Abstract
Cytolytic CD8 T cells play a crucial role in the control and elimination of virus-infected cells and are a major focus of HIV cure efforts. However, it has been shown that HIV-specific CD8 T cells are infrequently found within germinal centers (GCs), a predominant site of active and latent HIV infection. We demonstrate that HIV infection induces marked changes in the phenotype, frequency, and localization of CD8 T cells within the lymph node (LN). Significantly increased frequencies of CD8 T cells in the B cell follicles and GCs were found in LNs from treated and untreated HIV-infected individuals. This profile was associated with persistent local immune activation but did not appear to be directly related to local viral replication. Follicular CD8 (fCD8) T cells, despite compromised cytokine polyfunctionality, showed good cytolytic potential characterized by high ex vivo expression of granzyme B and perforin. We used an anti-HIV/anti-CD3 bispecific antibody in a redirected killing assay and found that fCD8 T cells had better killing activity than did non-fCD8 T cells. Our results indicate that CD8 T cells with potent cytolytic activity are recruited to GCs during HIV infection and, if appropriately redirected to kill HIV-infected cells, could be an effective component of an HIV cure strategy.
INTRODUCTION
Follicular CD4 T helper (TFH) cells, which are characterized by high expression of PD-1 and CXCR5 and reside in the germinal center (GC) of secondary lymphoid organs [lymph nodes (LNs) and spleen], serve as a major site for HIV replication (1–6). This is evidenced by the fact that they harbor high amounts of HIV gag DNA and support active replication of virus in vitro (7, 8). Simian immunodeficiency virus (SIV) infection in nonhuman primates mimics this situation in which TFH cells are a source of active virus replication (9, 10). Understanding the immune populations localized within the GC and their cytolytic potential is therefore of great interest, especially when considering novel ways to eradicate HIV or SIV. In most virus infections, local recruitment of cytolytic CD8 T cells to the site of active virus replication is a major mechanism leading to elimination of infected cells. Therefore, an analysis of the phenotype and function of bulk and virus-specific CD8 T cells within the LN, and particularly the GC, could provide critical information for the design of novel immunotherapies targeting HIV-infected CD4 T cells in this anatomical compartment.
There exists, within the B cell follicle, a population of CD8 T cells that express a CXCR5high phenotype (11–13). In HIV infection, the distribution of HIV-specific CD8 T cells between the bloodstream and the LNs is in continual flux and tends to shift from bloodstream to LN predominance during the course of infection (14–16). However, a better understanding of the role of CD8 T cells in LN immune reactions requires delineating their topology within the different compartments of the LN. There are conflicting data regarding the frequency of HIV-specific CD8 T cells within GCs. Early studies revealed the presence of cytolytic CD8 T cells within the GCs of LN tissues from HIV-infected people (17–19). Some studies suggested that there was accumulation of HIV-specific CD8 T cells with cytolytic function within the splenic GCs from HIV-infected individuals (4, 20). Furthermore, exogenously engineered and reinfused autologous HIV-specific CD8 T cells could traffic to LN and localize to the follicular area (21). On the other hand, tissue staining with HIV tetramers revealed a lower frequency of HIV-specific CD8 T cells within the GC compared to extra-follicular areas (1). In SIV-infected rhesus monkeys, control of viremia was significantly correlated with the frequency of SIV-specific CD8 T cells in the LN (22, 23). However, the localization of the SIV-specific CD8 T cells within the LN was not addressed in these studies.
The use of bispecific antibodies to mobilize and redirect the cytolytic activity of CD8 T cells in HIV and cancer has been previously described (24–28). We have recently shown that an engineered antibody combining the specificity of a broadly neutralizing antibody (VRC07) to HIV-1 (29) with a monoclonal antibody against CD3 exhibits potent killing activity against HIV-infected targets (30). The use of such bispecific antibodies could lead to viral control or elimination if sufficient CD8 T cells with appropriate cytolytic potential were resident within GCs. Here, we describe the phenotype, function, and localization of CD8 T cell populations within the LN. We found an accumulation of CD8 T cells within the follicular areas and particularly within the GCs during chronic HIV infection. Furthermore, using a bispecific (aCD3/VRC07) antibody, we demonstrate that these follicular CD8 (fCD8) T cells have increased capacity for in vitro killing of HIV-infected cells. Our data further justify the potential testing of such reagents as tools for elimination of HIV-infected cells in vivo.
RESULTS
fCD8 T cells accumulate in GCs in HIV-infected LNs
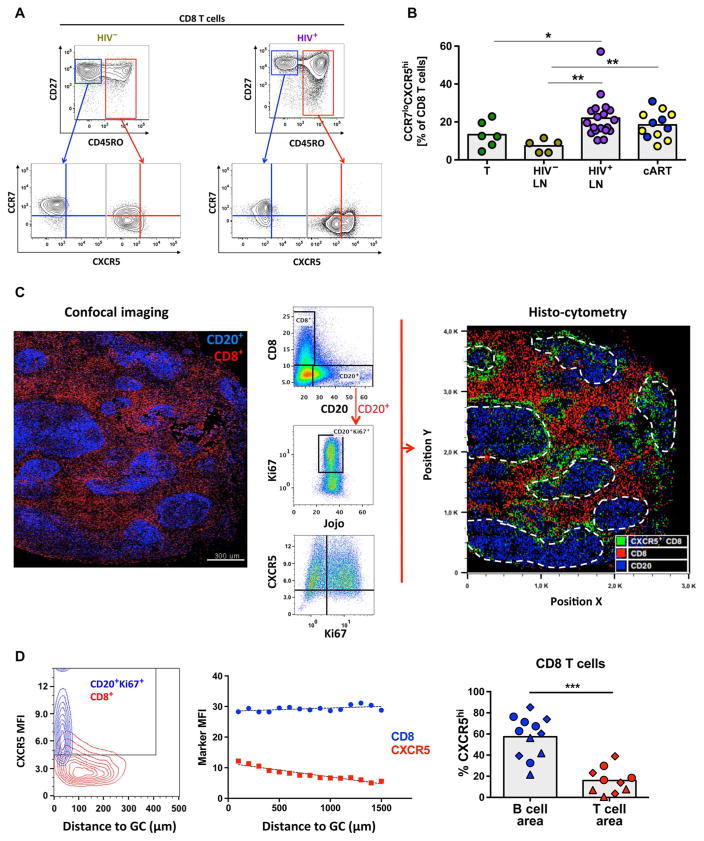
LN tissues from HIV− and HIV+ donors (table S1) and tonsils were analyzed. We characterized CD8 T cells with respect to naïve and memory subsets (CD27 and CD45RO) and the expression of CCR7 and CXCR5, chemokine receptors whose opposing actions play a major role in determining lymphocyte localization within LN (Fig. 1A and fig. S1A) (31). HIV infection, regardless of treatment status, was associated with an overall increased frequency of total and memory (CD27hi/loCD45ROhi) CD8 T cells in LNs (fig. S1, B and C). We then assessed the relative expression of CCR7 and CXCR5 on LN CD8 T cells and found an increased frequency of CCR7loCXCR5hi CD8 T cells in HIV+ LNs regardless of treatment status (Fig. 1B).
Fig. 1. Human LN fCD8 T cells express a CCR7loCXCR5hi phenotype.
(A) Gating strategy used to define CCR7loCXCR5hi CD8 T cells in flow cytometry data sets. Representative flow cytometry plots from one HIV− and one HIV+ LN are shown. (B) Pooled data showing the relative frequency (expressed as percentage of total CD8 T cells) of CD8 T cells expressing a CCR7loCXCR5hi phenotype. Cells from HIV− tonsils (n = 6), HIV− LNs (n = 5), viremic HIV+ LNs (n = 18), and short-term (n = 5, blue dots) or long-term (n = 7, yellow dots) cART-treated HIV+ LNs were analyzed. Experimental variables were analyzed by Mann-Whitney U test (shown; *P < 0.05, **P < 0.001) or Kruskal-Wallis analysis of variance (ANOVA) (P = 0.0025). (C) Histocytometry analysis of an HIV+ LN (HIV+ #22). The imaged area and distribution of CD8 and CD20 populations using a multicolor confocal assay are shown on the left. The imaging data were converted to flow cytometry type of data, and the gating scheme for the detection of particular populations is shown in the middle. CD20hiKi67hi B cells were used for the identification of GCs. The generated histo-cytometry image is shown on the right. The distribution of CD20 (blue), CD8 (red), and CXCR5hiCD8 (green) cells is shown (white dashed lines indicate GCs). (D) The mean fluorescence intensity (MFI) of CXCR5 on GC B cells and total CD8 T cells (HIV+ #22) with respect to their distance from the follicle is shown in the left panel. A similar analysis showing the MFI of CD8 and CD8-associated CXCR5 expression with respect to the distance from GC for another HIV+ LN (HIV+ #19) is shown in the middle panel. Pooled data showing the frequency of CXCR5hiCD8 T cells within individual B cell and T cell areas from viremic HIV+ LNs (n = 3) (right panel). Different symbols (circle, triangle, or diamond) represent different donors, whereas each dot represents a different B or T cell area. The Mann-Whitney U test was used for analysis; ***P < 0.0001.
We applied multiparameter histo-cytometry to further analyze the location of CD8 T cells with respect to their phenotype (32). Simultaneous expression of CD20 and Ki67 was used to identify GCs (fig. S2A). A representative example for an HIV+ LN is shown in Fig. 1C and fig. S2B. Most of the B cells express a CXCR5hi phenotype (Fig. 1, C and D). The expression of CXCR5 on LN CD8 T cells was inversely proportional to their distance from B cell follicles (Fig. 1D). We observed significantly higher frequencies of CXCR5hi CD8 T cells in GCs compared to T cell areas (57.4 ± 5.7 versus 15.9 ± 3.8, respectively; P < 0.0001) in viremic HIV+ LNs (n = 3) (Fig. 1D). On the basis of these data, we define the CCR7loCXCR5hi CD8 T cells as fCD8 T cells in agreement with previous data (11–13).
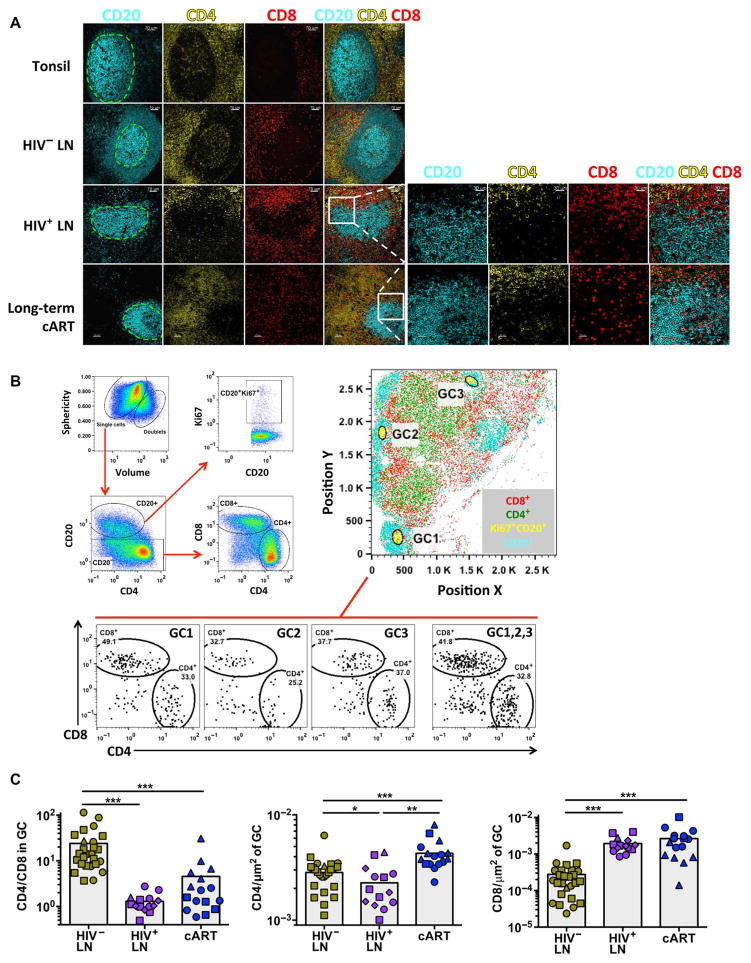
Imaging analysis confirmed an accumulation of fCD8 T cells in HIV+ LNs (Fig. 2A). Histocytometry (Fig. 2B) revealed a significant reversal in CD4/CD8 T cell ratio in GCs from viremic HIV+ compared to HIV− individuals (1.4 ± 0.16 versus 24.1 ± 5.2, CD4/CD8 ratio; P < 0.0001), which was due to both a decrease in CD4 T cells in the GCs and an increase in CD8 T cells in the GCs (Fig. 2C). Although combination antiretroviral therapy (cART) partially reversed this profile, the reversal was due to an increase in CD4 T cells in GCs, not a decrease in fCD8 T cells (Fig. 2C).
Fig. 2. fCD8 T cells accumulate in HIV-infected LNs.
(A) Confocal images showing the distribution of CD20, CD4, and CD8 T cells in representative tonsil and HIV− and HIV+ LNs (more than four samples were analyzed per group) and cART HIV+ LNs (three donors were analyzed). The green dashed lines indicate the GCs. Zoomed images of the highlighted area (white square) from a representative HIV+ LN, with or without treatment, show the presence of CD8 T cells within the GC (right panel). (B) Representative example of histo-cytometric analysis of an HIV+ LN (HIV+ #19). The gating scheme for the detection of particular populations is shown (left panel). The whole imaged area (position X, Y plot) and the position of three particular GCs (defined as CD20hiKi67hi) are shown (right panel). The relative frequency of CD4 and CD8 T cells within the individual and combined (GC1, 2, 3) GCs is shown (lower panel). (C) Pooled data of the CD4 and CD8 T cell frequencies within the GCs from HIV− (n = 3), viremic HIV+ (n = 4), and cART-treated HIV+ (n = 3) LNs. Different symbols (circle, square, triangle, or diamond) represent different donors, whereas each dot represents a different GC. Results show the CD4/CD8 T cell ratio within GCs (left panel) and the CD4 (middle panel) and CD8 (right panel) T cell frequencies normalized by the GC area. The Mann-Whitney U test was used for analysis; *P < 0.05, **P < 0.001, ***P < 0.0001.
Local viral replication does not determine fCD8 T cell accumulation
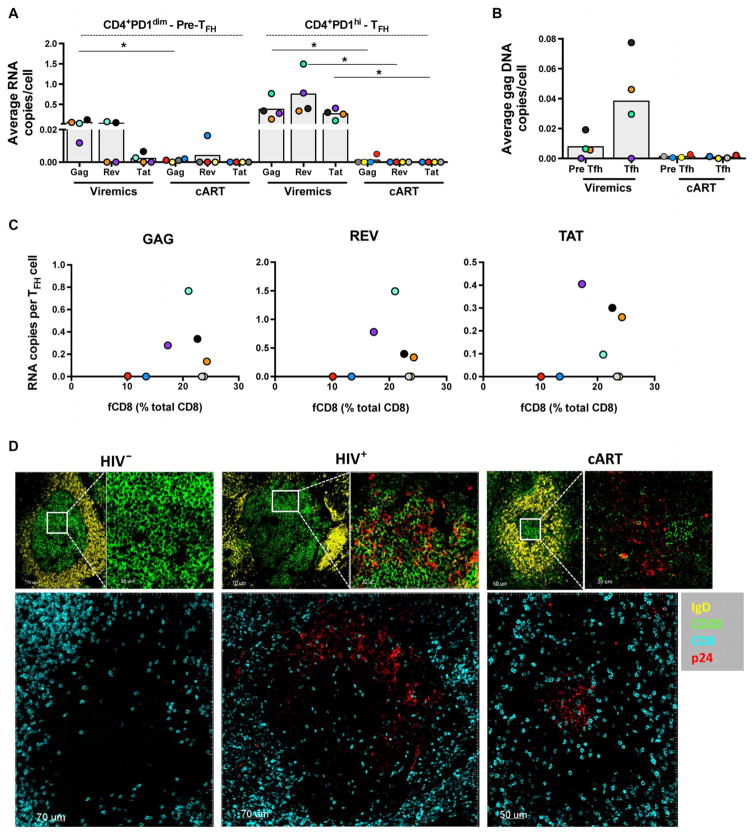
We next studied the role of local viral replication as a regulator of fCD8 T cell accumulation. High copy numbers of actively replicating virus, determined by gag and spliced rev and tat mRNA levels, were found in the TFH cell compartment, whereas lower replication was detected in the memory PD-1low/dim pre-TFH cell population (Fig. 3A). Significantly lower levels of actively transcribed virus were found in LNs from cART-treated compared to untreated viremic individuals (Fig. 3A, P = 0.0286). In line with the HIV RNA profile, a similar distribution of HIV DNA levels was found between the PD-1low/dim pre-TFH and TFH cell populations in LNs from cART-treated and viremic individuals (Fig. 3B). However, no correlation was found between the frequency of fCD8 T cells and viral transcription in TFH cells in the small number of viremic LNs tested (Fig. 3C). In addition, the duration of cART did not affect the frequency of fCD8 in LNs when individuals after short term (cART average, 8 months; 20.1% of fCD8, n = 5) or long term (cART average, 98 months; 17.2% of fCD8, n = 7) were compared. We further investigated the possible role of HIV antigens, assessed by the local expression of the p24 HIV antigen, as potential attractants for CD8 T cells in the GC. Despite similar frequencies of fCD8 T cells (Fig. 1B), a higher expression of p24 protein was found in GCs from viremic compared to cART-treated LNs (Fig. 3D and fig. S3A).
Fig. 3. No direct association between viral antigens and fCD8.
(A) Average copy number per cell of gag, rev, and tat RNA in sorted CD4+PD-1dim pre-TFH and CD4+PD-1high TFH cells from viremic (n = 4) and long-term cART-treated donors (n = 4). Each color represents a different donor. Mann-Whitney U test, *P < 0.05. (B) HIV gag DNA copies in sorted TFH CD4 T cells from viremic (n = 4) and long-term cART (n = 4) LNs and the frequency of the corresponding fCD8 are shown. (C) Relationship between the frequency of fCD8 and the average copy number per TFH cell of gag, rev, and tat RNA. (D) Representative confocal images (20×) showing the p24 antigen distribution in follicles from HIV−, HIV+, and long-term cART LNs. Follicular areas were defined by IgD and CD20 expression. Zoomed areas showing the distribution of p24+ cells within the GCs (IgDloCD20hi) are also shown. The distribution of CD8 T cells and p24+ cells in the follicles is shown in the lower panels.
The proportion of fCD8 and non-fCD8 T cells that were HIV-specific was investigated. Although no responses were found in several HIV+ LNs, our data show an accumulation of HIV-specific CD8 T cells in the extrafollicular compared to follicular areas (Fig. 4A), confirming the relative paucity of HIV-specific fCD8 T cells within GCs of HIV-infected individuals (1). These data suggest that local virus replication does not provide the direct trafficking signal for movement of fCD8 T cells to GCs during HIV infection.
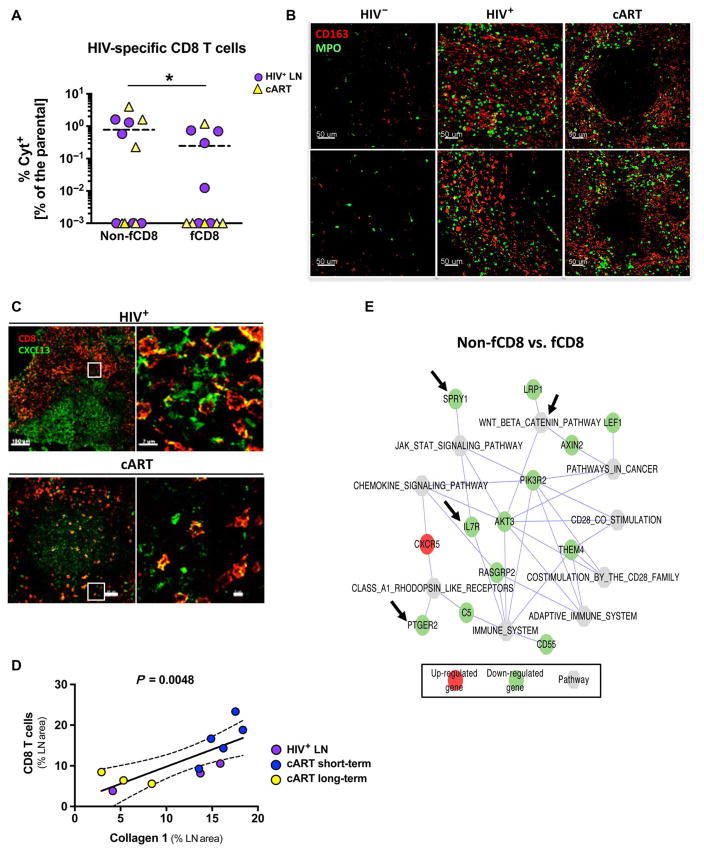
Fig. 4. Local immune activation is a driving force for fCD8 T cell dynamics.
(A) Frequency of gag-specific non-fCD8 and fCD8 T cells producing cytokines [interferon-γ (IFN-γ) and/or tumor necrosis factor–α (TNF-α) and/or macrophage inflammatory protein 1β (MIP-1β)] from HIV+ (n = 6) and long-term cART (n = 6) LNs. (B) Representative confocal images (20×) showing CD163+ monocyte infiltration and tissue activation [myeloperoxidase (MPO)] in HIV−, HIV+, and long-term cART LNs. Two representative B cell follicle areas from each tissue are shown. (C) Representative confocal images (40×) showing CD8 and CXCL13 distribution in HIV-infected LN with or without treatment. A follicular area and a zoomed area within each B cell follicle (white squares) are shown. (D) Correlation between the frequency (expressed as the LN area covered by CD8) of CD8 T cells within the LN and the amount of fibrosis (expressed as the LN area covered by collagen 1) in LNs from HIV-infected donors (three viremic, five short-term cART, and three long-term cART). (E) Network pathways and related genes analysis between non-fCD8 and fCD8 T cells generated by cytoscape software. Gene transcripts highlighted in red occurred at higher levels in fCD8 cells, and those in green occurred at higher levels in non-fCD8 cells. Specific genes or pathways mentioned in Results are highlighted with arrows.
Because local movement of cells within tissues is primarily mediated by chemokines and other chemoattractants (33), we assessed whether inflammatory signals could be operative in fCD8 T cell accumulation during HIV infection. Image analysis revealed an increased and sustained presence of monocytes/macrophages and myeloperoxidase-expressing cells (a surrogate of tissue immune activation) (34) within LNs from viremic and cART-treated subjects (Fig. 4B), indicative of ongoing immune activation. CXCL-13, the main ligand/chemoattractant for CXCR5 (35), was found in the GCs from both viremic and cART LNs, although its expression was more extensive in follicles from viremic LNs (Fig. 4C and fig. S3B). In addition, the vast majority of fCD8 T cells were positive for CXCL-13, indicative of chemokine bound on CXCR5 expressed by fCD8 T cells (Fig. 4C). Chronic HIV infection is characterized by extensive collagen deposition/fibrosis in the LNs (36, 37). Therefore, we investigated whether this process could act as a regulator of fCD8 T cell dynamics. Increased presence of collagen 1 was found in LNs from viremic individuals (including those recently treated) as compared to virologically suppressed individuals who were on long-term treatment (fig. S3C). Quantitative analysis of our imaging data showed a significant correlation between the frequency (expressed as the percentage of LN covered area) of CD8 T cells and collagen 1 (Fig. 4D, P = 0.0048). Flow cytometry analysis of all LNs available revealed a significant correlation between the frequency of total and fCD8 T cells (fig. S3C, P = 0.0021) and presumably between fCD8 T cells and collagen 1 deposition.
We further characterized the gene signature of fCD8 T cells by whole-transcriptome sequencing of sorted naïve, memory non-fCD8, and fCD8 T cells. Several genes were differentially expressed between naïve and memory nonfollicular or fCD8 T cells (fig. S4A). Notably, several genes (fig. S4B) and pathways (Fig. 4E) involved in cellular activation [that is, spry1 (38), ptger2 (39), and il-7r (40)] and memory differentiation [that is, β-catenin–related genes (41)] are down-regulated in fCD8 T cells, the effect of which would be to increase fCD8 T cell activation.
Cytokine polyfunctionality of fCD8 T cells in HIV infection is impaired
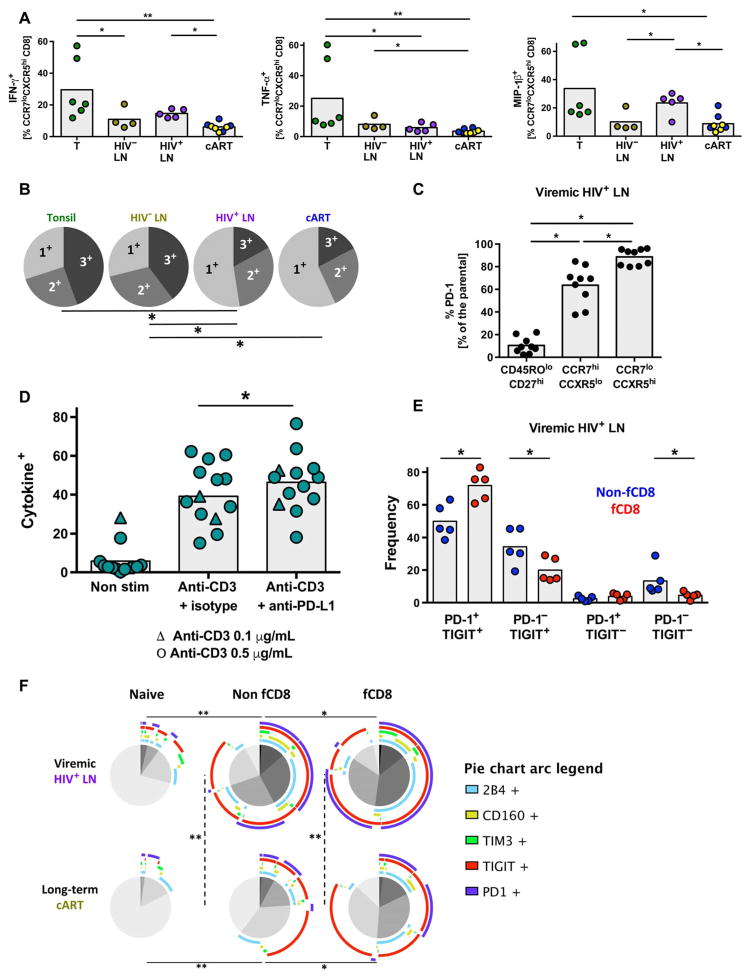
We tested the ability of fCD8 T cells to produce cytokines and chemokines after short in vitro anti-CD3 stimulation. The intrinsic ability of fCD8 T cells to produce the chemokine MIP-1β, a factor widely expressed by HIV-specific CD8 T cells (42), was greater in HIV+ compared to HIV− and cART-treated HIV+ LNs. (Fig. 5A). However, the polyfunctionality of fCD8 T cells (judged by the simultaneous expression of IFN-γ, TNF-α, and MIP-1β) was compromised in HIV infection (Fig. 5B). Next, the expression of several costimulatory/co-inhibitory receptors by HIV+ LN CD8 T cells was investigated (fig. S5A). We found significantly higher expression of PD-1, either as a frequency (P = 0.0039) or MFI (P = 0.0059), on fCD8, compared to non-fCD8, T cells (Fig. 5C and fig. S5B). This was confirmed by histo-cytometry (fig. S6A). The possible role of PD-1 as a negative regulator of fCD8 T cell function was investigated in vitro using tonsil-derived T cells. A relatively high frequency (30 to 35%) of tonsil-derived memory CD4 and CD8 T cells coexpress PD-L1 and PD-1 ex vivo. In vitro blocking of the PD-L1/PD-1 axis, using either an anti–PD-L1 or anti–PD-1 antibody in cultured tonsil-derived cells stimulated with anti-CD3, resulted in increased production of IFN-γ and TNF-α from fCD8 T cells (Fig. 5D and fig. S6, B and C), suggesting that the PD-1 expressed on fCD8 T cells is actively inhibiting their ability to produce cytokines.
Fig. 5. Expression of inhibitor receptors in fCD8 T cells.
(A) Relative frequency of IFN-γ–, TNF-α–, and MIP-1β–producing fCD8 T cells from tonsils (n = 6), HIV− LNs (n = 4), HIV+ viremic LNs (n = 5), and short-term (n = 4, blue dots) and long-term (n = 4, yellow dots) cART-treated LNs. Results are expressed as frequency of the parental fCD8 (CCR7loCXCR5hi) population. Mann-Whitney U test [shown; *P < 0.05, **P < 0.001 or ANOVA (P = 0.0006, P = 0.0005, and P = 0.0023 for IFN-γ, TNF-α, and MIP-1β, respectively)] tests were used for the analysis. (B) Simplified Presentation of Incredibly Complex Evaluations (SPICE) plots illustrating the functionality of fCD8 T cells. Each slice of the pie represents the fraction of the total response of CD8 T cells positive for a given number of cytokines (IFN-γ, TNF-α, and MIP-1β) produced, *P < 0.05. (C) Relative frequency of PD-1 expression among naïve CD8 (CD27hiCD45ROlo), non-fCD8 (CCR7hiCXCR5lo), fCD8 (CCR7loCXCR5hi), and CD4 TFH (PD-1hiCXCR5hi) T cells, Wilcoxon signed-rank test; *P < 0.05. ANOVA nonparametric test P < 0.0001. (D) Pooled data showing the frequency of total memory CD8 T cells producing cytokines (IFN-γ and/or TNF-α and/or IL-2) after a 5-day stimulation in the absence or presence of anti–PD-L1. Triangles, anti-CD3 (0.1 μg/ml); circles, anti-CD3 (0.5 μg/ml). Wilcoxon signed-rank test; *P < 0.05. ANOVA (Friedman test); P < 0.001. (E) Frequency of the PD-1 and TIGIT expression in non-fCD8 and fCD8 T cells from viremic LNs, Wilcoxon signed-rank test; *P < 0.05. (F) SPICE plots showing the coexpression of five different co-inhibitory receptors (PD-1, TIM3, TIGIT, CD160, and 2B4) in naïve, non-fCD8, and fCD8 from viremic (n = 5) and long-term cART-treated (n = 4) LNs. Arcs show the contribution of each different co-inhibitory receptor to the expression patterns. *P < 0.05, **P < 0.001.
We also assessed other potential negative regulators of fCD8 T cell function. Notably, we found that a significantly higher frequency of fCD8 T cells coexpressed PD-1 and TIGIT compared to non-fCD8 T cells (Fig. 5E, P = 0.0159). When the simultaneous expression of several inhibitory coreceptors was analyzed, fCD8 T cells were characterized by increased coexpression of inhibitory receptors compared to non-fCD8 T cells (Fig. 5F). Long-term cART was associated with relative persistence of TIGIT on nonfollicular and fCD8 T cells (Fig. 5F). Therefore, our data reveal that PD-1 and other costimulatory/co-inhibitory receptor expressions are increased on fCD8 T cells and may have an impact on fCD8 T cell function (43).
fCD8 T cells have high cytolytic potential that is retained after long-term cART
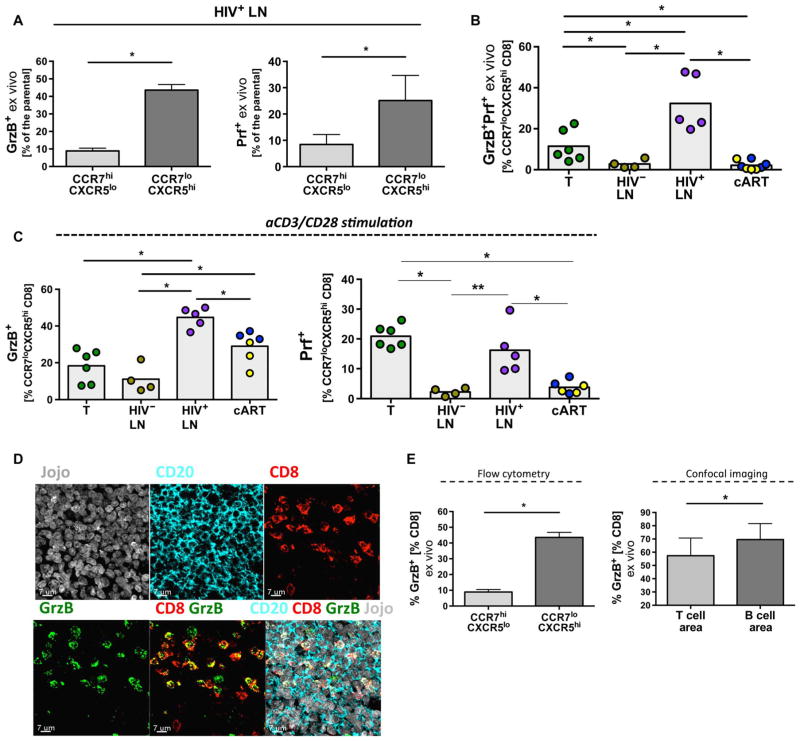
We estimated the cytolytic potential of fCD8 T cells by measuring the content of granzyme B (GrzB) and perforin (Prf) directly ex vivo or after in vitro stimulation. We applied the B-D48 anti-human Prf clone to measure de novo stimulation-induced Prf (fig. S7A) (44, 45). Higher ex vivo expression of GrzB and Prf was found in fCD8 T cells compared to CCR7hiCXCR5lo non-fCD8 T cells (Fig. 6A). Furthermore, we found a significantly higher frequency of fCD8 T cells that express GrzB and Prf directly ex vivo in LNs from viremic HIV+ compared to HIV− tissues (P = 0.0159) and tonsils (P = 0.0043) (Fig. 6B). The frequency of fCD8 T cells coexpressing GrzB and Prf was lower in cART-treated samples (Fig. 6B, P = 0.0016) owing to the loss of Prf expression. A higher frequency of fCD8 T cells from viremic and cART-treated HIV+ LN, compared to non-infected LNs and tonsils, express GrzB after short in vitro stimulation with anti-CD3 (Fig. 6C). Although significantly lower compared to viremic LNs (P = 0.0043), measurable Prf responses were also detected in fCD8 T cells from cART LNs after in vitro stimulation (Fig. 6C). However, prolonged in vitro stimulation (24 hours) induced similar amounts of soluble GrzB, Prf, and soluble Fas ligand (sFasL) in the supernatants from sorted tonsil fCD8 T cells and non-fCD8 T cells (fig. S7B).
Fig. 6. fCD8 T cells exhibit potent cytolytic ability.
(A) Frequency of CCR7hiCXCR5lo and CCR7loCXCR5hi CD8 T cells from HIV+ LNs (n = 5) expressing ex vivo GrzB or Prf. Mann-Whitney U test; *P < 0.05. (B) Frequency of fCD8 T cells co-expressing GrzB and Prf ex vivo. Results are shown as frequency of the parental fCD8 (CCR7loCXCR5hi) population, Mann-Whitney U test (shown; *P < 0.05), ANOVA (P = 0.0017) (blue dots, short-term cART; yellow dots, long-term cART). (C) Frequency of fCD8 T cells expressing GrzB or de novo synthesized Prf among the different tissues after short stimulation with aCD3/CD28. Results are shown as frequency of fCD8 T cells. Mann-Whitney U test (shown; *P < 0.05), ANOVA (P = 0.0018 and P = 0.0012 for GrzB and Prf, respectively). (D) Representative confocal images (63×) showing fCD8 T cells from a viremic HIV+ LN (GC area) expressing GrzB. Individual staining as well as merged (CD8/GrzB and CD20/CD8/GrzB/Jojo) images are shown. (E) The ratio of GrzB+CD8/CD8 in GCs and nonfollicular areas calculated by flow cytometry or imaging analysis from viremic LNs (n = 3) is shown, Mann-Whitney U test; *P < 0.05.
We confirmed that HIV infection was associated with an increased frequency of fCD8 T cells expressing GrzB using image analysis (Fig. 6D and fig. S8). By enumerating CD8 T cells in T cell and GC areas, we found that the highest frequency of CD8 T cells expressing GrzB were present within the GCs (Fig. 6E). Our data show that fCD8 T cells contain multiple effector molecules needed to carry out cytolysis.
fCD8 T cells can mediate redirected killing of HIV-infected T cells
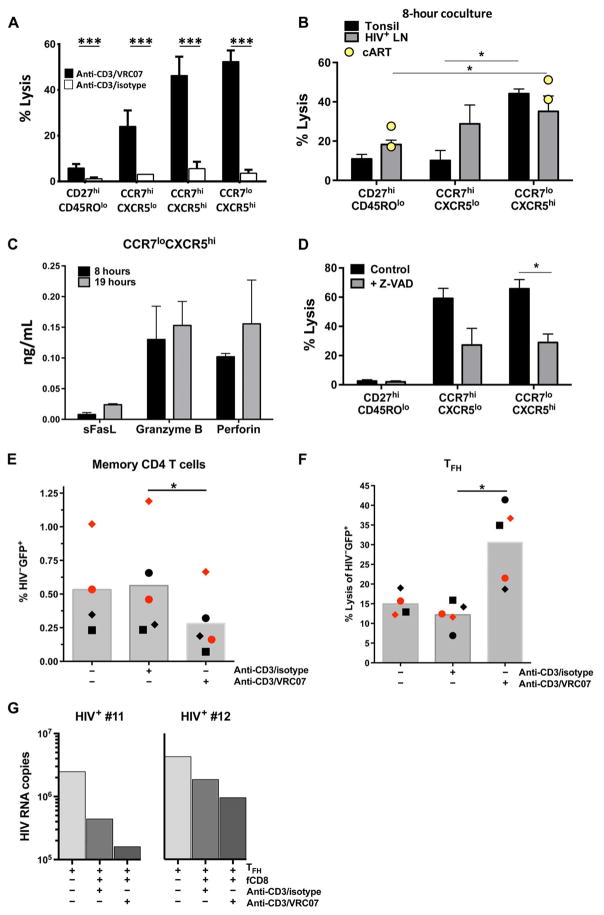
The presence of an increased number of GrzB+ CD8 T cells in the follicles of HIV-infected individuals raised the question of whether these cells could be harnessed to provide localized lytic activity against infected cells. We therefore investigated the in vitro capacity of sorted CD8 T cell populations from LNs and tonsils to mediate bispecific antibody–directed killing. We used a bispecific antibody that combines the specificity of a broadly neutralizing antibody (VRC07) to HIV-1 (29) with a monoclonal antibody against CD3 (anti-CD3/VRC07) (fig. S9A) to redirect fCD8 T cells to kill HIV-infected cells (30). Cytolysis was readily detected in the presence of the bispecific antibody (Fig. 7A and fig. S9B). The bispecific antibody–mediated lysis in this assay was HIV-specific, because only minimal killing activity was found in the presence of a control bispecific antibody lacking the VRC07 component (Fig. 7A). fCD8 T cells from tonsils and HIV-infected LNs showed higher bispecific antibody–directed killing activity compared to naïve or non-fCD8 T cells (Fig. 7, A and B), consistent with the higher cytolytic potential of fCD8 T cells compared to non-fCD8 T cells (Fig. 6A). Notably, fCD8 T cells from fully cART-suppressed HIV-infected LNs retain cytolytic ability (Fig. 7B). GrzB and Prf were readily detectable in the supernatants, suggesting that these cytolytic molecules mediate the killing in our assays (Fig. 7C). Furthermore, we found a significant reduction of the killing activity in the presence of the pan-caspase inhibitor ZVAD (Fig. 7D, P = 0.0047).
Fig. 7. fCD8 T cells mediate potent killing of HIV-infected cells via bispecific antibodies.
(A) Bispecific antibody–mediated killing activity of HIV-infected cells using an isotype antibody fragment (open bars) or the broadly neutralizing VRC07 anti-env antibody fragment (black bars) with an anti-CD3 fragment, Mann-Whitney U test; ***P < 0.001. (B) Accumulated data showing the bispecific antibody–mediated killing activity of sorted naïve CD8 (CD27hiCD45ROlo), non-fCD8 (CCR7hiCXCR5lo), and fCD8 (CCR7loCXCR5hi) from tonsils (n = 5), viremic HIV+ (n = 5) LNs, and long-term cART-treated HIV+ LNs (n = 2, yellow dots). HIV+ targets and sorted CD8 T cells were cocultured for 8 hours in the presence of the bispecific antibody, Mann-Whitney U test; *P < 0.05. (C) The concentration of sFasL, GrzB, and Prf in supernatants from the bispecific antibody–mediated killing assays after 8 hours (black bars) or 19 hours (gray bars) of incubation is shown. (D) Inhibition of the bispecific antibody–mediated killing activity by the pan-caspase inhibitor Z-VAD in a 19-hour in vitro killing assay. Sorted cells from six tonsils were used, Mann-Whitney U test; *P < 0.05. (E) Sorted primary memory CD4 T cells from HIV− PBMCs (n = 2) or (F) TFH cells from HIV− tonsils (n = 3) were infected in vitro, and the killing [judged by the frequency of GFP+ cells (left panel) or the expression of death markers-aqua, annexin V (right panel)] of cells bearing virus (GFP+) by autologous memory CD8 T cells after coculture (8 hours) in the absence or presence of bispecific antibodies is shown. Different symbols (circle, square, or diamond) represent different donors. Mann-Whitney U test (*P < 0.05). (G) Sorted TFH cells from viremic LNs (n = 2) were stimulated in vitro (anti-CD3), and the virus production (judged by the HIV gag-RNA copies) in the presence of autologous fCD8 T cells (with or without bispecific antibodies) is shown after 48 hours of coculture.
Next, we tested bispecific antibody–directed killing capacity against HIV-infected primary CD4 T cells. Sorted memory CD4 T cells from HIV− peripheral blood mononuclear cell (PBMC) preparations (n = 2) or TFH cells from HIV− tonsils (n = 3) were infected in vitro with HIV expressing a green fluorescent protein (GFP) reporter, and the killing activity of autologous memory CD8 T cells was measured using the infected CD4 T cells as targets. A significantly lower frequency of HIV-infected cells (GFP+, P = 0.0207) and increased expression of apoptotic markers within GFP+ target cells (P = 0.0270) were observed in the presence of CD8 T cells and anti-CD3/VRC07 antibody (Fig. 7, E and F, and fig. S9C). We further investigated the killing activity of the bispecific antibody and fCD8 T cells by measuring the effect upon virus production in a culture system using sorted primary TFH cells from viremic LNs (n = 2) (fig. S9D). Suppression of virus production was greatest in the presence of fCD8 T cells and anti-CD3/VRC07 (Fig. 7G). Anti-CD3/VRC07 alone did not result in viral suppression (fig. S9E), which indicates that the reduced viral release in the presence of anti-CD3/VRC07 and fCD8 T cells was not due to neutralization of the virus by anti-CD3/VRC07. Our data demonstrate that fCD8 T cells are armed with potent cytolytic activity that can be redirected to kill HIV-expressing targets.
DISCUSSION
The ability to eliminate HIV-infected cells in vivo is a major obstacle in the fight to cure HIV infection. Therapeutic vaccination and/or the use of checkpoint inhibitors to improve cytotoxic T lymphocyte (CTL) activity, and thereby augment clearance of HIV-producing cells, have been proposed as interventions that could accomplish better viral clearance in vivo as part of a shock-and-kill strategy (46, 47). For such strategies to be effective, CTLs would need access to the major sites of active HIV replication. Several lines of evidence suggest that the B cell follicle and particularly the GCs serve as major sites for active HIV replication and reactivation of the latent virus reservoir (1–5). Here, we demonstrate that chronic HIV infection is associated with trafficking of highly potent cytolytic CD8 T cells into B cell follicles and GCs. However, despite the presence of potent cytolytic CD8 T cells in follicles, these cells fail to adequately control or eliminate the virus.
Our studies provide a comprehensive analysis of the phenotype, function, localization, and gene signature of CD8 T cell populations in LNs and particularly those in the B cell follicle and GC. We applied several methods to investigate the localization of CD8 T cell populations in LN areas, including flow cytometry and quantitative confocal microscopy. Our quantitative analysis of the imaging data demonstrated a significant increase of fCD8 T cells in LNs from HIV+ individuals, consistent with our flow cytometry data. It was the combined application of multispectral confocal imaging and multi-parametric flow cytometry analysis (histo-cytometry) that allowed us to definitively and quantitatively assess T cell populations within anatomically distinct regions of the LN.
The factors that regulate LN CD8 T cell dynamics during HIV infection are not known. GCs are a site of active virus replication (7, 8), and as such, local viral expression could play a role in recruiting CD8 T cells into GCs. Because CD8 T cells do not traffic in response to cognate antigen, but rather to specific chemoattractants, it is likely that virus replication could initiate a cascade, including production of such chemoattractants, that leads to recruitment of bulk fCD8 T cells. However, the low viral expression assessed by p24 staining in the GCs especially in cART-treated donors could, at least in part, explain the relative paucity of HIV-specific CD8 T cells found in these areas.
Our data indicate that local inflammation and immune activation may serve in an antigen-nonspecific manner as the more relevant forces driving the accumulation of CD8 T cells in B cell follicles. This conclusion is supported by (i) the lack of evidence that local HIV antigens act as attractants for CD8 T cells in the GC; (ii) the presence of increased inflammatory cells and CXCL-13, the major ligand for CXCR5, in HIV+ LNs, particularly in the follicular/perifollicular areas; (iii) the significant correlation between CD8 T cells and collagen 1 deposition, a surrogate of inflammation and disease progression (48); (iv) the up-regulation of pathways associated with cell activation in the fCD8 compared to non-fCD8 T cells; and (v) the absence of preferential localization of HIV-specific CD8 T cells in follicular areas, in line with previous reports (1, 3). Contrary to CD4 T cells (36, 48), a positive correlation between LN CD8 T cells and collagen deposition was found, suggesting that CD4 T cells, but not CD8 T cells, have a distinct dependence on fibroblastic reticular cell–produced homeostatic cytokines [which are progressively lost with increased fibrosis (36)] and that alteration of the normal LN architecture affects CD4 and CD8 T cell populations differently. cART results in reduced immune activation although not to levels observed in HIV-uninfected individuals (49, 50). Our data show that although cART effectively shuts off local viral replication, local inflammatory environment and CXCL-13 levels persist compared to HIV− LNs, as does the accumulation of fCD8 T cells, lending further credence to the conclusion that inflammation, and not virus-specific signals, is primarily responsible for fCD8 T cell accumulation during HIV infection. Further longitudinal studies using LNs from individuals on cART are needed to dissect the cellular mechanisms regulating CD8 T cell trafficking in LN areas.
We found a relatively higher frequency of HIV-specific CD8 T cells expressing a nonfollicular rather than a follicular phenotype, but CD8 T cells were not completely excluded from the GC, as previously suggested (1, 3). Notably, a higher frequency of HIV-specific CD8 T cells in the extrafollicular area could, at least in part, contribute to the lower expression of HIV RNA and DNA in sorted non-TFH versus TFH CD4 cells, in line with previously described data in SIV-infected monkeys (10). Utilization of the SIV model could provide critical information regarding the cellular and molecular mechanisms governing the trafficking of CD8 T cells within the LN areas. Analysis of fCD8 T cells during different stages of infection (acute, early chronic, and late chronic) could provide additional information with respect to the relative impact of the viral replication and immune activation on fCD8 T cell maturation and dynamics.
fCD8 T cells were characterized by impaired cytokine and chemokine polyfunctionality, a profile associated with increased expression of several co-inhibitory surface receptors. Multiple negative regulators of T cell function, such PD-1 and TIGIT, were found on fCD8 T cells even in virally suppressed individuals. In vitro blocking of PD-1/PD-L1 axis increased the capacity of tissue-derived CD8 T cells for cytokine production, indicating that manipulation of checkpoint inhibitors could improve some functions of fCD8 T cells. Furthermore, the complex network of co-inhibitory receptors expressed on fCD8 T cells, even after long-term treatment, indicates that manipulation of multiple co-inhibitory signals may be needed for optimal recovery of their function.
In contrast to their impaired cytokine polyfunctionality, fCD8 T cells express potent cytolytic activity characterized by high ex vivo levels of cytolytic molecules in chronic and long-term treated HIV infection. Our imaging analysis further confirmed the accumulation of GrzB+ fCD8 T cells within the GCs in HIV-infected LNs. However, few of these cells are HIV-specific. To re-direct their killing activity, we used bispecific antibodies and demonstrated that fCD8 T cells have a potent ability to kill HIV-infected targets. Our data suggest that delivery of GrzB and/or Prf is the dominant killing mechanism in this system. We verified the cytolytic activity of fCD8 by using (i) in vitro HIV-infected cells (CEM cell line or sorted tonsil-derived primary TFH CD4 T cells) and (ii) sorted primary TFH cells from HIV+ LNs in our bispecific antibody killing assay. Although tonsils and LNs represent lymphoid organs of different anatomical position and cellularity, fCD8 T cells from both tissue types showed a similar pattern of bispecific antibody–directed killing activity further supporting their inherent cytolytic ability. Our data consistently demonstrated potent killing of HIV-infected cells by fCD8 T cells in the presence of bispecific antibodies.
The presence of follicular cytolytic CD8 T cells in mouse models of chronic infection and HIV was recently reported (12, 13). The expression of CXCR5 was necessary for their trafficking into follicles (12), although their development was critically regulated by the Bcl-6/Blimp1 axis (12, 13). In line with these data (13), we found a higher cytotoxic capacity of fCD8 compared to non-fCD8 T cells despite the expression of co-inhibitory receptors such as PD-1 and 2B4 (12, 13). In addition, when we blocked the PD-1/PD-L1 axis in vitro, we saw an increased capacity of fCD8 T cells to produce cytokines. This is consistent with the significantly higher therapeutic potential of fCD8 T cells that was observed when the PD-1/PD-L1 interaction was blocked in vivo (13).
One major limitation of our study is the absence of longitudinal LN samples that could provide definitive data regarding the role of tissue viral replication and local immune activation in the observed dynamics of fCD8 T cells. An accurate estimation of the disease duration for the tissues tested was also not possible. Therefore, analysis of fCD8 T cells during different stages of infection (acute, early chronic, and late chronic) and disease (progressors versus elite controllers) is needed. Furthermore, in the context of the current study, we were not able to investigate whether the dynamics we observed apply to LNs from different anatomical sites. Future studies using the SIV NHP model could provide critical information regarding these issues.
In line with previous reports (27, 30), our findings justify the further evaluation of bispecific antibodies for the elimination of HIV-infected cells. We should emphasize that this strategy overcomes the relatively low frequency of HIV-specific CD8 T cells in the follicle, the epitope specificity of CTLs within the B cell follicle, or even the potential presence of CTL escape variants within the latent reservoir of HIV (51). Our data indicate that fCD8 T cells persist, and continue to have strong cytolytic capacity, even after long-term suppressive cART. Hence, the use of bispecific antibodies could be considered as part of a shock-and-kill strategy in individuals on long-term suppressive ART. Further development of such a strategy could involve the testing of bispecific antibodies with differential anti-CD3 affinity as well as the inclusion of other specificities targeting surface receptors that are uniquely expressed on fCD8 T cells. The expression pattern of co-inhibitory receptors (PD-1 and TIGIT) on fCD8 T cells indicates that manipulation of checkpoint inhibitors could improve the poly-functionality of fCD8 T cells in the context of such a strategy. Our data demonstrate an accumulation of fCD8 T cells in chronic HIV and that these cells can act as potent effectors in conjunction with bispecific antibodies as part of a strategy to eliminate HIV-infected cells in vivo.
MATERIALS AND METHODS
Study design
Our objective was to characterize the dynamics of the LN fCD8 T cells in HIV. Tonsils and LNs from HIV− and HIV+ individuals were analyzed. The impact of antiretroviral treatment was investigated by analyzing LNs from short- and long-term treated donors. The phenotype, localization, functionality, gene signatures, and in vitro killing activity of LN CD8 T cell subsets were investigated by flow cytometry–based assays, quantitative multiplexed confocal imaging, and deep sequencing. We analyzed any available LN tissues we had access to. No blinding or randomization criteria were used and no outliers were excluded. The number of tissues analyzed is provided in the figure legends and in the respective sections in Supplementary Materials and Methods.
Subjects
Samples from viremic treatment naïve (n = 18, average age of 31 years old, CD4 counts = 561 cells/mm3, log10VL = 4.22 copies/ml) and short-term (n = 6, average age of 33 years old, CD4 counts = 390 cells/mm3, log10VL = 2.02 copies/ml, cART 1 to 28 months) and long-term (n = 4, average of 48 years old, CD4 counts = 700 cells/mm3, log10VL < 1.6 copies/ml, cART 2 to 17 years) cART-treated HIV+ donors were analyzed by flow cytometry. Tissues from five viremic donors (CD4 counts = 427 cells/mm3, log10VL = 4.81 copies/ml) and three cART donors (CD4 counts = 635 cells/mm3, log10VL < 1.66 copies/ml) were analyzed via confocal imaging and histo-cytometry, as previously described (32). Viral loads were measured on the same day as biopsies, and none of the patients included in this study had evidence of active opportunistic infections. Characteristics of all HIV-infected patients are shown in table S1. HIV-negative tissues were obtained from patients aged 46 to 78 years old undergoing surgical procedure for thyroid, breast cancer, or cervical lymphadenitis. They were all cancer-free and HIV-negative.
Imaging studies
Fixed, paraffin-embedded tissue sections were stained with titrated amounts of antibodies and imaged with a Leica SP8 or Nikon C2si confocal microscope. After correction of fluorochrome spillover, multiparameter images were further analyzed via the histo-cytometry pipeline (32).
Transcriptome analysis
Sorted tissue CD8 T cell subsets were subjected to whole-transcriptome sequencing analysis. Barcoded cDNA libraries were generated using the Illumina TruSeq platform, and flow cells were sequenced on an Illumina HiSeq 2000. Trimmomatic (version 0.22) was used to remove adapters and low-quality bases. The R package (“gplots”) was used for generation of heat maps, and the network analysis was generated by cytoscape software (version 2.8.3).
In vitro killing assays
The in vitro capacity of sorted tissue CD8 T cell subsets to kill HIV-infected cells was investigated using either an HIV-infected cell line (CEM) or infected primary CD4 T cells. Briefly, target infected cells were cocultured with effector CD8 T cells for 8 hours in the presence of anti-CD3/isotype or anti-CD3/VRC07 bispecific antibodies, and cell death was measured with a flow cytometry–based assay.
Statistics
Experimental variables were analyzed using the Friedman test, the non-parametric Mann-Whitney U test (for unpaired variables), the non-parametric Wilcoxon test (for paired variables), and the Kruskal-Wallis ANOVA (nonparametric) test. Bars depict mean (±SEM) values. P values <0.05 were considered significant. The GraphPad Prism statistical analysis program (GraphPad Software) was used throughout. Analysis and graphical representation of cytokine production in relation to CCR7 and CXCR5 expression were conducted using the data analysis program (SPICE, version 5.05013) provided by M. Roederer, National Institutes of Health (Bethesda, MD).
Detailed protocols for all experimental protocols applied are provided in the Supplementary Materials.
Supplementary Material
Acknowledgments
We thank the personnel of the Flow Cytometry Core at the Vaccine Research Center (VRC), National Institute of Allergy and Infectious Diseases (NIAID), NIH, the Children’s National Medical Center (for providing tonsil tissues), and the personnel of HUVR working with LN biopsies. We are grateful to J. Hu (Human Immunology Section, VRC, NIH) for providing critical help with the transcriptome analysis.
Funding: This research was supported by the Intramural Research Program of the VRC, NIAID, NIH, and CAVD grant (#OPP1032325) from the Bill and Melinda Gates Foundation (R.A.K.). This project was funded in part with federal funds from the National Cancer Institute, NIH, under contract no. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Part of the research was also supported by Redes Telemáticas de Investigación Cooperativa en Salud (RETICS; Red de SIDA, RD12/0017/0029) and Fondo de Investigacion Sanitaria (grants PI12/02283 and PI13/01912). E.R.-M. was supported by Fondo de Investigacion Sanitaria (grants CP08/00172 and CPII14/00025).
Footnotes
Author contributions: C.P. and R.A.K. conceived the study. C.P., S.F.-M., and M.Y.G. designed the experiments. C.P., S.F.-M., M.Y.G., J.P.C., A.P., C.D., A.C., J.H., D.A., S.A., P.M.D.R.E., R.P., E.M., K.L.B., E.B., and J.D.E. performed and analyzed experiments. E.R.-M., I.V., M.L., Y.A.-T., A.R., L.A.G.-H., S.M., G.R.-T., F.D., and G.P. provided human samples. A.B.M., D.C.D., M.R.B., R.N.G., and J.R.M. gave conceptual advice. J.R.M. provided the bispecific antibodies. C.P., S.F.-M., M.Y.G., and R.A.K. wrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The whole-transcriptome sequencing data were deposited at GenBank, Gene Expression Omnibus submission: GSE78522 (National Center for Biotechnology Information tracking system #17783318).
www.sciencetranslationalmedicine.org/cgi/content/full/9/373/eaag2285/DC1
Materials and Methods
Fig. S1. T cell dynamics in HIV-infected LNs.
Fig. S2. Representative example of the histo-cytometry analysis.
Fig. S3. Dynamics of fCD8 with respect to HIV antigen distribution and collagen deposition in the LN areas.
Fig. S4. Gene signatures of sorted tissue CD8 T cell populations.
Fig. S5. Expression of exhaustion markers in fCD8 T cells.
Fig. S6. Expression of PD-1 in fCD8 T cells.
Fig. S7. Functionality of fCD8 T cells.
Fig. S8. Localization of GrzB+ CD8 T cells in the GC.
Fig. S9. fCD8 T cells can mediate in vitro bispecific antibody killing.
Table S1. Demographic (age and gender) and clinical (CD4 counts, pVL, and treatment) data of the donors as well as the type of assay used for their tissue analysis are shown.
REFERENCES AND NOTES
- 1.Connick E, Mattila T, Folkvord JM, Schlichtemeier R, Meditz AL, Ray MG, McCarter MD, MaWhinney S, Hage A, White C, Skinner PJ. CTL fail to accumulate at sites of HIV-1 replication in lymphoid tissue. J Immunol. 2007;178:6975–6983. doi: 10.4049/jimmunol.178.11.6975. [DOI] [PubMed] [Google Scholar]
- 2.Embretson J, Zupancic M, Ribas JL, Burke A, Racz P, Tenner-Racz K, Haase AT. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature. 1993;362:359–362. doi: 10.1038/362359a0. [DOI] [PubMed] [Google Scholar]
- 3.Folkvord JM, Armon C, Connick E. Lymphoid follicles are sites of heightened human immunodeficiency virus type 1 (HIV-1) replication and reduced antiretroviral effector mechanisms. AIDS Res Hum Retroviruses. 2005;21:363–370. doi: 10.1089/aid.2005.21.363. [DOI] [PubMed] [Google Scholar]
- 4.Gratton S, Cheynier R, Dumaurier MJ, Oksenhendler E, Wain-Hobson S. Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc Natl Acad Sci USA. 2000;97:14566–14571. doi: 10.1073/pnas.97.26.14566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hufert FT, van Lunzen J, Janossy G, Bertram S, Schmitz J, Haller O, Racz P, von Laer D. Germinal centre CD4+ T cells are an important site of HIV replication in vivo. AIDS. 1997;11:849–857. doi: 10.1097/00002030-199707000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Pantaleo G, Graziosi C, Demarest JF, Butini L, Montroni M, Fox CH, Orenstein JM, Kotler DP, Fauci AS. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature. 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 7.Lindqvist M, van Lunzen J, Soghoian DZ, Kuhl BD, Ranasinghe S, Kranias G, Flanders MD, Cutler S, Yudanin N, Muller MI, Davis I, Farber D, Hartjen P, Haag F, Alter G, Schulze zur Wiesch J, Streeck H. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J Clin Invest. 2012;122:3271–3280. doi: 10.1172/JCI64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, Weatherall C, Bailey M, Alcantara S, De Rose R, Estaquier J, Wilson K, Suzuki K, Corbeil J, Cooper DA, Kent SJ, Kelleher AD, Zaunders J. Simian immunodeficiency virus infects follicular helper CD4 T cells in lymphoid tissues during pathogenic infection of pigtail macaques. J Virol. 2013;87:3760–3773. doi: 10.1128/JVI.02497-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, Morcock D, Swanson T, Legasse AW, Axthelm MK, Hesselgesser J, Geleziunas R, Hirsch VM, Edlefsen PT, Piatak M, Jr, Estes JD, Lifson JD, Picker LJ. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quigley MF, Gonzalez VD, Granath A, Andersson J, Sandberg JK. CXCR5+ CCR7− CD8 T cells are early effector memory cells that infiltrate tonsil B cell follicles. Eur J Immunol. 2007;37:3352–3362. doi: 10.1002/eji.200636746. [DOI] [PubMed] [Google Scholar]
- 12.Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, Zotos D, Fenix KA, Atnerkar A, Preston S, Chipman JG, Beilman GJ, Allison CC, Sun L, Wang P, Xu J, Toe JG, Lu HK, Tao Y, Palendira U, Dent AL, Landay AL, Pellegrini M, Comerford I, McColl SR, Schacker TW, Long HM, Estes JD, Busslinger M, Belz GT, Lewin SR, Kallies A, Yu D. CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol. 2016;17:1187–1196. doi: 10.1038/ni.3543. [DOI] [PubMed] [Google Scholar]
- 13.He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, Xu L, Chen X, Hao Y, Wang P, Zhu C, Ou J, Liang H, Ni T, Zhang X, Zhou X, Deng K, Chen Y, Luo Y, Xu J, Qi H, Wu Y, Ye L. Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature. 2016;537:412–428. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 14.Altfeld M, van Lunzen J, Frahm N, Yu XG, Schneider C, Eldridge RL, Feeney ME, Meyer-Olson D, Stellbrink HJ, Walker BD. Expansion of pre-existing, lymph node-localized CD8+ T cells during supervised treatment interruptions in chronic HIV-1 infection. J Clin Invest. 2002;109:837–843. doi: 10.1172/JCI14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellefsen K, Harari A, Champagne P, Bart PA, Sékaly RP, Pantaleo G. Distribution and functional analysis of memory antiviral CD8 T cell responses in HIV-1 and cytomegalovirus infections. Eur J Immunol. 2002;32:3756–3764. doi: 10.1002/1521-4141(200212)32:12<3756::AID-IMMU3756>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 16.Pantaleo G, Soudeyns H, Demarest JF, Vaccarezza M, Graziosi C, Paolucci S, Daucher MB, Cohen OJ, Denis F, Biddison WE, Sekaly RP, Fauci AS. Accumulation of human immunodeficiency virus-specific cytotoxic T lymphocytes away from the predominant site of virus replication during primary infection. Eur J Immunol. 1997;27:3166–3173. doi: 10.1002/eji.1830271213. [DOI] [PubMed] [Google Scholar]
- 17.Devergne O, Peuchmaur M, Crevon MC, Trapani JA, Maillot MC, Galanaud P, Emilie D. Activation of cytotoxic cells in hyperplastic lymph nodes from HIV-infected patients. AIDS. 1991;5:1071–1079. doi: 10.1097/00002030-199109000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Tenner-Rácz K, Rácz P, Dietrich M, Kern P, Janossy G, Veronese-Dimarzo F, Klatzmann D, Gluckman JC, Popovic M. Monoclonal antibodies to human immunodeficiency virus: Their relation to the patterns of lymph node changes in persistent generalized lymphadenopathy and AIDS. AIDS. 1987;1:95–104. [PubMed] [Google Scholar]
- 19.Tenner-Racz K, Racz P, Thomé C, Meyer CG, Anderson PJ, Schlossman SF, Letvin NL. Cytotoxic effector cell granules recognized by the monoclonal antibody TIA-1 are present in CD8+ lymphocytes in lymph nodes of human immunodeficiency virus-1-infected patients. Am J Pathol. 1993;142:1750–1758. [PMC free article] [PubMed] [Google Scholar]
- 20.Hosmalin A, Samri A, Dumaurier MJ, Dudoit Y, Oksenhendler E, Karmochkine M, Autran B, Wain-Hobson S, Cheynier R. HIV-specific effector cytotoxic T lymphocytes and HIV-producing cells colocalize in white pulps and germinal centers from infected patients. Blood. 2001;97:2695–2701. doi: 10.1182/blood.v97.9.2695. [DOI] [PubMed] [Google Scholar]
- 21.Brodie SJ, Lewinsohn DA, Patterson BK, Jiyamapa D, Krieger J, Corey L, Greenberg PD, Riddell SR. In vivo migration and function of transferred HIV-1-specific cytotoxic T cells. Nat Med. 1999;5:34–41. doi: 10.1038/4716. [DOI] [PubMed] [Google Scholar]
- 22.Fukazawa Y, Park H, Cameron MJ, Lefebvre F, Lum R, Coombes N, Mahyari E, Hagen SI, Bae JY, Reyes MD, III, Swanson T, Legasse AW, Sylwester A, Hansen SG, Smith AT, Stafova P, Shoemaker R, Li Y, Oswald K, Axthelm MK, McDermott A, Ferrari G, Montefiori DC, Edlefsen PT, Piatak M, Jr, Lifson JD, Sékaly RP, Picker LJ. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nat Med. 2012;18:1673–1681. doi: 10.1038/nm.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mudd PA, Martins MA, Ericsen AJ, Tully DC, Power KA, Bean AT, Piaskowski SM, Duan L, Seese A, Gladden AD, Weisgrau KL, Furlott JR, Kim Y-I, Veloso de Santana MG, Rakasz E, Capuano S, III, Wilson NA, Bonaldo MC, Galler R, Allison DB, Piatak M, Jr, Haase AT, Lifson JD, Allen TM, Watkins DI. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature. 2012;491:129–133. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berg J, Lötscher E, Steimer KS, Capon DJ, Baenziger J, Jäck HM, Wabl M. Bispecific antibodies that mediate killing of cells infected with human immunodeficiency virus of any strain. Proc Natl Acad Sci USA. 1991;88:4723–4727. doi: 10.1073/pnas.88.11.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chamow SM, Zhang DZ, Tan XY, Mhatre SM, Marsters SA, Peers DH, Byrn RA, Ashkenazi A, Junghans RP. A humanized, bispecific immunoadhesin-antibody that retargets CD3+ effectors to kill HIV-1-infected cells. J Immunol. 1994;153:4268–4280. [PubMed] [Google Scholar]
- 26.Bargou R, Leo E, Zugmaier G, Klinger M, Goebeler M, Knop S, Noppeney R, Viardot A, Hess G, Schuler M, Einsele H, Brandl C, Wolf A, Kirchinger P, Klappers P, Schmidt M, Riethmuller G, Reinhardt C, Baeuerle PA, Kufer P. Tumor regression in cancer patients by very low doses of a T cell–engaging antibody. Science. 2008;321:974–977. doi: 10.1126/science.1158545. [DOI] [PubMed] [Google Scholar]
- 27.Sung JAM, Pickeral J, Liu L, Stanfield-Oakley SA, Lam CYK, Garrido C, Pollara J, LaBranche C, Bonsignori M, Moody MA, Yang Y, Parks R, Archin N, Allard B, Kirchherr J, Kuruc JD, Gay CL, Cohen MS, Ochsenbauer C, Soderberg K, Liao HX, Montefiori D, Moore P, Johnson S, Koenig S, Haynes BF, Nordstrom JL, Margolis DM, Ferrari G. Dual-affinity re-targeting proteins direct T cell-mediated cytolysis of latently HIV-infected cells. J Clin Invest. 2015;125:4077–4090. doi: 10.1172/JCI82314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sloan DD, Lam CYK, Irrinki A, Liu L, Tsai A, Pace CS, Kaur J, Murry JP, Balakrishnan M, Moore PA, Johnson S, Nordstrom JL, Cihlar T, Koenig S. Targeting HIV reservoir in infected CD4 T cells by dual-affinity re-targeting molecules (DARTs) that bind HIV envelope and recruit cytotoxic T cells. PLOS Pathog. 2015;11:e1005233. doi: 10.1371/journal.ppat.1005233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudicell RS, Kwon YD, Ko S-Y, Pegu A, Louder MK, Georgiev IS, Wu X, Zhu J, Boyington JC, Chen X, Shi W, Yang Z-y, Doria-Rose NA, McKee K, O’Dell S, Schmidt SD, Chuang G-Y, Druz A, Soto C, Yang Y, Zhang B, Zhou T, Todd J-P, Lloyd KE, Eudailey J, Roberts KE, Donald BR, Bailer RT, Ledgerwood J, Mullikin JC, Shapiro L, Koup RA, Graham BS, Nason MC, Connors M, Haynes BF, Rao SS, Roederer M, Kwong PD, Mascola JR, Nabel GJ NISC Comparative Sequencing Program. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J Virol. 2014;88:12669–12682. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pegu A, Asokan M, Wu L, Wang K, Hataye J, Casazza JP, Guo X, Shi W, Georgiev I, Zhou T, Chen X, O’Dell S, Todd J-P, Kwong PD, Rao SS, Yang Z-y, Koup RA, Mascola JR, Nabel GJ. Activation and lysis of human CD4 cells latently infected with HIV-1. Nat Commun. 2015;6:8447. doi: 10.1038/ncomms9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haynes NM, Allen CDC, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 32.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: A method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37:364–376. doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi H, Kastenmüller W, Germain RN. Spatiotemporal basis of innate and adaptive immunity in secondary lymphoid tissue. Annu Rev Cell Dev Biol. 2014;30:141–167. doi: 10.1146/annurev-cellbio-100913-013254. [DOI] [PubMed] [Google Scholar]
- 34.Pallikkuth S, Micci L, Ende ZS, Iriele RI, Cervasi B, Lawson B, McGary CS, Rogers KA, Else JG, Silvestri G, Easley K, Estes JD, Villinger F, Pahwa S, Paiardini M. Maintenance of intestinal Th17 cells and reduced microbial translocation in SIV-infected rhesus macaques treated with interleukin (IL)-21. PLOS Pathog. 2013;9:e1003471. doi: 10.1371/journal.ppat.1003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ansel KM, Ngo VN, Hyman PL, Luther SA, Förster R, Sedgwick JD, Browning JL, Lipp M, Cyster JG. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 2000;406:309–314. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- 36.Zeng M, Smith AJ, Wietgrefe SW, Southern PJ, Schacker TW, Reilly CS, Estes JD, Burton GF, Silvestri G, Lifson JD, Carlis JV, Haase AT. Cumulative mechanisms of lymphoid tissue fibrosis and T cell depletion in HIV-1 and SIV infections. J Clin Invest. 2011;121:998–1008. doi: 10.1172/JCI45157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schacker TW, Reilly C, Beilman GJ, Taylor J, Skarda D, Krason D, Larson M, Haase AT. Amount of lymphatic tissue fibrosis in HIV infection predicts magnitude of HAART-associated change in peripheral CD4 cell count. AIDS. 2005;19:2169–2171. doi: 10.1097/01.aids.0000194801.51422.03. [DOI] [PubMed] [Google Scholar]
- 38.Collins S, Waickman A, Basson A, Kupfer A, Licht JD, Horton MR, Powell JD. Regulation of CD4+ and CD8+ effector responses by Sprouty-1. PLOS ONE. 2012;7:e49801. doi: 10.1371/journal.pone.0049801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B, Qu L, Yan S. Cyclooxygenase-2 promotes tumor growth and suppresses tumor immunity. Cancer Cell Int. 2015;15:106. doi: 10.1186/s12935-015-0260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omilusik KD, Best JA, Yu B, Goossens S, Weidemann A, Nguyen JV, Seuntjens E, Stryjewska A, Zweier C, Roychoudhuri R, Gattinoni L, Bird LM, Higashi Y, Kondoh H, Huylebroeck D, Haigh J, Goldrath AW. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J Exp Med. 2015;212:2027–2039. doi: 10.1084/jem.20150194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gattinoni L, Ji Y, Restifo NP. Wnt/β-catenin signaling in T-cell immunity and cancer immunotherapy. Clin Cancer Res. 2010;16:4695–4701. doi: 10.1158/1078-0432.CCR-10-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto T, Price DA, Casazza JP, Ferrari G, Nason M, Chattopadhyay PK, Roederer M, Gostick E, Katsikis PD, Douek DC, Haubrich R, Petrovas C, Koup RA. Surface expression patterns of negative regulatory molecules identify determinants of virus-specific CD8+ T-cell exhaustion in HIV infection. Blood. 2011;117:4805–4815. doi: 10.1182/blood-2010-11-317297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hersperger AR, Makedonas G, Betts MR. Flow cytometric detection of perforin upregulation in human CD8 T cells. Cytometry A. 2008;73:1050–1057. doi: 10.1002/cyto.a.20596. [DOI] [PubMed] [Google Scholar]
- 45.Makedonas G, Hutnick N, Haney D, Amick AC, Gardner J, Cosma G, Hersperger AR, Dolfi D, Wherry EJ, Ferrari G, Betts MR. Perforin and IL-2 upregulation define qualitative differences among highly functional virus-specific human CD8+ T cells. PLOS Pathog. 2010;6:e1000798. doi: 10.1371/journal.ppat.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin AR, Siliciano RF. Progress toward HIV eradication: Case reports, current efforts, and the challenges associated with cure. Annu Rev Med. 2016;67:215–228. doi: 10.1146/annurev-med-011514-023043. [DOI] [PubMed] [Google Scholar]
- 47.Barouch DH, Deeks SG. Immunologic strategies for HIV-1 remission and eradication. Science. 2014;345:169–174. doi: 10.1126/science.1255512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Estes JD. Pathobiology of HIV/SIV-associated changes in secondary lymphoid tissues. Immunol Rev. 2013;254:65–77. doi: 10.1111/imr.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klatt NR, Chomont N, Douek DC, Deeks SG. Immune activation and HIV persistence: Implications for curative approaches to HIV infection. Immunol Rev. 2013;254:326–342. doi: 10.1111/imr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Regidor DL, Detels R, Breen EC, Widney DP, Jacobson LP, Palella F, Rinaldo CR, Bream JH, Martínez-Maza O. Effect of highly active antiretroviral therapy on biomarkers of B-lymphocyte activation and inflammation. AIDS. 2011;25:303–314. doi: 10.1097/QAD.0b013e32834273ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng K, Pertea M, Rongvaux A, Wang L, Durand CM, Ghiaur G, Lai J, McHugh HL, Hao H, Zhang H, Margolick JB, Gurer C, Murphy AJ, Valenzuela DM, Yancopoulos GD, Deeks SG, Strowig T, Kumar P, Siliciano JD, Salzberg SL, Flavell RA, Shan L, Siliciano RF. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 2015;517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70:46–58. doi: 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 53.Cooper A, Garcia M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 54.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, Dewar RL, Planta A, Liu S, Metcalf JA, Mellors JW, Coffin JM. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2003;41:4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 56.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, Gordon I, Casazza J, Conan-Cibotti M, Migueles SA, Tressler R, Bailer RT, McDermott A, Narpala S, O’Dell S, Wolf G, Lifson JD, Freemire BA, Gorelick RJ, Pandey JP, Mohan S, Chomont N, Fromentin R, Chun T-W, Fauci AS, Schwartz RM, Koup RA, Douek DC, Hu Z, Capparelli E, Graham BS, Mascola JR, Ledgerwood JE VRC 601 Study Team. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med. 2015;7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandler NG, Bosinger SE, Estes JD, Zhu RTR, Tharp GK, Boritz E, Levin D, Wijeyesinghe S, Makamdop KN, del Prete GQ, Hill BJ, Timmer JK, Reiss E, Yarden G, Darko S, Contijoch E, Todd JP, Silvestri G, Nason M, Norgren RB, Jr, Keele BF, Rao S, Langer JA, Lifson JD, Schreiber G, Douek DC. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.