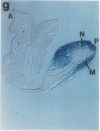
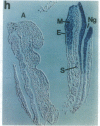
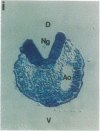
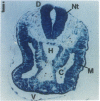
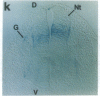
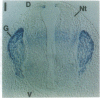
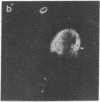
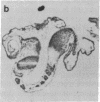
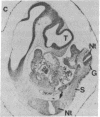
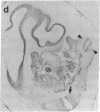
Abstract
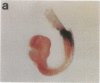
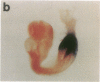
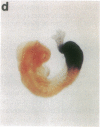
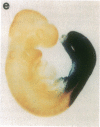
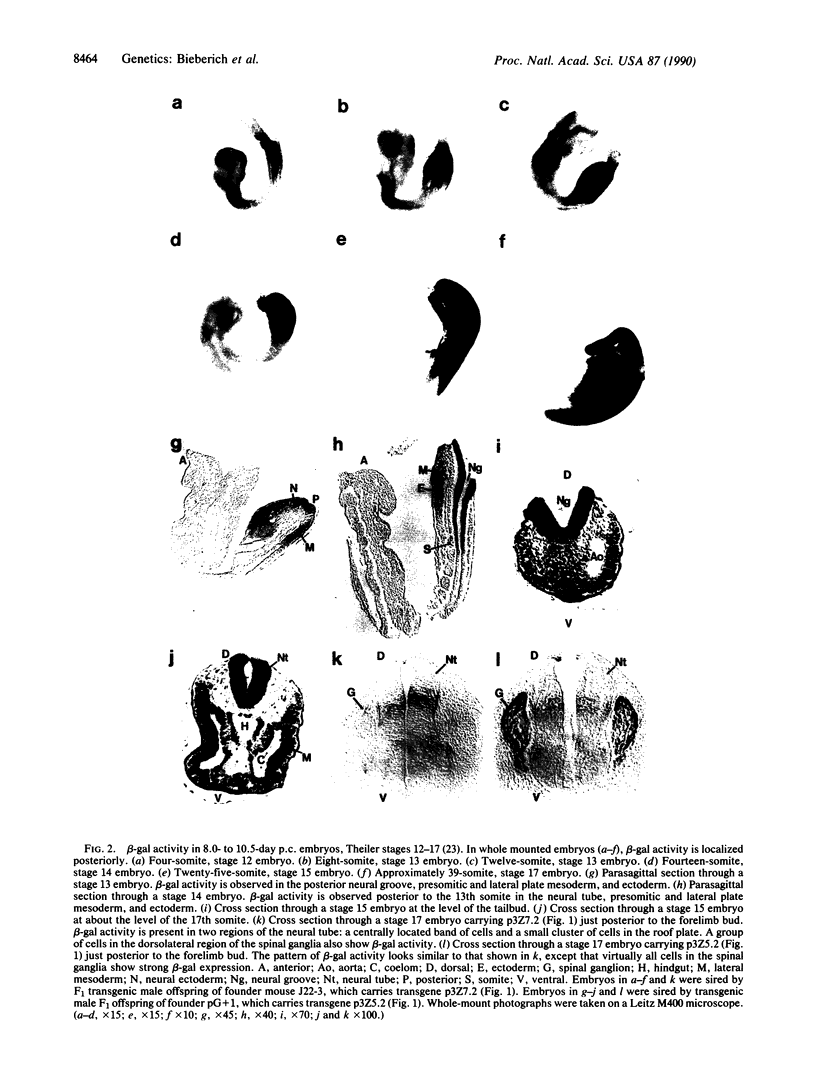
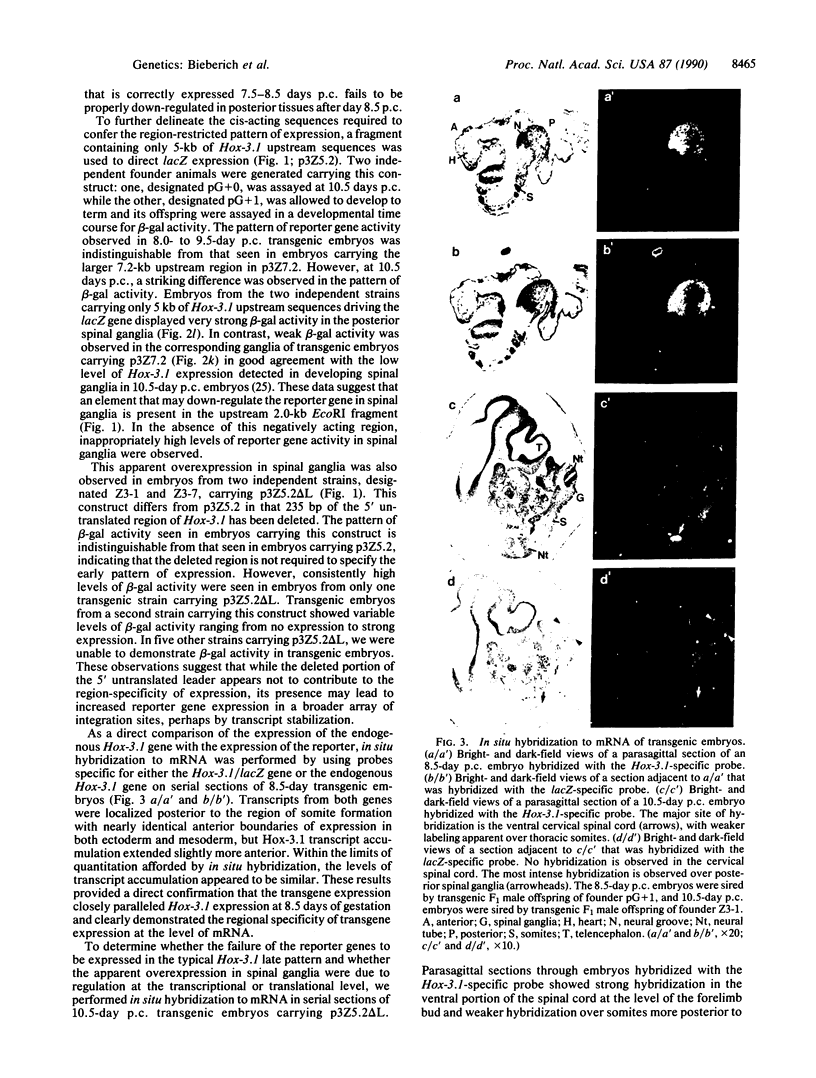
The region-specific patterns of expression of mouse homeobox genes are considered important for establishing the embryonic body plan. A 5-kilobase (kb) DNA fragment from the Hox-3.1 locus that is sufficient to confer region-specific expression to a beta-galactosidase reporter gene in transgenic mouse embryos has been defined. The observed reporter gene expression pattern closely parallels endogenous Hox-3.1 expression in 8- to 9.5-day postcoitum (p.c.) embryos. At 10.5 days p.c. and later, the pattern of beta-galactosidase activity diverges from the Hox-3.1 pattern, and an inappropriately high level of reporter gene expression is observed in posterior spinal ganglia. Inclusion of an additional 2 kb of upstream sequences is sufficient to suppress this aberrant expression in the developing spinal ganglia. Together, these results show that the control of early Hox-3.1 expression is complex, involving at least one positively acting and one negatively acting element.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987 Sep;101(1):1–22. [PubMed] [Google Scholar]
- Awgulewitsch A., Bieberich C., Bogarad L., Shashikant C., Ruddle F. H. Structural analysis of the Hox-3.1 transcription unit and the Hox-3.2--Hox-3.1 intergenic region. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6428–6432. doi: 10.1073/pnas.87.16.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awgulewitsch A., Jacobs D. Differential expression of Hox 3.1 protein in subregions of the embryonic and adult spinal cord. Development. 1990 Mar;108(3):411–420. doi: 10.1242/dev.108.3.411. [DOI] [PubMed] [Google Scholar]
- Awgulewitsch A., Utset M. F., Hart C. P., McGinnis W., Ruddle F. H. Spatial restriction in expression of a mouse homoeo box locus within the central nervous system. 1986 Mar 27-Apr 2Nature. 320(6060):328–335. doi: 10.1038/320328a0. [DOI] [PubMed] [Google Scholar]
- Bogarad L. D., Utset M. F., Awgulewitsch A., Miki T., Hart C. P., Ruddle F. H. The developmental expression pattern of a new murine homeo box gene: Hox-2.5. Dev Biol. 1989 Jun;133(2):537–549. doi: 10.1016/0012-1606(89)90056-0. [DOI] [PubMed] [Google Scholar]
- Breier G., Dressler G. R., Gruss P. Primary structure and developmental expression pattern of Hox 3.1, a member of the murine Hox 3 homeobox gene cluster. EMBO J. 1988 May;7(5):1329–1336. doi: 10.1002/j.1460-2075.1988.tb02948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. B. Zebra patterns in fly embryos: activation of stripes or repression of interstripes? Cell. 1990 Jan 12;60(1):9–16. doi: 10.1016/0092-8674(90)90711-m. [DOI] [PubMed] [Google Scholar]
- Dressler G. R., Gruss P. Anterior boundaries of Hox gene expression in mesoderm-derived structures correlate with the linear gene order along the chromosome. Differentiation. 1989 Sep;41(3):193–201. doi: 10.1111/j.1432-0436.1989.tb00747.x. [DOI] [PubMed] [Google Scholar]
- Duboule D., Baron A., Mähl P., Galliot B. A new homeo-box is present in overlapping cosmid clones which define the mouse Hox-1 locus. EMBO J. 1986 Aug;5(8):1973–1980. doi: 10.1002/j.1460-2075.1986.tb04452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D., Dollé P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989 May;8(5):1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fienberg A. A., Utset M. F., Bogarad L. D., Hart C. P., Awgulewitsch A., Ferguson-Smith A., Fainsod A., Rabin M., Ruddle F. H. Homeo box genes in murine development. Curr Top Dev Biol. 1987;23:233–256. doi: 10.1016/s0070-2153(08)60627-4. [DOI] [PubMed] [Google Scholar]
- Gaunt S. J. Mouse homeobox gene transcripts occupy different but overlapping domains in embryonic germ layers and organs: a comparison of Hox-3.1 and Hox-1.5. Development. 1988 May;103(1):135–144. doi: 10.1242/dev.103.1.135. [DOI] [PubMed] [Google Scholar]
- Gehring W. J., Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- Gordon J. W., Ruddle F. H. Gene transfer into mouse embryos: production of transgenic mice by pronuclear injection. Methods Enzymol. 1983;101:411–433. doi: 10.1016/0076-6879(83)01031-9. [DOI] [PubMed] [Google Scholar]
- Graham A., Papalopulu N., Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989 May 5;57(3):367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft G. B., Greaves D. R., Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987 Dec 24;51(6):975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Hart C. P., Awgulewitsch A., Fainsod A., McGinnis W., Ruddle F. H. Homeo box gene complex on mouse chromosome 11: molecular cloning, expression in embryogenesis, and homology to a human homeo box locus. Cell. 1985 Nov;43(1):9–18. doi: 10.1016/0092-8674(85)90007-8. [DOI] [PubMed] [Google Scholar]
- Holland P. W., Hogan B. L. Expression of homeo box genes during mouse development: a review. Genes Dev. 1988 Jul;2(7):773–782. doi: 10.1101/gad.2.7.773. [DOI] [PubMed] [Google Scholar]
- Le Mouellic H., Condamine H., Brûlet P. Pattern of transcription of the homeo gene Hox-3.1 in the mouse embryo. Genes Dev. 1988 Jan;2(1):125–135. doi: 10.1101/gad.2.1.125. [DOI] [PubMed] [Google Scholar]
- Levine M., Hoey T. Homeobox proteins as sequence-specific transcription factors. Cell. 1988 Nov 18;55(4):537–540. doi: 10.1016/0092-8674(88)90209-7. [DOI] [PubMed] [Google Scholar]
- Püschel A. W., Balling R., Gruss P. Position-specific activity of the Hox1.1 promoter in transgenic mice. Development. 1990 Mar;108(3):435–442. doi: 10.1242/dev.108.3.435. [DOI] [PubMed] [Google Scholar]
- Tuggle C. K., Zakany J., Cianetti L., Peschle C., Nguyen-Huu M. C. Region-specific enhancers near two mammalian homeo box genes define adjacent rostrocaudal domains in the central nervous system. Genes Dev. 1990 Feb;4(2):180–189. doi: 10.1101/gad.4.2.180. [DOI] [PubMed] [Google Scholar]
- Utset M. F., Awgulewitsch A., Ruddle F. H., McGinnis W. Region-specific expression of two mouse homeo box genes. Science. 1987 Mar 13;235(4794):1379–1382. doi: 10.1126/science.2881353. [DOI] [PubMed] [Google Scholar]
- Wolgemuth D. J., Behringer R. R., Mostoller M. P., Brinster R. L., Palmiter R. D. Transgenic mice overexpressing the mouse homoeobox-containing gene Hox-1.4 exhibit abnormal gut development. Nature. 1989 Feb 2;337(6206):464–467. doi: 10.1038/337464a0. [DOI] [PubMed] [Google Scholar]
- Zakany J., Tuggle C. K., Patel M. D., Nguyen-Huu M. C. Spatial regulation of homeobox gene fusions in the embryonic central nervous system of transgenic mice. Neuron. 1988 Oct;1(8):679–691. doi: 10.1016/0896-6273(88)90167-5. [DOI] [PubMed] [Google Scholar]