Abstract
The AlkB protein is a repair enzyme that uses an α-ketoglutarate/Fe(II)-dependent mechanism to repair alkyl DNA adducts. AlkB has been reported to repair highly susceptible substrates, such as 1-methyladenine and 3-methylcytosine, more efficiently in ss-DNA than in ds-DNA. Here, we tested the repair of weaker AlkB substrates 1-methylguanine and 3-methylthymine, and found that AlkB prefers to repair them in ds-DNA. We also discovered AlkB and its human homologs, ABH2 and ABH3, are able to repair the aforementioned adducts when the adduct is present in a mismatched base pair. These observations demonstrate the strong adaptability of AlkB on repairing various adducts in different environments.
Keywords: AlkB, ds-DNA repair, 1-methylguanine, 3-methylthymine, triple quadrupole-TOF mass spectrometry
INTRODUCTION
Nuclear DNA is constantly exposed to damage from exogenous and endogenous processes, generating a variety of DNA adducts in the genome.1–3 If those adducts occur in a single stranded (ss) DNA region, such as at a replication fork, they may cause mutations or replication blocks during DNA synthesis. If the lesions occur in a double stranded (ds) context, they may additionally change local DNA architecture, leading to an unstable DNA duplex, or possibly disrupt the proper recognition of specific sites by sequence-specific DNA binding proteins, such as transcription factors.1,2 To avoid these adverse effects from DNA adducts, organisms have developed an array of DNA repair pathways that are able to protect cells against lesions in both ss- and ds-DNA. One of these enzymes, the adaptive response protein AlkB of E. coli, has been reported to repair alkyl DNA adducts, such as 1-methyladenine (m1A) and 3-methylcytosine (m3C), in both ss- and ds-contexts, although it prefers repairing lesions in single stranded substrates.4–9
The AlkB protein was discovered to be a dioxygenase that uses an α-ketoglutarate/Fe(II)-dependent mechanism to oxidize the aberrant alkyl groups, ultimately restoring the undamaged DNA bases (Figure 1a).4,5 Different homologs of the E. coli AlkB protein exist in prokaryotic and eukaryotic species; nine such homologs exist in human cells (ABH1-8 and FTO).4,5 Among the nine AlkB homologs, ABH1 functions as an apyrimidinic/apurinic lyase and nucleic acid demethylase.6,7 ABH48,9 and ABH710,11 modify protein substrates and ABH812–16 is a tRNA methyltransferase and hydroxylase. ABH517–21 and FTO22–25 have been demonstrated to work on N6-methyladenine (m6A) in RNA or DNA. ABH22,26–30 and ABH331–34 are DNA repair enzymes; and the function of ABH6 remains to be established. Since the discovery of this class of enzymes, a variety of DNA adducts have been identified as substrates for AlkB and its mammalian homologs, ABH2 and ABH3, both in vitro and in vivo.1,10–13 The adducts include all of the seven N-methyl lesions occurring at the Watson-Crick (W-C) base-pairing face of the four nucleobases.13 The seven adducts include m1A, m3C, m6A, N4-methylcytosine (m4C), 1-methylguanine (m1G), N2-methylguanine (m2G), and 3-methylthymine (m3T). Among these lesions, m6A, m4C and m2G are exocyclic adducts whose structures afford the opportunity to avoid disruption of W-C base-pairing by allowing the methyl group to swivel away from the H-bond interface. By contrast, m1A/m3C/m1G/m3T have the methyl group on the nucleobase ring, which unavoidably will interfere with hydrogen-bond pairing if left unrepaired. AlkB has also been reported to repair other DNA adducts, such as 3-ethylcytosine (e3C), N2-ethylguanine, 1,N6-ethenoadenine (εA), 3,N4-ethenocytosine (εC), 1,N2-ethenoguanine (1,N2-εG), 1,N6-ethanoadenine (EA), 3,N4-α-hydroxyethanocytosine (HEC), 3,N4-α-hydroxypropanocytosine (HPC), N2-furan-2-yl-methylguanine, N2-tetrahydrofuran-2-yl-methylguanine, α-hydroxypropanoguanine, γ-hydroxypropanoguanine, and malondialdehydeguanine.35–43 The substrate scope and repair efficiency of the AlkB family enzymes have been reviewed by several papers.1,5,44,45
Figure 1.
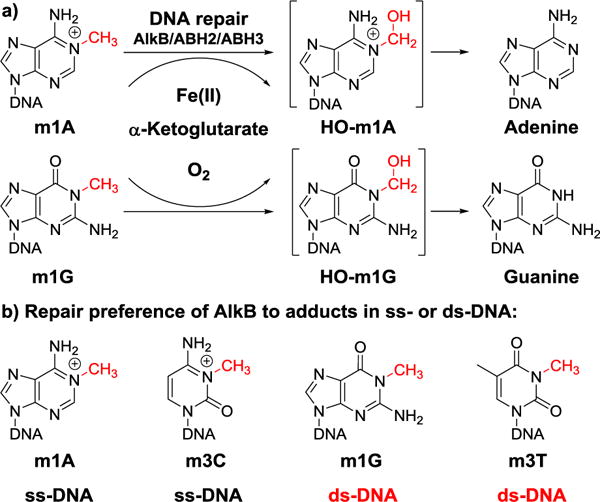
a) Repair mechanism of the AlkB family enzymes on alkyl DNA lesions. Adducts m1A and m1G are used here as examples to represent strong and weak substrates of AlkB, respectively. b) The four DNA adducts studied in this work and their strand preference (ss- vs ds-DNA) for AlkB to act on them.
EXPERIMENTAL PROCEDURES
Oligonucleotide synthesis
Sixteen-mer oligonucleotides (oligos) with the sequence 5′-GAAGACCTXGGCGTCC-3′ containing the lesions at X position were made by using solid-phase phosphoramidite chemistry36,37,40–42 on a MerMade-4 Oligonucleotide Synthesizer. The complementary 17mer oligos were synthesized with the sequence of 5′-TGGACGCCYAGGTCTTC-3′, where Y represents the position incorporating the regular bases A, C, G and T. For the ss-23mer unrelated DNA, the sequence was 5′-AAAGCTTCTGCAATCAGGTTCAG-3′. The oligos were purified by reverse-phase HPLC with two solvents. Solvent A was 100 mM 1:1 triethylamine-acetic acid (TEAA) in water and solvent B was 100% acetonitrile. The column was Phenomenex DNAPac PA-100 Semi-Preparative (9 × 250 mm, 5μm). The concentration of DNA was determined by UV absorbance at 260nm. The extinction coefficient (ε) of a certain adduct is calculated as its unmodified counterpart due to the negligible variation between the values in the context of a 16-mer DNA. The oligos were characterized by HPLC-electrospray ionization (ESI) triple quadrupole-TOF mass spectrometry (MS) (AB Sciex). Solvent A was 10mM ammonium acetate in water and solvent B was 100% acetonitrile. The column was Phenomenex Luna C 18 column (4.6 × 100 mm; 5μm). The calculated and observed monoisotopic MW and m/z value of the oligos are shown in Table S1.
Expression and purification of the AlkB, ABH2 and ABH3 proteins
The AlkB gene was cloned into a pET28a+ vector (EMD Millipore) and then transformed into E. coli Rosetta2(DE3)pLysS (EMD Millipore) cells for expression.40,41 The expressed protein was purified by affinity column chromatography, HisTrap HP (GE Healthcare Life Sciences). Thrombin (Sigma-Aldrich, 0.005U/10μg protein) was used to digest His-tag containing AlkB protein overnight. Cation-exchange column chromatography, HiTrap SP HP (GE Healthcare Life Sciences) was used for further purification. The final purified protein was concentrated by Amicon® Ultra Centrifugal Filters (EMD Millipore) and stored in the AlkB storage buffer (10 mM Tris, 100 mM NaCl, 1 mM 2-mercaptoethanol, 10% glycerol, pH 8.0). The ABH2 and ABH3 proteins were purified using the same procedure, with the exception that the E. coli cell used for expression was BL21(DE3)pLysS (EMD Millipore) instead of Rosetta2(DE3)pLysS. The ABH2 and ABH3 proteins were stored in the ABH storage buffer (50 mM N-[Tris(hydroxymethyl)methyl]-3-aminopropanesulfonic acid, 300 mM NaCl, 10% glycerol, and 1 mM 2-mercaptoethanol, pH 8.0). Protein standard was purchased from Bio-Rad.
Enzymatic reaction
All reactions were performed at 37 ℃ for 1h in a reaction buffer containing 70 μM Fe(NH4)2(SO4)2·6H2O, 0.93 mM α-ketoglutarate, 1.86 mM ascorbic acid, and 46.5 mM HEPES (pH 8.0).37,39,40 The reactions were quenched by adding 10 mM EDTA followed by heating at 95 ℃ for 5 min. Samples were then analyzed by HPLC or HPLC-electrospray ionization triple quadrupole-TOF MS (AB Sciex). Typically, the purified AlkB, ABH2 and ABH3 proteins were incubated with 5 μM DNA oligos in the presence of all cofactors in a 20 μL reaction volume. The amount for these enzymes were variable on different substrates as shown in Table S3–5. For the double-stranded DNA substrates, 1.5 equivalents (7.5 μM) of the 17mer complementary oligo containing A/C/G/T opposite to the adduct was annealed by heating the mixture at 80 ℃ for 10 min and then cooled down slowly to room temperature; the rest of the reaction was under similar conditions to ss-DNA repair reaction. For the initial velocity measurements, the reactions were stopped at 0, 0.5, 1, 4, and 10min for m1A and m3C and at 0, 4, 8, and 12 min for m1G and m3T. For the reactions with excessive amount of unrelated DNA, each reaction was carried out with 5μM substrate and additional 15μM ss-23mer DNA. The corresponding concentrations of the enzymes were listed in Table S5. Each reaction was carried out in triplicate.
LC-MS analyses
Oligonucleotide analyses were performed on AB Sciex triple quadrupole-TOF mass spectrometer. ESI was conducted using a needle voltage on 4.0 kV. Nitrogen gas was used with a setting of drying 40 L/min and a heated capillary at 600 ℃. Liquid chromatographic separation was performed using a Phenomenex Luna C 18 column (4.6 × 100 mm; 5μm) at a flow rate 0.4 mL/min. Solvent A was 10mM ammonium acetate in water and solvent B was 100% acetonitrile.40,41 A linear gradient was carried out under the following conditions: 2.0% of B for 0.5 min, 2.0 to 17.4% of B over 11 min, 17.4 to 60.0% of B over 0.1 min, 60.0% of B for 2 min, 60.0 to 2.0% of B over 0.1 min, then 2.0 % B over 3.3 min. The LC analyses were carried out with the temperature of the column oven set at 40 ℃. Data analyses were performed with the AB Sciex Analyst TF software 1.7. The relative amount of the starting material and product was quantified by integrating the peak areas of the corresponding oligos in the mass spectrum. In each reaction, we treated 16mer oligo containing the starting material had the same ionization efficiency as the 16mer product due to the negligible variation between the structures in the context of a 16-mer DNA.
Statistical analyses
All statistical analyses were carried out by IBM SPSS Statistics 23 and Microsoft Excel 2013. Statistical significance for data between two groups were performed with Student’s two-tailed t test. P-value <0.05 was considered as statistically significant.
RESULTS
AlkB has been reported to repair strong substrates, such as m1A and m3C, more efficiently in ss-DNA than in ds-DNA.4–9 However, the strand preference of AlkB for some of its weak substrates, such as m1G and m3T, remained unanswered. In this work, we selected four lesions to investigate the repair efficiency of AlkB in both ss- and ds-DNA (Figure 1b). Because these adducts are mutagenic to varying degrees, causing misincorporation of non-authentic pairing bases during replication, we also tested the ds-DNA conditions with all four DNA bases opposite to each adduct in the complementary strand to demonstrate AlkB’s repair capacity under mismatch conditions. ABH2 and ABH3 have been reported to prefer repairing DNA adducts in ds- and ss-DNA, respectively, and were also tested on those lesions.
The ability of AlkB family enzymes to repair the four methylated bases was tested in vitro using a previously reported experimental procedure.12,13 AlkB and its human homologs were purified from E. coli cells (Figure S3–5). Oligos containing the four adducts were synthesized by incorporating each adduct into a 16mer oligo in a site-specific manner and their purity and identity were characterized by LC-MS (Figure S6 and S8–11).11 For each lesion, experiments were conducted both in the presence and absence of the repair enzyme with all necessary cofactors, and the reaction products were analyzed by high resolution triple quadrupole-TOF MS (Figure 2). To test repair efficiency in ds-DNA, each adduct was first annealed to 17mer complementary oligos (Figure S7 and S12–15) with all four DNA bases opposite the lesion. To avoid the interference of the complementary oligo with the 16mer starting material in the MS spectra, a 17mer complementary oligo was chosen because it has ~300 Daltons difference in MW than the 16mer oligo. The 16mer lesion-bearing oligos demonstrated a good signal in the −4 charge envelope of the MS spectra (Figure 2). To illustrate the MS analysis, the MW of the 16mer containing an m1G lesion is calculated as 4918.87 Daltons (Table S1); its monoisotopic peak (all 12C, 14N, 16O, etc.) in the −4 charge state has a calculated mass/charge (m/z) of 1228.71. Experimentally, we observed a peak at an m/z of 1228.70 (Figure 2c). The area underneath the peak envelopes of the starting material and product were used to quantify the conversion of the reaction (Figure 2).
Figure 2.
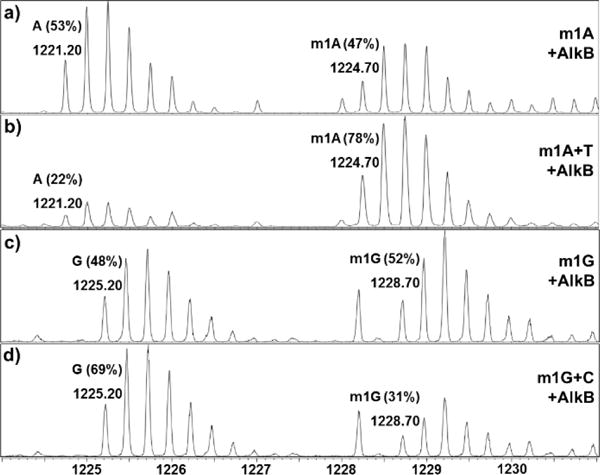
High resolution triple quadrupole -TOF MS analyses of AlkB repairing different alkyl adducts in ss- and ds-DNA. Data represent the starting material and product in their −4 charge envelopes, with the observed m/z values of their monoisotopic (all 12C, 14N, 16O, etc.) peaks labeled above each envelope. The percentage of the starting material and product in each reaction is also labeled above the corresponding peak envelopes. The peak near m/z 1228.20 in panel c and d is from a non-DNA impurity. a) AlkB + ss-m1A; b) AlkB + ds-m1A:T; c) AlkB + ss-m1G; and d) AlkB + ds-m1G:C.
For each lesion-containing oligo, 5μM starting material was used in the repair reaction. To accurately compare the efficiency of repair for each lesion, the concentration of each enzyme was optimized to ensure no reaction proceeded to 100% conversion (Table S3). Acting upon an m1A lesion in ss-DNA, AlkB converted 53% of the starting material to a 16mer product containing A at the lesion site (Figure 2a and 3a). When m1A was annealed to the 17mer oligo containing complementary base T, the theoretical perfect match in a repaired state, only 22% of m1A was repaired. For m1A paired with mismatched bases, such as A/G/C, the reactions also showed lower repair ratios than in ss-DNA (Figure 3a and Table S2). Similarly, AlkB was demonstrated to repair m3C better in ss-DNA than ds-DNA with the exception that m3C:A mismatched pair provided a slightly higher repair efficiency (Figure 3b and Table S2). These results were consistent with the previous observations of AlkB’s repair of m1A and m3C in ss- and ds-DNA.4–9
Figure 3.
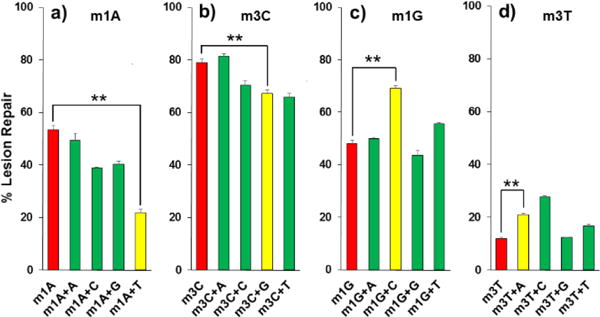
Repair efficiency of AlkB on different 16mer adducts in ss- and ds-DNA. For each lesion, the extent of repair in ss-DNA is colored in red, the extent of repair in ds-DNA with non-mutagenic pairing is colored in yellow, and the extent of repair in ds-DNA with mismatch bases is colored in green. Detailed information of enzyme concentrations is listed in Table S3. The error bars represent the standard deviation from triplicate experiments. The significance of the difference between ss-DNA and ds-DNA with non-mutagenic pairing was tested using the Student’s two-tailed t test. ** indicates p-value <0.01. a) m1A; b) m3C; c) m1G; and d) m3T.
After testing the strong substrates of AlkB (m1A and m3C), the two weak substrates m1G and m3T were investigated. For m1G, 48% of the adduct was repaired in ss-DNA, but 69% was repaired when m1G was put opposite to C (Figure 2c, 2d, 3c, and Table S2). For the mismatch reactions, complementary base A (50%) and T (56%) also showed higher repair ratios than the single-stranded reaction. For m3T repair, the ss-DNA condition (12%) provided the least efficient repair among all conditions (Table S2). The perfect match of m3T with A was repaired 21% and the mismatch with C yielded 28% conversion. Because m1A and m3C are the strong substrates of the three enzymes and m1G and m3T are the weak substrates of the proteins, the concentrations of the enzymes for different reactions varied significantly from 0.08–5.0 μM in the steady state studies (Table S3). The concentrations were optimized to make sure that the repair ratios were significantly different between ss- and ds-repair. The results from m1G and m3T reactions demonstrated AlkB repairs them more efficiently in ds-perfect match conditions than in ss-DNA in the steady state studies. To further support this conclusion, we also measured the initial velocity of the repair reactions on the adducts in ss-DNA and in the perfect match ds-DNA (Table 1). The results confirmed the preferential repair of m1G and m3T in ds-DNA, and the differences of repair efficiency between ss- and ds-conditions were statistically significant. On the other hand, the initial rates of repair on m1A and m3C were higher in ss-DNA than ds-DNA (Table 1).
Table 1.
Initial velocity measurements of AlkB on the four DNA substrates in ss- and perfect match ds-DNA. Each reaction was carried out in triplicate. The significance of the difference between ss- and ds-DNA reactions was tested using the Student’s two-tailed t test. The p-values are statistically significant.
| ss-DNA [μM·min−1] |
ds-DNA [μM·min−1] |
p-value | |
|---|---|---|---|
| m1A | 1.17 ± 0.05 | 1.03 ± 0.02 | 0.012 |
| m3C | 1.13 ± 0.01 | 0.84 ± 0.02 | <0.001 |
| m1G | 0.57 ± 0.03 | 0.88 ± 0.02 | <0.001 |
| m3T | 0.03 ± 0.01 | 0.10 ± 0.01 | 0.001 |
The current studies on AlkB’s repair of the four DNA ad-ducts were carried out in a 16mer sequence context, which is different from the situation in cell where adducts are more likely to be present in parts per thousand or less. To study the repair of AlkB with an excessive amount of non-damaged DNA, the repair reactions were performed by adding of an extra 15 μM unrelated ss-23mer DNA oligos. This condition allowed us to decrease the lesion:normal base ratio from 1:15 to 1:84. The repair efficiency also supported the previous conclusions that AlkB prefers to repair m1G and m3T in ds-DNA and m1A and m3C in ss-DNA with the repair ratio differences statistically significant (Figure S16 and Table S5).
We also tested the repair of these four adducts by AlkB’s human homologs ABH2 and ABH3. For the repair ratios of the four lesions with ABH2, the double stranded condition for the perfect match base pairs and most of the mismatch base pairs were better repaired than ss-DNA, except the m3C:T mismatched pair (54%), which demonstrated a slightly lower repair efficiency than m3C in ss-DNA (60%, Figure S1 and Table S2). For the repair of m1A and m3C by ABH3, ss-DNA repair was consistently more effective than in ds-DNA (Figure S2 and Table S2). However, we were not able to detect the reaction products of m1G and m3T because they showed very little repair susceptibility, even when ABH3 was present at a very high concentration (5 μM, Table S3). These results were consistent with previous observations that ABH2 prefers ds repair and ABH3 prefers ss repair.28,29,46–50
DISCUSSION
In the above section, we demonstrated that AlkB is able to repair DNA adducts in both ss- and ds-DNA. AlkB repairs m1A and m3C better in ss-DNA than in ds-DNA for most strand conditions. However, the preference of repairing m1G and m3T is the opposite: AlkB prefers to repair these lesions in perfect match and most of the mismatch ds-DNA. AlkB has been reported to repair those four DNA adducts both in vitro and in vivo.36,39 It has been demonstrated that m1A and m3C are strong substrates for AlkB because both are positively charged under physiological pH conditions.35,36 In AlkB deficient cells, m1A and m3C behave as strong replication blocks: only ~12% of them could be bypassed by the replication polymerase.36 m1A is not mutagenic and m3C is ~ 30% mutagenic with the predominant mutations being C to T and C to A. The existence of AlkB could completely repair m1A and m3C in cell, thus fully alleviating their toxicity and mutagenicity. AlkB can also efficiently repair e3C, εA and EA both in vitro and in vivo.36–38,40 On the other hand, m1G, m3T, 1,N2-εG and εC are weak substrates for AlkB with substantial cytotoxic and mutagenic signatures in both AlkB positive and negative cells.36,37,43 For example, m1G and m3T are only bypassed less than 10% of the replications in the absence of the AlkB protein. The presence of AlkB only slightly increases the bypass of these two adducts. For mutagenicity, m1G is ~80% mutagenic with G to T, G to A and G to C mutations without the repair of AlkB. Similarly, m3T is ~60% mutagenic with T to A and T to C mutations. The mutagenicity of m1G and m3T decreases with the presence of AlkB. The cellular results of AlkB’s repair on those four adducts support the current observation that m1A and m3C are strong substrates of AlkB, and m1G and m3T are weak substrates. If these two weak substrates are left unrepaired in cell, they will not only strongly block replication but also cause a significant increment in the percentage and type of mutations. Based on these observations, we hypothesize that AlkB could efficiently repair strong substrates, such as m1A and m3C, in ss-DNA before they encounter a polymerase. In contrast, AlkB cannot efficiently repair weak substrates, such as m1G and m3T, when they exist in ss-DNA. AlkB may have evolved to repair the weak substrates better in a ds-DNA context. This preference would avoid mutations to a certain degree, or in combination with mismatch repair, could potentially prevent mutations pre- and post-replication.
Previously, Zhu et al. identified an active site region and key amino acid residues that are responsible for the repair of m1A by AlkB and the repair of m6A by FTO and ABH5.51 By swapping the active site sequences between the two types of demethylases, they found the enhanced activity of not only AlkB on repairing m6A but also FTO and ABH5 on m1A. Similarly, Chen et al. achieved preferential ss-repair by ABH2 and ds-repair by ABH3 (the opposite strand preference for both of the enzymes) by swapping the recognition residues in their structures.27 AlkB has been reported to repair different DNA adducts in both ss- and ds-DNA. Further structural analysis should provide valuable information on AlkB’s strand preference of m1A/m3C and m1G/m3T. Besides the strand preference, we also demonstrated that all three enzymes can repair lesions in ds-DNA under mismatch conditions; some of the mismatches are even better repaired than the perfect match. These observations demonstrate the strong adaptability of AlkB on repairing various adducts under different strand and sequence contexts. The current studies utilized pH 8.0 and an iron concentration that would provide a steady state measurement. However, Maciejewska et. al have shown that AlkB’s repair efficiency depends on the pKa of substrates as well as optimal Fe(II) ion concentration.35 They demonstrated the optimal repair of m3C and HPC was achieved at pH 7.5, and the optimal repair of HEC was achieved at pH 5.8. Correspondingly, the adducts would be positively charged under the above pH conditions. The preference to positively charged adducts is possibly due to the interaction with the negatively charged aspartate residue (Asp135) in the catalytic center of AlkB. On the other hand, the poor repair efficiency on m1G and m3T could be explained, at least partially, that the methylated nitrogen atoms in those two adducts do not exist in cationic form under any pH condition (not limited to physiologically relevant pH range).35,42 The molecular and structural mechanisms of AlkB’s preference on ss- or ds-DNA warrant further investigation.
Supplementary Material
Acknowledgments
The authors want to thank the RI-INBRE program, its directors Prof. Zahir Shaikh and Prof. David Rowley, and the staff Dr. Al Bach, Ms. Kim Andrews and Ms. Patricia Murray for their kind help. We also want to thank Prof. Bingfang Yan, Prof. Bongsup Cho, Prof. Gongqin Sun, Prof. Fatemeh Akhlaghi, Dr. James C. Delaney, Dr. Lifang Xu, Ms. Sravani Adusumalli, Mr. Aram Babcock, Mr. Ang Cai, Ms. Yixin Cui, and Ms. Zhengxi Wei for their support and helpful discussions.
Funding Sources
This work was supported by an Institutional Development Award from the National Institute of General Medical Sciences of the National Institutes of Health under grant number 2 P20 GM103430, and a Medical Research Funds grant from the Rhode Island foundation (to D. L.). This work was also supported, in whole or in part, by National Institutes of Health Grants P01 CA26731, R37 CA080024, and P30 ES002109 (to J. M. E.). C. L. D. is a Howard Hughes Medical Institute Investigator.
ABBREVIATIONS
- m1A
1-methyladenine
- m3C
3-methylcytosine
- m1G
1-methylguanine
- m3T
3-methylthymine
- m6A
N6-methyladenine
- e3C
3-ethylcytosine
- εA
1,N6-ethenoadenine
- εC
3,N4-ethenocytosine
- 1
N2-εG, 1,N2-ethenoguanine
- EA
1,N6-ethanoadenine
- HEC
3,N4-α-hydroxyethanocytosine
- HPC
3,N4-α-hydroxypropanocytosine
- ESI
electrospray ionization
- TOF
time-of-flight, MS, mass spectrometry
- ss
single stranded
- ds
double stranded
Footnotes
Supporting Information
Five supplementary tables and sixteen supplementary figures (in PDF). This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
References
- 1.Yi C, He C. DNA repair by reversal of DNA damage. Cold Spring Harb Perspect Biol. 2013;5:a012575. doi: 10.1101/cshperspect.a012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu D, Samson LD. Direct repair of 3,N(4)-ethenocytosine by the human ALKBH2 dioxygenase is blocked by the AAG/MPG glycosylase. DNA Repair. 2012;11:46–52. doi: 10.1016/j.dnarep.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shrivastav N, Li D, Essigmann JM. Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-research0007. RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair. 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Silvestrov P, Müller TA, Clark KN, Hausinger RP, Cisneros GA. Homology modeling, molecular dynamics, and site-directed mutagenesis study of AlkB human homolog 1 (ALKBH1) J Mol Graph Model. 2014;54:123–130. doi: 10.1016/j.jmgm.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westbye MP, Feyzi E, Aas PA, Vågbø CB, Talstad VA, Kavli B, Hagen L, Sundheim O, Akbari M, Liabakk NB, Slupphaug G, Otterlei M, Krokan HE. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem. 2008;283:25046–25056. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li MM, Nilsen A, Shi Y, Fusser M, Ding YH, Fu Y, Liu B, Niu Y, Wu YS, Huang CM, Olofsson M, Jin KX, Lv Y, Xu XZ, He C, Dong MQ, Rendtlew Danielsen JM, Klungland A, Yang YG. ALKBH4-dependent de-methylation of actin regulates actomyosin dynamics. Nat Commun. 2013;4:1832. doi: 10.1038/ncomms2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjørnstad LG, Zoppellaro G, Tomter AB, Falnes PØ, Andersson KK. Spectroscopic and magnetic studies of wild-type and mutant forms of the Fe(II)- and 2-oxoglutarate-dependent decarboxylase ALKBH4. Biochem J. 2011;434:391–398. doi: 10.1042/BJ20101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu D, Jordan JJ, Samson LD. Human ALKBH7 is required for alkylation and oxidation-induced programmed necrosis. Genes Dev. 2013;27:1089–1100. doi: 10.1101/gad.215533.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, He Q, Feng C, Liu Y, Deng Z, Qi X, Wu W, Mei P, Chen Z. The atomic resolution structure of human AlkB homolog 7 (ALKBH7), a key protein for programmed necrosis and fat metabolism. J Biol Chem. 2014;289:27924–27936. doi: 10.1074/jbc.M114.590505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu D, Brophy JAN, Chan CTY, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol. 2010;30:2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y, Dai Q, Zhang W, Ren J, Pan T, He C. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew Chem Int Ed Engl. 2010;49:8885–8888. doi: 10.1002/anie.201001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Born E, Vågbø CB, Songe-Møller L, Leihne V, Lien GF, Leszczynska G, Malkiewicz A, Krokan HE, Kirpekar F, Klungland A, Falnes PØ. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun. 2011;2:172. doi: 10.1038/ncomms1173. [DOI] [PubMed] [Google Scholar]
- 15.Pastore C, Topalidou I, Forouhar F, Yan AC, Levy M, Hunt JF. Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification. J Biol Chem. 2012;287:2130–2143. doi: 10.1074/jbc.M111.286187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zdżalik D, Vågbø CB, Kirpekar F, Davydova E, Puścian A, Maciejewska AM, Krokan HE, Klungland A, Tudek B, van den Born E, Falnes PØ. Protozoan ALKBH8 oxygenases display both DNA repair and tRNA modification activities. PloS One. 2014;9:e98729. doi: 10.1371/journal.pone.0098729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu C, Liu K, Tempel W, Demetriades M, Aik W, Schofield CJ, Min J. Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J Biol Chem. 2014;289:17299–17311. doi: 10.1074/jbc.M114.550350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen F, Huang W, Huang JT, Xiong J, Yang Y, Wu K, Jia GF, Chen J, Feng YQ, Yuan BF, Liu SM. Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J Clin Endocrinol Metab. 2015;100:E148–154. doi: 10.1210/jc.2014-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thalhammer A, Bencokova Z, Poole R, Loenarz C, Adam J, O’Flaherty L, Schödel J, Mole D, Giaslakiotis K, Schofield CJ, Hammond EM, Ratcliffe PJ, Pollard PJ. Human AlkB homologue 5 is a nuclear 2-oxoglutarate dependent oxygenase and a direct target of hypoxia-inducible factor 1α (HIF-1α) PloS One. 2011;6:e16210. doi: 10.1371/journal.pone.0016210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RPG, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng C, Liu Y, Wang G, Deng Z, Zhang Q, Wu W, Tong Y, Cheng C, Chen Z. Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J Biol Chem. 2014;289:11571–11583. doi: 10.1074/jbc.M113.546168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Mou S, Cai Y. FTO gene variant and risk of overweight and obesity among children and adolescents: a systematic review and meta-analysis. PloS One. 2013;8:e82133. doi: 10.1371/journal.pone.0082133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X, Yang Y, Sun BF, Zhao YL, Yang YG. FTO and obesity: mechanisms of association. Curr Diab Rep. 2014;14:486. doi: 10.1007/s11892-014-0486-0. [DOI] [PubMed] [Google Scholar]
- 25.Chandola U, Das R, Panda B. Role of the N6-methyladenosine RNA mark in gene regulation and its implications on development and disease. Brief Funct Genomics. 2014;14:169–179. doi: 10.1093/bfgp/elu039. [DOI] [PubMed] [Google Scholar]
- 26.Li P, Gao S, Wang L, Yu F, Li J, Wang C, Li J, Wong J. ABH2 couples regulation of ribosomal DNA transcription with DNA alkylation repair. Cell Rep. 2013;4:817–829. doi: 10.1016/j.celrep.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 27.Chen B, Liu H, Sun X, Yang CG. Mechanistic insight into the recognition of single-stranded and double-stranded DNA substrates by ABH2 and ABH3. Mol Biosyst. 2010;6:2143–2149. doi: 10.1039/c005148a. [DOI] [PubMed] [Google Scholar]
- 28.Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yi C, Jia G, Hou G, Dai Q, Zhang W, Zheng G, Jian X, Yang CG, Cui Q, He C. Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase. Nature. 2010;468:330–333. doi: 10.1038/nature09497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nay SL, Lee DH, Bates SE, O’Connor TR. Alkbh2 protects against lethality and mutation in primary mouse embryonic fibroblasts. DNA Repair. 2012;11:502–510. doi: 10.1016/j.dnarep.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dango S, Mosammaparast N, Sowa ME, Xiong LJ, Wu F, Park K, Rubin M, Gygi S, Harper JW, Shi Y. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol Cell. 2011;44:373–384. doi: 10.1016/j.molcel.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koike K, Ueda Y, Hase H, Kitae K, Fusamae Y, Masai S, Inagaki T, Saigo Y, Hirasawa S, Nakajima K, Ohshio I, Makino Y, Konishi N, Yamamoto H, Tsujikawa K. anti-tumor effect of AlkB homolog 3 knockdown in hormone- independent prostate cancer cells. Curr Cancer Drug Targets. 2012;12:847–856. doi: 10.2174/156800912802429283. [DOI] [PubMed] [Google Scholar]
- 33.Calvo JA, Meira LB, Lee CYI, Moroski-Erkul CA, Abolhassani N, Taghizadeh K, Eichinger LW, Muthupalani S, Nordstrand LM, Klungland A, Samson LD. DNA repair is indispensable for survival after acute inflammation. J Clin Invest. 2012;122:2680–2689. doi: 10.1172/JCI63338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundheim O, Vågbø CB, Bjørås M, Sousa MML, Talstad V, Aas PA, Drabløs F, Krokan HE, Tainer JA, Slupphaug G. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006;25:3389–3397. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maciejewska AM, Poznanski J, Kaczmarska Z, Krowisz B, Nieminuszczy J, Polkowska-Nowakowska A, Grzesiuk E, Kusmierek JT. AlkB dioxygenase preferentially repairs protonated substrates: specificity against exocyclic adducts and molecular mechanism of action. J Biol Chem. 2013;288:432–441. doi: 10.1074/jbc.M112.353342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat Struct Mol Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 38.Frick LE, Delaney JC, Wong C, Drennan CL, Essigmann JM. Alleviation of 1,N6-ethanoadenine geno-toxicity by the Escherichia coli adaptive response protein AlkB. Proc Natl Acad Sci U S A. 2007;104:755–760. doi: 10.1073/pnas.0607377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li D, Delaney JC, Page CM, Chen AS, Wong C, Drennan CL, Essigmann JM. Repair of DNA Alkylation Damage by the Escherichia coli Adaptive Response Protein AlkB as Studied by ESI-TOF Mass Spectrometry. J Nucleic Acids. 2010;2010:369434. doi: 10.4061/2010/369434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li D, Delaney JC, Page CM, Yang X, Chen AS, Wong C, Drennan CL, Essigmann JM. Exocyclic carbons adjacent to the N6 of adenine are targets for oxidation by the Escherichia coli adaptive response protein AlkB. J Am Chem Soc. 2012;134:8896–8901. doi: 10.1021/ja3010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D, Fedeles BI, Shrivastav N, Delaney JC, Yang X, Wong C, Drennan CL, Essigmann JM. Removal of N-Alkyl Modifications from N(2)-Alkylguanine and N(4)-Alkylcytosine in DNA by the Adaptive Response Protein AlkB. Chem Res Toxicol. 2013;26:1182–1187. doi: 10.1021/tx400096m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh V, Fedeles BI, Li D, Delaney JC, Kozekov ID, Kozekova A, Marnett LJ, Rizzo CJ, Essigmann JM. Mechanism of repair of acrolein- and malondialdehyde-derived exocyclic guanine adducts by the α-ketoglutarate/Fe(II) dioxygenase AlkB. Chem Res Toxicol. 2014;27:1619–1631. doi: 10.1021/tx5002817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang S, Fedeles BI, Wu J, Delaney JC, Li D, Zhao L, Christov PP, Yau E, Singh V, Jost M, Drennan CL, Marnett LJ, Rizzo CJ, Levine SS, Guengerich FP, Essigmann JM. Next-generation sequencing reveals the biological significance of the N(2),3-ethenoguanine lesion in vivo. Nucleic Acids Res. 2015;43:5489–5500. doi: 10.1093/nar/gkv243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng G, Fu Y, He C. Nucleic acid oxidation in DNA damage repair and epigenetics. Chem Rev. 2014;114:4602–4620. doi: 10.1021/cr400432d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM. The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J Biol Chem. 2015;290:20734–20742. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falnes PØ, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 47.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 48.Aas PA, Otterlei M, Falnes PO, Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, Krokan HE. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 49.Falnes PØ, Bjørås M, Aas PA, Sundheim O, Seeberg E. Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:3456–3461. doi: 10.1093/nar/gkh655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy TW, Bhagwat AS. Kinetic studies of Escherichia coli AlkB using a new fluorescence-based assay for DNA demethylation. Nucleic Acids Res. 2007;35:e147. doi: 10.1093/nar/gkm1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu C, Yi C. Switching demethylation activities between AlkB family RNA/DNA demethylases through exchange of active-site residues. Angew Chem Int Ed Engl. 2014;53:3659–3662. doi: 10.1002/anie.201310050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


