration and invasion [6]. Our studies revealed that AMPI-109 has a marked ability to block TNBC cell migration and invasion, attributable in-part to its ability to downregulate levels of the metastasis-promoting phosphatase, PRL-3 [6]. We found that AMPI-109 treatment, knock down of PRL-3 or catalytic impairment of PRL-3 activity, blocked the ability of TNBC cells to migrate, whereas overexpression of PRL-3 markedly enhanced both the rate and absolute degree of cell migration in a scratch-wound assay [6]. Moreover, we demonstrated that PRL-3 exerted analogous effects on TNBC cell invasion through Matrigel [6]. These studies suggest that PRL-3 is involved in controlling precursor events for TNBC metastasis. However, the molecular mechanism for how PRL-3 promotes the motility and invasiveness of TNBC cells is unclear.
In order to metastasize, cancer cells must lose attachment with neighboring tumor cells and adopt the ability to migrate, attach to, and invade through the epithelial basement membrane. This is a complex process primarily orchestrated through the formation, stabilization and remodeling of focal adhesion (FA) complexes composed of integrins, Src, FAK, ERK and numerous adaptor proteins and downstream effectors, such as RhoGTPases, that collectively regulate epithelial-to-mesenchymal transition (EMT) and trigger activation of cell migration and invasion programs [7, 8]. Assembly and disassembly of FA sites within invadopodia control cell migration and invasion by mediating actin assembly and contraction, thereby promoting cell adhesion to various extracellular substrates on which cancer cells can migrate on or invade through [7, 8]. Therefore, the identification of key regulators to FA site signaling is critical in order to determine strategies to prevent activation of these metastatic-enabling properties.
Here, we describe PRL-3 modulation of the FA pathway as a potential mechanism by which PRL-3 controls these pro-metastatic phenotypes. Altering PRL-3 expression had a significant impact on both protein expression and activation levels of a number of FA pathway effectors including the proto-oncogene tyrosine-protein kinase, c-src (Src), ERK, and several RhoGTPases involved in actin cytoskeletal restructuring. We also investigated the role of the matrix metalloproteinase (MMP), MMP-10, which we identified as being upregulated following overexpression of PRL-3. We found that MMP-10 upregulation following forced PRL-3 overexpression coincides with preferential TNBC cell attachment to and degradation of laminin, which is a major basement membrane component in breast tissue and a selective substrate for degradation by MMP-10. Moreover, PRL-3 overexpressing TNBC cells were capable of invading through laminin-rich Matrigel through an MMP-10 dependent mechanism.
Collectively, these data represent new molecular insight on how PRL-3 activates cell migration and invasion programs in TNBC as precursor events to metastasis – the major driver of TNBC-associated deaths.
2. Materials and methods
2.1. Materials
AMPI-109 was synthesized as previously described [9].
2.2. Plasmids, transfection and viral transduction
PRL-3 cDNA expression vector was purchased from Origene (Cat. # SC308739). Transfections were carried out using Mirus TransIT LT1 reagent according to manufacturer’s instructions (Mirus Bio). Individual pLKO.1 lentiviral shRNA clones were purchased from the University of Colorado Cancer Center Functional Genomics Shared Resource. The RNAi Consortium identifiers are: TRCN0000010661 (shPRL-3 #1), TRCN0000355597 (shPRL-3 #2), TRCN0000378843 (shMMP-10 #1), TRCN0000372935 (shMMP-10 #2). Transduced cells were selected in medium containing 2.5 μg/mL puromycin. Specificity of PRL-3 knock down was determined by qRT-PCR. Both PRL-3 shRNAs (#1 and #2) exerted specific knock down of PRL-3 and did not reduce RNA levels of either PRL-1 or PRL-2.
2.3. Cell culture and immunoblot analysis
Cell lines were obtained from the University of Colorado Cancer Center Tissue Culture Shared Resource. BT-20 and MDA-MB-468 cells were cultured in DMEM/F-12 medium (Corning #10-092-CV) containing 10% fetal bovine serum. SUM-159 cells were cultured in HAM’s F-12 medium (Corning #10-080-CV) containing 5% fetal bovine serum, 1 μg/mL hydrocortisone and 5 μg/mL insulin. All cell lines were authenticated by short tandem repeat DNA profiling performed by the UCCC DNA Sequencing and Analysis Core. Western blot analysis was conducted according to our previous protocol [10]. Antibodies used in the study were: PRL-3 (Cat. # ab82568, Abcam), p-Src (Y416) (Cat. #2101, Cell Signaling), Src (36D10) (Cat. #2109, Cell Signaling), p-ERK 1/2 (T202/Y204) (Cat. #4377, Cell Signaling), ERK 1/2 (44/42) (Cat. #4695, Cell signaling), RhoA (67B9) (Cat. #2117, Cell Signaling), Rac1/2/3 (Cat. #2465, Cell Signaling), MMP-10 (Cat. #SC-9941, Santa Cruz), β-actin (Cat. # A5441, Sigma-Aldrich).
2.4. Immunofluorescence analysis
Immunofluorescence staining was performed as previously described [11] using green Alexa Fluor 488 phalloidin staining for F-actin (Cat. #A12379, Thermo Fisher), β-actin antibody for both filamentous and monomer actin forms (Cat. # A5441, Sigma-Aldrich) and nuclear DAPI stain (Cat. #P-36931, Thermo Fisher).
2.5. MMP array
A human MMP antibody array kit was purchased from Abcam (Cat. # ab134004). BT-20 cells were transiently transfected with PRL-3 cDNA expression vector 48 hours prior to cell lysis and the array developed according to the manufacturer’s protocol. Membranes were developed using enhanced chemiluminescence (Perkin Elmer) and autoradiography.
2.6. Cell adhesion and spreading assay
We utilized the impedance-based xCELLigence Real-Time Cell Analysis system (ACEA Biosciences) for the detection of BT-20 and SUM159 TNBC cell adhesion and spreading on the following substrates: Laminin (Cat. #L4544, Sigma-Aldrich), Elastin (Cat. #E1625-5G, Sigma-Aldrich), Fibronectin (Cat. #F1141, Signa-Aldrich) and Collagen (Cat. #C2124, Sigma-Aldrich). Briefly, each substrate was diluted to 10 μg/mL in appropriate TNBC cell media and added to wells on a 96X E-Plate (ACEA Biosciences) and incubated for 1 hour at 37°C. The coated plates were then washed with PBS and incubated in 0.5% BSA solution in PBS for 20 minutes at 37°C. Wells were washed again with PBS and 5,000 cells were added per well. Cell adhesion and spreading was measured as changes in impedance with the RT-CES system every 3 minutes for 3 hours. The assay expresses impedance in arbitrary cell index (AU) units. The cell index at each time point is defined as (Rn−Rb)/15; where Rn is the cell-electrode impedance of the well when it contains cells and Rb is the background impedance of the well with the media alone.
2.7. Cell invasion assay
Cell invasion was assessed by IncuCyte Zoom Kinetic Live Cell Imaging (Essen BioScience). 96 well ImageLock Plates (Essen Bioscience) were coated overnight with 100 μg/mL Matrigel Basement Membrane Matrix (Cat. # 356231, Corning). The following day, Matrigel was aspirated and 25,000 cells were plated per well and allowed to adhere prior to making a scratch with the 96-pin WoundMaker (Essen Bioscience). 50 μL of Matrigel Matrix (8 mg/mL) was then added to the wells, and covered in 100 μL media containing the appropriate treatments (refer to figure 6 legend for details). Migration and invasion were quantified using the “Relative Wound Density” metric generated by IncuCyte software.
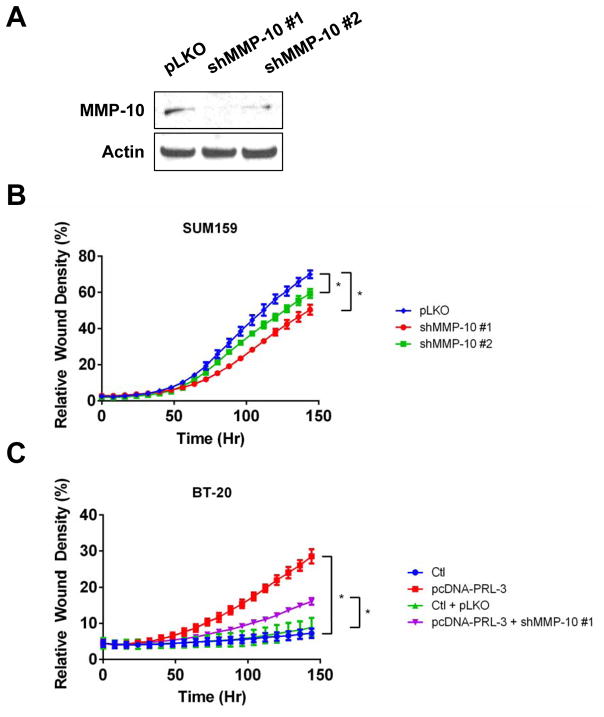
Figure 6. PRL-3 driven cell invasion is mediated through MMP-10.
(A). MMP-10 levels in SUM159 cells transduced with non-silencing control (pLKO) and two shRNAs against MMP-10 (shMMP-10 #1 and #2). (B). Real-time kinetic monitoring of invasion through Matrigel in SUM159 cells stably expressing two shRNA clones to MMP-10 (red and green) or non-silencing control (blue). (C). Real-time kinetic monitoring of invasion through Matrigel using BT-20 control cells (Ctl; blue or Ctl + pLKO; green), transiently transfected to overexpress PRL-3 (pcDNA-PRL-3; red) or overexpressing PRL-3 in BT-20 cells with MMP-10 knocked down (pcDNA-PRL-3 + shMMP-10 #1; purple). Invasive score was determined by the ability of cells to invade through a Matrigel wound (Relative Wound Density). * = p-value <0.05 as determined by Student t test on last time-point. The experiments were repeated three times.
2.8. Reverse phase protein array analysis
The results outlined in Table 1 are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. Protein expression values from the “Provisional TCGA Invasive Breast Carcinoma” dataset as assessed by the reverse phase protein array (RPPA) tool on the Memorial Sloan-Kettering Cancer Center’s cBio Cancer Genomics Portal. Protein expression values are indicated by the Z-score (mean RPPA score/std) and were obtained using a Z-score threshold of +/− 2.0.
Table 1.
Protein expression changes in invasive human breast carcinoma samples exhibiting PRL-3 copy number amplification (>2 copies).
| Protein Expression Z-score (RPPA) | |||
|---|---|---|---|
| Protein | PRL-3 Unaltered | PRL-3 Amplified | P-value |
| AR | 0.15 | −0.47 | 8.33E-08 |
| GATA3 | 0.12 | −0.37 | 3.15E-05 |
| ESR1 | 0.11 | −0.32 | 2.41E-04 |
| PRKAA1 | 0.09 | −0.28 | 3.48E-04 |
| STK11 | 0.09 | −0.28 | 3.00E-03 |
| BCL2 | 0.08 | −0.25 | 3.00E-03 |
| MAPK9 | 0.08 | −0.25 | 4.00E-03 |
| PGR | 0.07 | −0.22 | 7.00E-03 |
| SRC | −0.08 | 0.23 | 6.00E-03 |
| CCNE1 | −0.08 | 0.25 | 4.00E-03 |
| SYK | −0.09 | 0.26 | 2.00E-03 |
| CCNB1 | −0.10 | 0.29 | 3.35E-04 |
Green indicates co-upregulation of proteins with PRL-3 and red indicates proteins downregulated with PRL-3 amplification. Significance values were calculated based on Student t test.
3. Results
3.1. PRL-3 copy number amplification positively correlates with high levels of Src protein in human breast cancers
To identify PRL-3 associated pathways potentially involved in migration and invasion phenotypes, we analyzed the provisional cancer genome atlas (TCGA) invasive breast carcinoma cohort and looked for proteins with expression patterns that correlated with PRL-3 amplification. We reasoned that the identification of proteins upregulated along with PRL-3 might lead to the discovery of pathways converging with PRL-3 signaling. We identified a number of proteins that were either downregulated or upregulated with PRL-3 amplification in human breast tumors, including Src (Table 1).
Src is the prototypic member of a family of non-receptor tyrosine kinases that play critical roles in signal transduction pathways involved with cell proliferation, differentiation, survival, motility and invasion [12–18]. Considering the aberrant activation of Src in a number of cancer types including breast cancer, drug therapy aimed at inhibiting Src and/or some of its downstream effectors is considered to be a potentially useful and promising clinical strategy [19–25]. Therefore, we sought to determine whether AMPI-109 treatment or PRL-3 knock down alter Src expression and/or activity in TNBC cells.
3.2. AMPI-109 treatment and knock down of PRL-3 reduces Src, ERK signaling
Src is activated at FA sites following engagement by a variety of different cellular receptors including immune response receptors, integrins, receptor tyrosine kinases, G protein-coupled receptors as well as cytokine receptors [12–18]. The recruitment of Src to integrin complexes at FA sites, for example, is thought to lead to auto-phosphorylation of Src at tyrosine 416 (Y416). Activated Src plays a number of direct and indirect roles at activating downstream pathways, including ERK, for propagating changes in cytoskeletal organization necessary for cell motility. Indeed, studies have demonstrated that Src-mediated migration and invasion of breast cancer and hepatocellular carcinoma cells is controlled through downstream activation of the ERK pathway [26–28]. Therefore, we treated BT-20 and MDA-MB-468 TNBC cells with AMPI-109 or transduced cells with two different shRNAs to PRL-3 and examined Src and ERK activation (phosphorylation) by immunblot analysis.
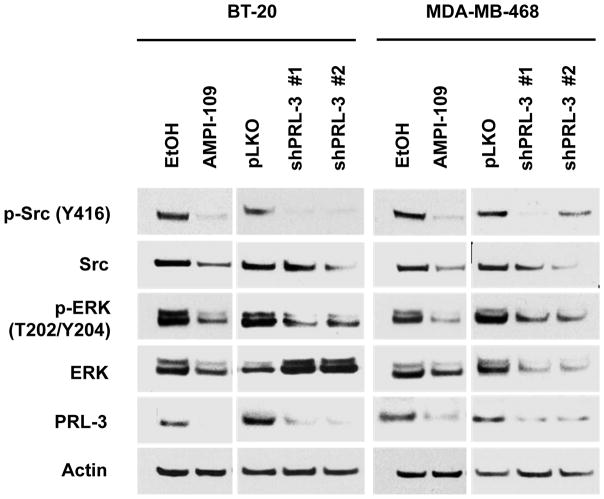
We observed that AMPI-109 significantly blocked activation of Src at tyrosine 416 (Y416) and ERK at threonine 202 and tyrosine 204 (T202/Y204) (Fig. 1). We also observed decreases in total levels of Src and ERK with AMPI-109 treatment. Additionally, knock down of PRL-3 resulted in deactivation of Src and ERK signaling, but in contrast to AMPI-109 treatment, largely had no effect on total levels of Src or ERK in BT-20 cells suggesting other targets for AMPI-109 likely exist (Fig. 1).
Figure 1. AMPI-109 treatment and PRL-3 knock down inactivate Src, ERK signaling in TNBC cells.
Immunoblot analysis of changes in activation of Src and ERK proteins as assessed by phosphorylation of Src at tyrosine 416 (Y416) and ERK at threonine 202 (T202) and tyrosine 204 (Y204) in vehicle control (EtOH) or AMPI-109 (100 nM) treated BT-20 and MDA-MB-468 TNBC cells. Right panels for each cell line depict changes in Src and ERK activation in PRL-3 knock down (shPRL-3 #1 and shPRL-3 #2) compared to non-silencing control cells (pLKO). The experiment was repeated twice and representative images shown.
3.3. Knock down of PRL-3 or treatment with AMPI-109 decreases RhoA and Rac1/2/3 GTPase levels and associates with remodeling of actin networks in TNBC cells
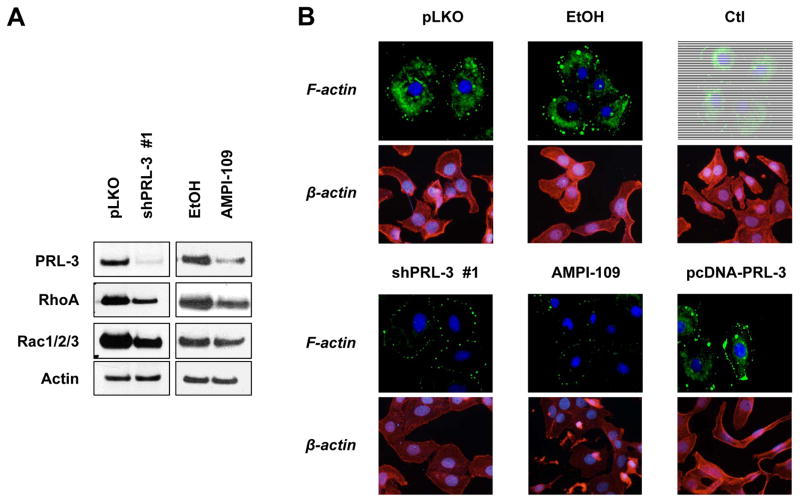
We next determined whether AMPI-109 and PRL-3 also affect common FA site transducers, downstream of Src and ERK. RhoGTPases function as downstream mediators of the FA site and are responsible for executing changes in the actin cytoskeleton that lead to cell restructuring and migration [29–34]. We observed decreases in RhoA and Rac1/2/3 RhoGTPase levels following knock down of PRL-3 or AMPI-109 treatment (Fig. 2A). We did not observe changes in Cdc42, RhoB or RhoC (data not shown). Additionally, knock down of PRL-3 and AMPI-109 treatment led to a significant reduction of filamentous actin (F-actin) networks as determined by phalloidin staining (Fig. 2B). Conversely, overexpression of PRL-3 lead to moderate cytoplasmic F-actin staining and pronounced clusters of actin that spatially coincided with regions of membrane extension or protrusion.
Figure 2. Knock down of PRL-3 and treatment with AMPI-109 alters RhoGTPase expression and filamentous actin structure.
(A). Western blot of BT-20 TNBC cells transduced with non-silencing control (pLKO) or shRNA to PRL-3 (shPRL-3 #1), or treated with vehicle control (EtOH) or AMPI-109 (100 nM) for 6 hours. Individual panels represent PRL-3 protein, RhoA, Rac1/2/3 and β-actin protein levels. (B). Immunofluorescence images of BT-20 cells transduced with non-silencing control (pLKO), PRL-3 shRNA (shPRL-3 #1), treated with vehicle control (EtOH), AMPI-109 (100 nM) or transiently transfected with scrambled DNA (Ctl) or PRL-3 expression vectors (pcDNA-PRL-3). F-actin (green) represents filamentous actin as determined by phalloidin staining. β-actin (red) represents the total actin monomer pool using an antibody to β-actin. DAPI = blue. The experiment was repeated twice and representative images shown.
Collectively, these data indicate that AMPI-109 and PRL-3 impinge on FA members and RhoGTPase networks to alter physical cellular components, such as F-actin, that are necessary for cell migration. These data suggest that PRL-3 may also then play a key role in controlling the cells ability to adhere to the extracellular matrix (ECM) – as changes in the internal cytoskeleton would likely influence extracellular adhesion dynamics.
3.4. PRL-3 overexpression increases levels of MMP-10 and promotes TNBC cell invasion by enhancing adhesion to and degradation of laminin
Understanding tumor cell adhesion to ECM substrates is critical in order to identify how cancer cells breach the basement membrane and invade neighboring normal tissue to metastasize. Migrating cells rely on attachment to the ECM via formation and maintenance of integrin clusters at FA sites. Collectively, these FA sites mediate attachment to basement membrane substrates such as laminin, elastin, collagen and fibronectin and serve as anchor or attachment points for migrating cells. This interaction incorporates extracellular mechanical contact with the substrate as a trigger event for intracellular signaling networks, such as Src, ERK and RhoGTPases [35]. In this regard, cell attachment to a specific substrate is important for spatial orientation of epithelial cells and serves as a general “sensor” for the cells local environment. In order for cells to invade through their specific substrate environment, MMPs need to be expressed and activated to degrade the substrate [36]. Because we have observed PRL-3 is involved in manipulating intracellular pathways activated by FA contact with the ECM, a key mechanistic question is whether increased PRL-3 expression alters the ability of TNBC cells to adhere to and degrade particular ECM substrates.
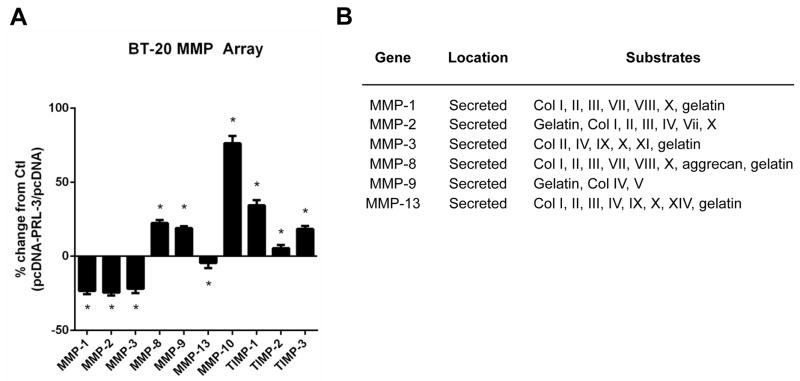
To examine this, we overexpressed PRL-3 in BT-20 TNBC cells in order to identify MMPs potentially associated with the PRL-3 signaling axis. Utilizing a human MMP protein array, we identified a list of MMPs, whose total protein expression levels changed in response to PRL-3 overexpression (Fig. 3A). This data suggested that MMP-10, the most highly upregulated MMP after PRL-3 overexpression, may be one critical MMP secreted to act upon and degrade the ECM. Interestingly, the preferred non-collagen type IV substrates for MMP-10 include laminin, fibronectin and elastin (Fig. 3B) suggesting that BT-20 cells may preferentially adhere to and degrade one or more of these three ECM components to facilitate cell invasion.
Figure 3. Overexpression of PRL-3 alters MMP levels in BT-20 TNBC cells.
(A). BT-20 cells were transiently transfected with an expression vector encoding PRL-3 (pcDNA-PRL-3) or scrambled control (pcDNA). Percent change in MMP expression from pcDNA control baseline depicted. (B). Table indicating the locations and substrate activities of MMPs.
We therefore characterized the ability of BT-20 cells to adhere to and spread on each of these three substrates. We pre-coated cell culture plates with elastin, laminin and fibronectin matrix and allowed BT-20 cells to adhere to each. We monitored cell attachment and extent of cell spreading on the various substrates by utilizing the real-time dynamic xCELLigence impedance-based adhesion and spreading platform. We observed that BT-20 cells exhibit a diverse adhesion profile to each of the three substrates (Fig. 4A). Fibronectin was the least preferred non-collagen substrate for BT-20 attachment, while elastin and laminin exhibited similar degrees of spreading at 2 hours. Interestingly, BT-20 cells contracted or decreased in size when initially exposed to laminin as indicated by the negative cell index values between 0.5 and 1 hours, but then recover and increase their spreading capacity on laminin at a higher rate relative to elastin (Fig. 4A) – reaching equivalent impedance to elastin at 2 hours.
Figure 4. BT-20 TNBC cells preferentially adhere to and spread on elastin and laminin basement membrane substrates.
(A). BT-20 cells transfected with empty cDNA expression vector (Ctl) were plated in suspension and allowed to adhere to plates pre-coated with elastin (red), laminin (green) or fibronectin (blue). (B). BT-20 cells were transfected with PRL-3 cDNA expression vector (pcDNA-PRL-3) or empty vector (Ctl) and immunoblots analysis performed 24 hours after transfection. Individual panels represent PRL-3 protein and β-actin protein levels. (C–E). BT-20 cells transfected with PRL-3 cDNA (pcDNA-PRL-3; blue, same cells as in (A)) or empty vector (Ctl; red) were plated in suspension and allowed to adhere to fibronectin (C), elastin (D) or laminin (E). Each substrate was plated at 10 μg/mL and cell impedance was monitored for 2 hours after plating. * = p-value <0.05 as determined by Student t test on last time-point. The experiments were repeated three times.
We next examined the effects of overexpressing PRL-3 on cell attachment to each of these three substrates (Fig. 4B). By doing so, we reasoned that we may identify differences in binding to specific substrates; reflecting on key aspects of PRL-3 biology. We observed that overexpression of PRL-3 (pcDNA-PRL-3) in BT-20 cells had no effect on the extent of cell attachment or spreading when exposed to fibronectin. However, these cells did appear to require more time sensing the local ECM environment before attaching and spreading (Fig. 4C; 1 versus 0.5 hours). In contrast, BT-20 cells overexpressing PRL-3 exhibited a modest and improved ability to adhere to and spread on elastin (Fig. 4D). Overexpression of PRL-3 in BT-20 cells exposed to laminin, however, significantly enhanced both the rate and magnitude of cell adhesion and spreading (Fig. 4E), strongly suggesting that PRL-3 functions to enhance BT-20 cell attachment to laminin, one of the most abundant ECM component in the basal lamina of human breast tissues.
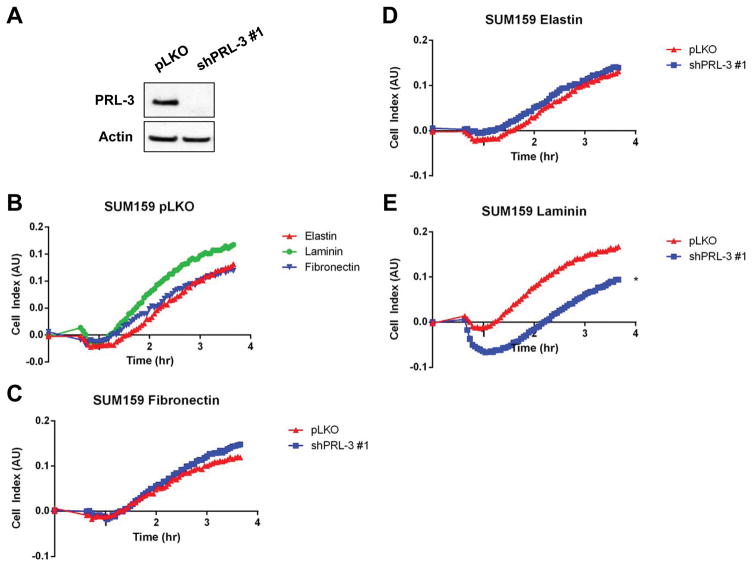
To further study the relationship between PRL-3 and TNBC cell attachment to laminin, we examined the effect of PRL-3 knock down in a TNBC cell line with high PRL-3 levels, SUM159 (Fig. 5A). Specifically, we were interested in examining whether reducing PRL-3 levels would reverse cell attachment to laminin. Interestingly, control SUM159 TNBC cells exhibited a similar preference pattern for ECM substrate binding as BT-20 cells with respect to laminin and elastin, but also fibronectin (Fig. 5B).
Figure 5. SUM159 TNBC cells preferentially adhere to, and spread on, elastin, fibronectin and laminin basement membrane substrates.
(A). PRL-3 levels following transduction of SUM159 cells with non-silencing control (pLKO) or PRL-3 shRNA (shPRL-3 #1) were assessed by immunoblot. (B). SUM159 pLKO cells were plated in suspension and allowed to adhere to elastin (green), fibronectin (brown) or laminin (dark green). (C–E). SUM159 cells were transduced with non-silencing control shRNA (pLKO; Same cells as in (B)) or shRNA to PRL-3 (shPRL-3 #1) and selected for three days prior to being plated in suspension and allowed to adhere to fibronectin (C), elastin (D) or laminin (E). Each substrate was plated at 10 μg/mL and cell impedance was monitored for 3.5 hours after plating. * = p-value <0.05 as determined by Student t test on last time-point. The experiments were repeated three times.
Following knock down of PRL-3 in SUM159 cells, we observed no changes in binding profiles to fibronectin (Fig. 5C) or elastin (Fig. 5D) but did observe, as hypothesized, a substantial impairment in the binding affinity and spreading on laminin (Fig. 5E).
We previously reported a role for PRL-3 in enhancing TNBC cell invasiveness through Matrigel [6]. Here, we demonstrate a potentially important role for TNBC cell interaction with a key basement membrane substrate, laminin. By knocking down PRL-3 in SUM159 cells, invasive potential could be diminished, and conversely, by overexpressing PRL-3 in BT-20 cells, invasion rate and absolute degree could be significantly enhanced [6]. However, these data did not take into account the identification of MMP-10 as being important for PRL-3 activity.
Therefore, we chose to examine the effects of knocking down MMP-10 on SUM159 cell invasion, reasoning that it would phenocopy the effect of knocking down PRL-3. We observed a reduction in the ability of SUM159 cells to invade through Matrigel after knocking down MMP-10 using two different shRNAs (Fig. 6A and B), thus phenocopying the effects of knocking down PRL-3 [6]. However, since overexpression of PRL-3 upregulates MMP-10, and MMP-10 exerts specific action on degrading laminin, a key question is whether knock down of MMP-10 could reverse the invasive potential driven by PRL-3 overexpression. After confirming that overexpression of PRL-3 enhances BT-20 cell invasion (Fig. 6C), we chose to overexpress PRL-3 in BT-20 cells with MMP-10 knocked down. PRL-3 was not as effective in driving cell invasion in the absence of MMP-10 (Fig. 6C), suggesting that PRL-3 driven invasiveness is mediated, at least in part, through MMP-10 activity.
4. Discussion
There is currently a pressing need to understand which signaling pathways promote TNBC cell migration and invasion so that new strategies aimed at targeting key regulators of these processes may be identified for therapeutic exploitation. PRL-3 was first identified as a metastasis-associated phosphatase through studies that demonstrated high PRL-3 levels in metastatic colorectal carcinoma samples, but low in non-metastatic colorectal carcinoma or normal colonic epithelial cells [37]. Similarly, our immunohistochemical evaluation of human breast cancers revealed that PRL-3 expression positively correlates with regional disease and distant metastases in TNBC [6]. However, a mechanistic explanation for how PRL-3 promotes acquisition of migratory and invasive programs essential for TNBC metastasis remained unclear.
Here, we describe a relationship between PRL-3 and activation of the FA Src-ERK pathway. Our data augment existing reports that indicate Src is preferentially localized and active at the cell plasma membrane in TNBCs versus other breast cancer subtypes [38] and that basal and TNBC tumors exhibit sensitivity to dasatinib, an orally bioavailable inhibitor of Src and abl kinases [39]. While the direct link between PRL-3 and the Src-ERK pathway remains unknown, these data strongly suggest there may be rationale for exploring combinatorial inhibition of Src and PRL-3 in TNBC models of migration and invasion to reduce metastatic potential.
We also discovered that changes in PRL-3 expression, or AMPI-109 treatment, exert an influence on protein expression levels of downstream members in the FA pathway, including RhoA and Rac1/2/3. These data are important because RhoGTPases are key mediators in the control of actin nucleation, filament assembly, stabilization and disassembly, mechanically controlling the cells ability to migrate and invade by forming cell protrusions or invadopodia. Indeed, coinciding with the decrease in RhoA and Rac1/2/3 expression after PRL-3 knock down was the absence of a detectable filamentous actin network. These data suggest that one terminal effect of PRL-3 knock down or AMPI-109 treatment, mediated through the FA Src-ERK-RhoGTPase pathway may be the disruption of actin assembly, extension or stabilization thereby eliminating the necessary “hardware” or infrastructure for TNBC cells to migrate and invade.
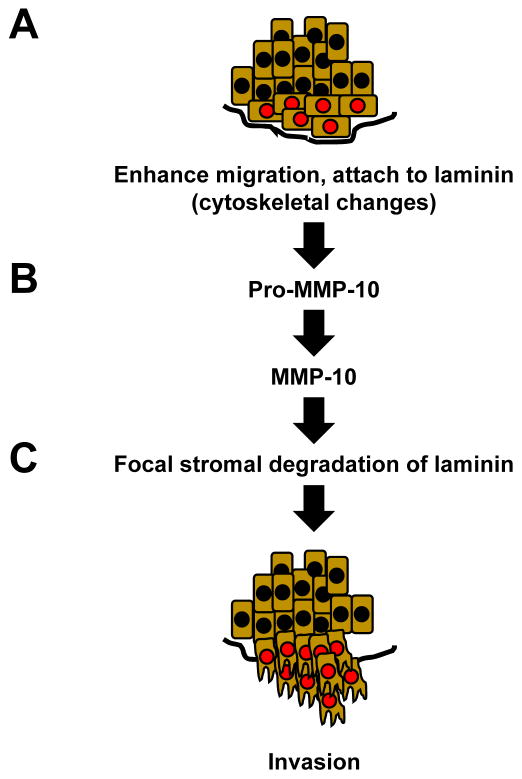
Because cancer cells must also degrade the local basement membrane to metastasize, we expanded our analysis on the role of PRL-3 in the metastatic cascade by studying the impact PRL-3 expression has on TNBC cell specificity for adhesion to and degradation of ECM substrates. We identified that PRL-3 expression in TNBC cells significantly alters TNBC cell adhesion and spreading on laminin, which is abundantly expressed in human breast tissue. Diaz et al. observed that out of 212 breast tumors, laminin protein staining was present in a linear pattern surrounding all normal ducts and lobules and around all ducts involved by ductal carcinoma in situ (DCIS) [40]. Deposition of laminin was also found immediately adjacent to invasive tumor cells and a negative correlation between the extent of extracellular laminin deposition and ER and HER2 positivity was found [40], suggesting that peri-cellular deposition of laminin may be higher in ER and HER2 negative tumors, such as TNBCs. We also observed that TNBC cells were more efficient at invading through laminin-rich Matrigel when PRL-3 was overexpressed and that this process was mediated by upregulation of MMP-10, a key metalloproteinase with specific action on laminin. Importantly, these data also suggest that PRL-3 ability to invoke a migration and invasion program may be limited by the surrounding levels of extracellular laminin. It is therefore conceivable that areas of “invasive heterogeneity” may exist in a tumor despite the levels of PRL-3 expression – which may not confer an invasive advantage in areas without laminin deposition. A working model incorporating the data presented herein is depicted in Figure 7.
Figure 7. Model depicting the role of PRL-3 in the metastatic cascade at the primary TNBC tumor site.
(A) PRL-3 expression engages the FA pathway to enhance migration through cytoskeletal changes that lead to cell attachment to laminin leading to (B) upregulation of MMP-10 and subsequent focal stromal degradation of the basement membrane to (C) facilitate invasion.
Taken together, these data provide a preliminary framework for further analyses on the interplay between PRL-3 and the FA pathway (Src-ERK-RhoGTPase), actin dynamics, and the relationship between PRL-3 expression and the upregulation of MMP-10. While another group has preliminarily reported the in vivo effects of knocking down PRL-3 on primary TNBC tumor growth [41], studies examining the effects of PRL-3 knock down or AMPI-109 treatment on invasive and metastatic endpoints in TNBC animal models are urgently needed. Our laboratory is currently pursuing these studies. Significantly, because AMPI-109 reduces PRL-3 levels [6], AMPI-109 treatment or blockade of PRL-3 using selective inhibitors may be a viable strategy for decreasing the metastatic potential of TNBC cells.
Acknowledgments
The authors thank the Colorado Cancer Center Protein Production/Monoclonal Antibody/Tissue Culture, DNA Sequencing and Analysis, Functional Genomics and Flow Cytometry Shared Resources for their services (supported by P30 CA046934).
6. Funding
This work was funded by a research grant from the Colorado Cancer League and gift funds from David Paradice of Paradice Investment Management.
Footnotes
7. Conflicts of interest
The authors disclose no conflicts of interest.
References
- 1.Polyak K, Metzger FO. SnapShot: breast cancer. Cancer Cell. 2012;22:562–562. doi: 10.1016/j.ccr.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Berrada N, Delaloge S, Andre F. Treatment of triple-negative metastatic breast cancer: toward individualized targeted treatments or chemosensitization? Ann Oncol. 2010:vii30–35. doi: 10.1093/annonc/mdq279. [DOI] [PubMed] [Google Scholar]
- 4.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 5.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 6.Gari HH, Gearheart CM, Fosmire S, De Gala GD, Fan Z, Torkko KC. Genome-wide functional genetic screen with the anticancer agent AMPI-109 identifies PRL-3 as an oncogenic driver in triple-negative breast cancers. Oncotarget. 2016;7:15757–15771. doi: 10.18632/oncotarget.7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu Rev Cell Dev Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- 8.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochim Biophys Acta. 2004;5:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Ray R, Lambert JR. 1alpha,25-Dihydroxyvitamin D3-3beta-bromoacetate, a potential cancer therapeutic agent: synthesis and molecular mechanism of action. Bioorg Med Chem Lett. 2011;21:2537–2540. doi: 10.1016/j.bmcl.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert JR, Eddy VJ, Young CD, Persons KS, Sarkar S, Kelly JA. A vitamin D receptor-alkylating derivative of 1alpha,25-dihydroxyvitamin D3 inhibits growth of human kidney cancer cells and suppresses tumor growth. Cancer Prev Res. 2010;12:1596–1607. doi: 10.1158/1940-6207.CAPR-10-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacamo R, Chen Y, Wang Z, Ma W, Zhang M, Spaeth EL. Reciprocal leukemia-stroma VCAM-1/VLA-4-dependent activation of NF-kB mediates chemoresistance. Blood. 2014;123:2691–2702. doi: 10.1182/blood-2013-06-511527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irby RB, Yeatman TJ. Role of Src expression and activation in human cancer. Oncogene. 2000;19:5636–5642. doi: 10.1038/sj.onc.1203912. [DOI] [PubMed] [Google Scholar]
- 13.Ishizawar R, Parsons SJ. c-Src and cooperating partners in human cancer. Cancer Cell. 2004;6:209–214. doi: 10.1016/j.ccr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Bromann PA, Korkaya H, Courtneidge SA. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene. 2004;23:7957–7968. doi: 10.1038/sj.onc.1208079. [DOI] [PubMed] [Google Scholar]
- 15.Shupnik MA. Crosstalk between steroid receptors and the c-Src-receptor tyrosine kinase pathways: implications for cell proliferation. Oncogene. 2004;23:7979–7989. doi: 10.1038/sj.onc.1208076. [DOI] [PubMed] [Google Scholar]
- 16.Burgess DJ. Breast cancer: SRC hits the mark. Nat Rev Cancer. 2011;11:314–315. doi: 10.1038/nrc3062. [DOI] [PubMed] [Google Scholar]
- 17.Elsberger B. Translational evidence on the role of Src kinase and activated Src kinase in invasive breast cancer. Crit Rev Oncol Hematol. 2014;89:343–351. doi: 10.1016/j.critrevonc.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Guarino M. Src signaling in cancer invasion. J Cell Physiol. 2009;223:14–26. doi: 10.1002/jcp.22011. [DOI] [PubMed] [Google Scholar]
- 19.Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol. 2009;6:587–595. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- 20.Wheeler DL, Iida M, Dunn EF. The role of Src in solid tumors. Oncologist. 2009;14:667–678. doi: 10.1634/theoncologist.2009-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn RS. Targeting Src in breast cancer. Ann Oncol. 2008;19:1379–1386. doi: 10.1093/annonc/mdn291. [DOI] [PubMed] [Google Scholar]
- 22.Martin GS. Fly Src: the Yin and Yang of tumor invasion and tumor suppression. Cancer Cell. 2006;9:4–6. doi: 10.1016/j.ccr.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 23.McCarthy N. Signaling: SRC and survival. Nat Rev Cancer. 2012;12:80–81. doi: 10.1038/nrc3208. [DOI] [PubMed] [Google Scholar]
- 24.Sgroi DC. Breast cancer SRC activity: bad to the bone. Cancer Cell. 2009;16:1–2. doi: 10.1016/j.ccr.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Tan M, Li P, Klos KS, Lu J, Lan KH, Nagata Y. ErbB2 promotes Src synthesis and stability: novel mechanisms of Src activation that confer breast cancer metastasis. Cancer Res. 2005;65:1858–1867. doi: 10.1158/0008-5472.CAN-04-2353. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Teng Y, Zhang Y, Liu J, Xu L, Qu J. C-Src-mediated RANKL-induced breast cancer cell migration by activation of the ERK and Akt pathway. Oncol Lett. 2012;3:395–400. doi: 10.3892/ol.2011.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun CK, Man K, Ng KT, Ho JW, Lim ZX, Cheng Q. Proline-rich tyrosine kinase 2 (Pyk2) promotes proliferation and invasiveness of hepatocellular carcinoma cells through c-Src/ERK activation. Carcinogenesis. 2008;29:2096–2105. doi: 10.1093/carcin/bgn203. [DOI] [PubMed] [Google Scholar]
- 28.Tang ZN, Zhang F, Tang P, Qi XW, Jiang J. RANKL-induced migration of MDA-MB-231 human breast cancer cells via Src and MAPK activation. Oncol Rep. 2011;26:1243–1250. doi: 10.3892/or.2011.1368. [DOI] [PubMed] [Google Scholar]
- 29.Hodgson L. Regulation of RhoGTPases in motility: A fine balancing act. Cell Adh Migr. 2014;8:525. doi: 10.1080/19336918.2014.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jimenez-Dalmaroni MJ, Heasman J, Wylie C. Control of cortical actin assembly and cadherin-catenin localization by RhoGTPases. Dev Biol. 2011;356(1):137. [Google Scholar]
- 31.Lane J, Martin TA, Jiang WG. HGF and RhoGTPases in cancer cell motility. Current Signal Transduction Therapy. 2011;6(2):173–179. [Google Scholar]
- 32.Lazer G, Katzav S. Guanine nucleotide exchange factors for RhoGTPases: good therapeutic targets for cancer therapy? Cell Signal. 2011;23:969–979. doi: 10.1016/j.cellsig.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 33.Moissoglu K, McRoberts KS, Meier JA, Theodorescu D, Schwartz MA. Rho GDP dissociation inhibitor 2 suppresses metastasis via unconventional regulation of RhoGTPases. Cancer Res. 2009;69:2838–2844. doi: 10.1158/0008-5472.CAN-08-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mokady D, Meiri D. RhoGTPases - A novel link between cytoskeleton organization and cisplatin resistance. Drug Resist Updat. 2015;19:22–32. doi: 10.1016/j.drup.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. Focal adhesion: Transmembrane junctions between the extracellular matrix and the cytoskeleton. Ann Rev Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- 36.Westermarck J, Kahari V. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999;13:781–792. [PubMed] [Google Scholar]
- 37.Saha S, Bardelli A, Buckhaults P, Velculescu VE, Rago C, St Croix B. A phosphatase associated with metastasis of colorectal cancer. Science. 2001;294:1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 38.Tryfonopoulos D, Walsh S, Collins DM, Flanagan L, Quinn C, Corkery B. Src: a potential target for the treatment of triple-negative breast cancer. Ann Oncol. 2011;22:2234–2240. doi: 10.1093/annonc/mdq757. [DOI] [PubMed] [Google Scholar]
- 39.Finn RS, Dering J, Ginther C, Wilson CA, Glaspy P, Tchekmedyian N. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/”triple-negative” breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105:319–326. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- 40.Diaz LK, Cristofanilli M, Zhou X, Welch KL, Smith TL, Yang Y. B4 integrin subunit gene expression correlates with tumor size and nuclear grade in early breast cancer. Modern Pathology. 2005;18:1165–1175. doi: 10.1038/modpathol.3800411. [DOI] [PubMed] [Google Scholar]
- 41.Den Hollander P, Rawls K, Tsimelzon A, Shepherd J, Mazumdar A, Hill J. Phosphatase PTP4A3 Promotes Triple-Negative Breast Cancer Growth and Predicts Poor Patient Survival. Cancer Res. 2016;76:1942–1953. doi: 10.1158/0008-5472.CAN-14-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]