Abstract
Mesenchymal stem cells (MSCs) are an excellent source for numerous cellular therapies due to their simple isolation, low immunogenicity, multipotent differentiation potential and regenerative secretion profile. However, over-expanded MSCs show decreased therapeutic efficacy. This shortcoming may be circumvented by identifying methods that promote self-renewal of MSCs in culture. HMGA2 is a DNA-binding protein that regulates self-renewal in multiple types of stem cells through chromatin remodeling, but its impact on human bone marrow-derived MSCs is not known. Using an isolation method to obtain pure MSCs within 9 days in culture, we show that expression of HMGA2 quickly decreases during early expansion of MSCs, while let-7 microRNAs (which repress HMGA2) are simultaneously increased. Remarkably, we demonstrate that FGF-2, a growth factor commonly used to promote self-renewal in MSCs, rapidly induces HMGA2 expression in a time-and concentration-dependent manner. The signaling pathway involves FGF-2 receptor 1 (FGFR1) and ERK1/2, but acts independent from let-7. By silencing HMGA2 using shRNAs, we demonstrate that HMGA2 is necessary for MSC proliferation. However, we also show that over-expression of HMGA2 does not increase cell proliferation, but rather abrogates the mitogenic effect of FGF-2, possibly through inhibition of FGFR1. In addition, using different methods to assess in vitro differentiation, we show that modulation of HMGA2 inhibits adipogenesis, but does not affect osteogenesis of MSCs. Altogether, our results show that HMGA2 expression is associated with highly proliferating MSCs, is tightly regulated by FGF-2, and is involved in both proliferation and adipogenesis of MSCs.
Keywords: BONE MARROW STROMAL CELLS, MESENCHYMAL STEM CELLS, HIGH MOBILITY GROUP A2, FIBROBLAST GROWTH FACTOR 2, SELF-RENEWAL
Multipotent bone marrow stromal cells/mesenchymal stem cells (MSCs) are good candidates for multiple cellular therapies due to both their differentiation potential and their paracrine activity that promotes wound repair, induces angiogenesis, and modulates the immune system, among others [Caplan and Correa, 2011]. Currently, more than 350 clinical trials are using MSCs for a wide plethora of conditions, encompassing tissue regeneration, immunomodulation, graft enhancement, and other indications [Trounson and McDonald, 2015]. The key to success of these trials relies on the ease by which human MSCs can be culture-expanded [Mendicino et al., 2014] and their capacity to evade the immune system [Ankrum et al., 2014], thus enabling allogeneic transplantation from a single donor into multiple patients without tissue matching. However, during standard culture conditions, proliferation of MSCs decreases over time; MSCs undergo senescence over long-term culture (several weeks) [Shibata et al., 2007; Isern et al., 2013; Mendicino et al., 2014], but are also susceptible to subtle changes in gene expression, even during very early stages of cell expansion [Isern et al., 2013]. Importantly, recent reports suggest that the therapeutic efficacy of MSCs is decreased with over-expansion of the cells, weeks before cellular senescence manifests [von Bahr et al., 2012; Wuchter et al., 2015]. A common strategy to improve MSC expansion, is by supplementing the culture medium with basic fibroblast growth factor (bFGF or FGF-2), which promotes proliferation of the cells while maintaining their stem cell properties (i.e., self-renewal) [Tsutsumi et al., 2001; Jiang et al., 2002; Solchaga et al., 2005; Ng et al., 2008].
High mobility group A-2 (HMGA2) is a chromatin binding protein widely expressed during embryogenesis, undetectable in most adult tissues and associated with multiple types of cancer. HMGA2 affects gene expression by binding to DNA, modifying its conformation and consequently affecting the binding of transcription factors [Fusco and Fedele, 2007]. In many types of stem cells, including neural stem cells (NSC) [Cimadamore et al., 2013], hematopoietic stem cells (HSC) [Copley et al., 2013], embryonic stem cells (ESC) [Melton et al., 2010], and induced pluripotent stem cells (iPSC) [Worringer et al., 2014] HMGA2 expression is critical to promote self-renewal. In addition, it has been shown that the microRNA (miRNA) family let-7 directly binds to HMGA2 mRNA, repressing protein translation [Mayr et al., 2007]. Hence, maintenance of self-renewal relies on low let-7 levels and high expression of HMGA2. Among all miRNA expressed in MSCs, the let-7 family is strongly represented [Clark et al., 2014]. Ten mature let-7 members have been identified in humans [Roush and Slack, 2008], from which at least seven are highly expressed in MSCs. Thus, we hypothesized that MSCs would alter their proliferation and differentiation potential by modulation of the let-7/HMGA2 axis. Even more, since FGF-2 promotes self-renewal in MSCs, we examined if exposure to FGF-2 would affect expression of HMGA2.
In this work, we show that during early stages of expansion of MSCs, HMGA2 expression is rapidly decreased while let-7 miRNAs are increased. Interestingly, FGF-2 induces a rapid up-regulation of HMGA2, but the mechanism is independent from let-7. Furthermore, silencing and over-expressing HMGA2 during proliferation and differentiation assays, demonstrate a cross-talk with FGF-2 signaling, where, in general, HMGA2 levels have to be tightly regulated for normal cell function.
MATERIALS AND METHODS
MSCs ISOLATION
MSCs were isolated using two different methods. For studies related to early expansion of MSCs (Fig. 1), a CD45 negative selection was applied to bone marrow mononuclear cells (MNC). Briefly, bone marrow aspirates from healthy human donors (StemExpress, Placerville, CA) were diluted with equal volume of phosphate-buffered saline (PBS) and centrifuged over Ficoll (GE Healthcare, Waukesha, WI) for 30 min at 700g. Then MNC were incubated with a magnetic bead-coupled antibody against the pan-hematopoietic marker CD45, following manufacturers’ instructions (Miltenyi Biotec, Auburn, CA). Enrichment of isolated cells (CD45 negative) was confirmed by flow cytometry, by incubation of MNC with an R-phycoerythrin-conjugated antibody against CD45 (BD Biosciences, San Jose, CA) for 30 min at room temperature. CD45− MNC were then plated into tissue culture flasks using standard culture medium, which was Minimum Essential Medium α (MEM α, Thermo Fisher, Carlsbad, CA) supplemented with 10% Fetal Bovine Serum (FBS, Atlanta Biologicals, Lawrenceville, GA). Culture medium was changed three times a week.
Fig. 1.
HMGA2 expression declines during in vitro MSCs expansion. MSCs were isolated by CD45 negative selection (see Methods). (A) Representative dot plot of flow cytometry data showing cell size (forward scatter, FS) versus CD45 expression. (B) After CD45 depletion, MSCs were plated into tissue culture flasks. Population doublings were calculated based on cell counts performed at the indicated time intervals using a hemocytometer (N = 4). (C) and (D) HMGA2 and Nestin mRNA levels, as detected by RT-PCR (N = 3). (E) and (F) let-7 family members detected by RT-PCR. (E) shows highly abundant let-7 members (–a, –b, and –e) while (F) shows low-expressed let-7 (–c, –d, and –f) (N = 4). *P <0.05.
For all other experiments, MSCs were obtained following a standard isolation protocol [Beegle et al., 2015; Lakatos et al., 2015]. Here, bone marrow-derived MNC were isolated using a density gradient centrifugation as described above and then MNC were directly plated into tissue culture flasks. MSCs were then expanded for at least three to five passages prior to experimentation. Under these culture conditions, a passage is defined as three to four population doublings, occurring within 5–7 days.
For all experiments involving supplementation with FGF-2 (Thermo Fisher), MSCs were cultured in standard culture medium or differentiation medium, as described below. For differentiation assays, supplementation of FGF-2 occurred only during medium changes (every 2–3 days). For experiments with inhibitors U0126 and PD173074 (both Sigma–Aldrich, St. Louis, MO), supplementation of inhibitors was done one hour prior to the addition of FGF-2 (1 ng/ml).
CELL PROLIFERATION ASSAYS
To determine the proliferation rate (population doubling (PD)/day) of MSCs during early expansion, cells were plated into tissue culture flasks at 1,000 cells per cm2. At the end of each incubation time (3 or 4 days after seeding) cells were lifted with trypsin and counted using Trypan blue exclusion dye (Thermo Fisher) and a hemocytometer, where PD = log(2)[cell number after incubation time/seeded cell number]. For proliferation assays addressing the effect of HMGA2, MSCs were plated in 96-well plates at 500 cells/well. At indicated time points, cells were incubated for 2 h in medium containing MTT dye solution (3-(4,5-dimethyl-2-Thiazolyl)-2,5-Diphenyl-2H-Tetra-zolium bromide; Promega, Sunnyvale, CA). Then, Solubilization/Stop solution (Promega) was added, and samples were incubated for one additional hour. Finally, optical density was measured at 570 nm (with correction at 650 nm) to quantify formation of formazan, which is proportional to the number of living cells.
RT-PCR
Total RNA was extracted using a Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions. RNA purity and quantity were measured by NanoDrop (Thermo Fisher), where an absorbance ratio (A260/A280) of 1.8–2.0 was considered appropriate for further analysis. Reverse transcription was performed with 1 μg of DNA-depleted RNA, using Taqman Reverse Transcription Reagents (Life Technologies, Grand Island, NY). mRNA levels were measured by real-time PCR using TaqMan Fast Universal MasterMix (Life Technologies), and TaqMan primers and probes for following genes: HMGA2 (Hs00971725_m1), Nestin (Hs04187831_g1), and FGFR1 (Hs00915142_m1). GAPDH (Hs02758991_g1) was used as an invariant housekeeping gene. For measurement of adipogenic markers, mRNA levels were quantified using SYBRGreen Universal MasterMix (Life Technologies) and the following primers: adipsin fwd [Isenmann et al., 2009]: 5′-GACACCAT CGACCACGAC-3′, adipsin rev: 5′-CCACGTCGCA-GAGAGTTC-3′, fabp4 fwd: 5′-AACCTT AGATGGGGGTGTCC-3′, fabp4 rev: 5′-GTGGAAGTGACGCCTTTCAT-3′, Gapdh fwd [Beegle et al., 2015]: 5′ACAGTCAGCCGCATCTTC3′, Gapdh rev: 5′CTCCGACCTTCAC CTTCC-3′.
To measure miRNA levels, total RNA was extracted as described above, but the reverse transcription was performed using TaqMan MicroRNA Reverse Transcription Kit (Life Technologies) with specific stem-loop primers for each miRNA. qPCR was then performed using TaqMan Universal Mastermix (Life Technologies) and the following TaqMan Small RNA Assays (Life Technologies): hsa-let-7a (#377), hsa-let-7b (#2619), hsa-let-7c (#379), hsa-let-7d (#2283), hsa-let-7e (#2406), hsa-let-7f (#382), hsa-miR-10b (#388), hsa-miR-16 (#391), hsa-miR-23b (#400), hsa-miR-24 (#402), hsa-miR-29b (#413), hsa-miR-181a (#480). U6 small nucleolar RNA (#1973) was used as an invariant control small RNA.
WESTERN BLOTTING ANALYSIS
MSCs were seeded at 10,000 cells/cm2 and incubated for 24 h in standard culture medium. Cells were then washed twice with PBS and lysed in ice-cold RIPA assay buffer (Thermo Fisher) containing a protease and phosphatase inhibitor cocktail (Thermo Fisher). Cell lysates were mixed with Laemmli buffer (Bio-Rad, Hercules, CA) containing β-mercaptoethanol, and boiled at 95°C for 5 min and subjected to SDS–PAGE. Proteins were transferred onto PVDF membranes and incubated overnight with primary antibodies against HMGA2 (1:2,000, Cell Signaling, Danvers, MA) and β-actin (1:5,000, Sigma–Aldrich). Secondary antibodies (1:5,000) conjugated to horseradish peroxidase (Thermo Fisher) were then added for 1 h at room temperature, and proteins were detected and photographed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher) and Image Lab software (Bio-Rad).
LENTIVIRAL VECTORS AND MSCs TRANSDUCTION
To over-express HMGA2 or LIN28 (LIN28A), we used the construct pCCLc-MNDU3-X-PGK-eGFP-WPRE, where X is HMGA2, LIN28 or no transgene (control vector). HMGA2 was TOPO-cloned (TOPO-TA cloning kit, Thermo Fisher) from MSCs cDNA using the following primers: HMGA2-fwd: 5′-ATGAGCGCACGCGGTGAG-3′ and HMGA2-rev: 5′-CACAAGAGTCTGCCGAAGAGGACTAG-3′. The insert was then sub-cloned into the final lentiviral construct using EcoRI restriction sites. LIN28 was sub-cloned from the plasmid pSin-EF2-LIN28-Pur (plasmid 16580, Addgene, Cambridge, MA) originally developed in James Thomson’s laboratory [Yu et al., 2007].
To silence HMGA2, two different short hairpin RNA (shRNAs) were used. The interference RNA to target HMGA2 mRNA was designed using siDesignCenter (dharmacon.gelifesciences.com) and is shown below underlined. Two different loop regions (shown in italics) were used to create the respective shRNAs [Schopman et al., 2010]. shHMGA2-1: 5′-AAAAAAGGGAAGAGGAGGAG-GAATTTTTCTCTTGGTAAAATTCCTCCTCCTCTTCCCGGTGTTTC-GTCCTTTCCACAAG-3′ and shHMGA2-2: 5′-AAAAA AGGGAAGAGGAGGAGGAATTTTCCAGTGTTGAGAGGAAAATTC-CTCCTCCTCTTCCCGGTGTTTCGTCCTTTCCACAAG-3′. A scramble shRNA with no specific mRNA target was used as control (shControl). These three shRNAs were firstly cloned using a TOPO-TA cloning kit (Thermo Fisher) and then sub-cloned into a lentiviral construct with the form: pCCLc-U6-Y-PGK-dTomato, where Y represents the insertion site for the respective shRNA. All constructs were packaged into third generation lentiviral vectors using Lenti-X 293T cells (Clontech Laboratories, Mountain View, CA). MSCs were transduced with 20 μg/ml protamine sulfate (MP Biomedicals, Santa Ana, CA) with sufficient lentivirus to generate 93–95% GFP/dTomato positive MSCs, 3 days after transduction.
OSTEOGENIC AND ADIPOGENIC DIFFERENTIATION
Osteogenic and adipogenic differentiation were performed as previously described [Fierro et al., 2011; Beegle et al., 2015], with an initial cell density of 10,000 MSCs/cm2 with regular media changes every 2–3 days. Osteogenic medium was standard culture medium supplemented with 0.2 mM ascorbic acid, 0.1 μM dexamethasone, and 20 mM β-glycerolphosphate. Adipogenic medium was standard culture medium with 0.5 mM isobutylmethylxanthine, 50 μM indomethacin, and 0.5 μM dexamethasone. For osteogenesis quantification, alkaline phosphatase (ALP) activity was measured at day 8 by lifting the cells with trypsin and extracting protein using a lysis buffer consisting of 1.5 mM Tris-HCl solution supplemented with 1.0 mM ZnCl2, 1.0 mM MgCl2, and 1% Triton X–100. Lysates were then centrifuged at 12,000g for 10 min and incubated with p-nitrophenylphosphate liquid substrate solution (Sigma–Aldrich) for 30 min. Released p-nitrophenolate was determined using a spectrophotometer at 405 nm. As loading control, total protein concentration was determined with Coomassie Brilliant Blue staining and measured at 595 nm. Matrix mineralization was determined at day 14 using Alizarin Red S indicator (ARS; Ricca Chemicals, Arlington, TX). Cells were fixed with 10% v/v formalin solution for 15 min, washed once with PBS, and stained for 20 min with 1% w/v ARS over gentle shaking. Samples were then incubated with 10% v/v acetic acid for 30 min, the cell layer scraped, vortexed for 30 s, and centrifuged at 12,000g for 10 min. Optic density of the supernatants was measured at 405 nm.
For adipogenesis, cells were fixed at day 14 with 10% v/v formalin for 15 min, rinsed once with PBS, and stained for 30 min with Oil Red O solution (Electron Microscopy Sciences, Hatfield, PA). Cells were then washed three times with PBS and incubated with 4% Tween 20 (#9005-64-5, Affymetrix) in isopropanol for 5 min, in order to release the dye. Optic density of supernatants was then measured at 490 nm.
DATA PRESENTATION AND STATISTICAL ANALYSIS
Results are shown as averages of biological replicates with standard error of the mean as error bars. The number of biological replicates (N; MSCs derived from different donors) is shown in the respective figure legends. Each biological replicate was measured in triplicate. Significant differences were firstly assessed using one-way analysis of variance (ANOVA), followed by a paired Student’s t-test with Bonferroni correction, to calculate a P-value for the difference in between a specific condition and its respective control. As indicated in the respective figure legends, *P <0.05, **P <0.005, and ***P <0.0005.
RESULTS
MSCs RAPIDLY CHANGE DURING EARLY CELL EXPANSION
MSCs are rare cells within the bone marrow and the standard isolation protocols require culture for a few weeks (i.e., two to three passages) to achieve a pure cell population free of hematopoietic components. In order to obtain pure MSCs within a few days, we performed a negative selection for the pan-hematopoietic marker CD45 on human bone marrow-derived mononuclear cells, as previously described [Isern et al., 2013]. This isolation allowed an initial seeding population of 95% CD45 negative cells (Fig. 1A). MSCs were then expanded following standard culture conditions (see Methods), under which no signs of senescence, such as cell proliferation arrest, morphological changes or expression of β-galactosidase and p16, are detectable for at least additional 7–10 passages (not shown). Interestingly, we found that even during the first days of MSCs in culture, a significant decrease in proliferation rate occurs, from 0.9 PD/day from days 9 to 12, versus 0.6 PD/day from days 16 to 20 (Fig. 1B).
Coinciding with this decline in the proliferation rate of MSCs, we observed a strong reduction in HMGA2 mRNA levels (Fig. 1C), while the intermediate filament nestin showed a more gradual reduction, as previously described [Isern et al., 2013] (Fig. 1D). HMGA2 is well known to be post-transcriptionally regulated by let-7 [Mayr et al., 2007] and indeed, the decay of HMGA2 (around day 15) coincides with an increase in expression of all let-7 members measured (Fig. 1E and 1F). However, let-7 levels gradually decreased over time while HMGA2 persisted at low levels, implying that other mechanisms are involved in the suppression of HMGA2 as well. We investigated methylation of CpG islands in the promoter region of HMGA2 as a potential mechanism for this. Bisulfite sequencing results showed an increase in methylation in 3 out of 6 sites identified (Supplementary Fig. S1). However, overall methylation levels were under 10%, suggesting that additional regulatory mechanisms must be involved in the down-regulation of HMGA2 during the early expansion of MSCs.
FGF-2 REGULATES HMGA2 EXPRESSION IN MSCS
Previously, Ayoubi et al. [1999] showed that FGF-1 stimulates HMGA2 expression in 3T3-L1 preadipocytes. Since FGF-2 induces proliferation and promotes self-renewal of MSCs [Tsutsumi et al., 2001], we next examined whether FGF-2 could affect HMGA2 expression in MSCs. Indeed, addition of FGF-2 increased HMGA2 mRNA and protein levels in a dose-dependent manner (Fig. 2A and D). This response was rapid, since robust up-regulation of HMGA2 mRNA was detected after 3 h exposure to FGF-2 (Fig. S2A). We next added PD173074 to block FGF-2 receptor 1 (FGFR1), the most abundantly expressed FGF receptor in human MSCs [Delorme et al., 2008; Lai et al., 2011] and the small molecule UO126 to block downstream mitogen-activated protein kinase (MAPK or ERK1/2). Both inhibitors significantly reduced the effect of FGF-2 on HMGA2 levels, at both mRNA and protein level (Fig. 2B and E), suggesting that FGF-2 increases HMGA2 expression in an FGFR1 and ERK1/2-dependent manner.
Fig. 2.
FGF-2 induces HMGA2 expression. (A) and (D) MSCs were cultured for 24 h in the presence of increasing concentrations of FGF-2, which enhanced HMGA2 expression in a dose-dependent manner, both at mRNA (N = 8) and protein levels (N = 3). (B) and (E) Similarly, MSCs were cultured for 24 h with 1 ng/ml FGF-2, in either the absence or presence of the ERK1/2-inhibitor U0126 (20 μM) or the selective FGFR1 inhibitor PD173074 (50 nM). For both, mRNA and protein levels, N = 4. (C) and (F) 3–5 days after transduction of MSCs with either Control or LIN28 lentiviral vectors, FGF-2 (1 ng/ml) was added to the culture medium and MSCs cultured for additional 24 h. Both mRNA (C; N = 6) and protein (F; N = 2) show that LIN28 and FGF-2 had an additive effect on HMGA2 induction. *P <0.05, **P <0.005, and ***P <0.0005.
LIN28 is a highly conserved RNA-binding protein that selectively blocks the biogenesis of let-7 [Viswanathan et al., 2008; Hagan et al., 2009]. To test whether the regulation of HMGA2 by FGF-2 is let-7 dependent, we silenced let-7 miRNAs by over-expressing LIN28. Both, supplementation of FGF-2 and over-expression of LIN28 increased HMGA2 (Fig. 2C and 2F). However, FGF-2 and LIN28 combined acted in an additive manner, suggesting that the regulatory mechanisms of FGF-2 and LIN28 are mutually independent. Even more, FGF-2 did not affect let-7 levels (Fig. 3A), while, as expected, over-expression of LIN28 significantly reduced all let-7 measured, but no other miRNAs (Fig. 3B). Altogether, these results demonstrate that FGF-2 induces HMGA2 expression in MSCs via FGFR1 and the ERK1/2 signaling pathway, but independently from let-7.
Fig. 3.
Addition of FGF-2 does not affect the levels of let-7 family members. (A) MSCs were cultured for 2 days in the presence FGF-2 (10 ng/ml). Then, total RNA was extracted and miRNA levels were measured by RT-PCR (N = 4). (B) Similarly, MSCs were transduced to over-express LIN28, and after 3–4 days, miRNA levels were measured. LIN28 over-expression specifically reduces the let-7 family members but has no effect on other miRNA (N = 4). *P <0.05, **P <0.005, and ***P <0.0005.
HMGA2 REGULATES PROLIFERATION OF MSCs
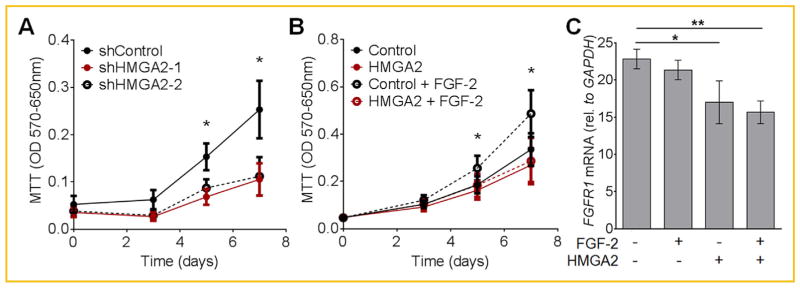
To test the hypothesis that ectopic expression of HMGA2 would promote self-renewal in MSCs, we investigated the function of HMGA2 using loss- and gain-of-function experiments, by either silencing or over-expressing HMGA2. To silence HMGA2, MSCs were transduced with either a scramble shRNA (sh-Control) or two different shRNAs targeting HMGA2. Both shRNAs targeting HMGA2 inhibited cell proliferation to a similar degree (Fig. 4A), while cellular viability was not affected (not shown). Over-expression of HMGA2 did not increase cell proliferation and instead abrogated the mitogenic effect of FGF-2 (Fig. 4B). Interestingly, over-expression of HMGA2 significantly reduced FGFR1 expression (Fig. 4C). Hence, HMGA2 may “desensitize” MSCs to FGF-2 by disrupting the expression of its receptor. These results suggest that HMGA2 is necessary but not sufficient to induce proliferation of MSCs. In addition, over-expression of HMGA2 limits FGF-2–mediated cell proliferation, by reducing the expression of FGFR1.
Fig. 4.
Silencing HMGA2 represses cell proliferation while over-expression of HMGA2 suppresses the mitogenic effect of FGF-2 in MSCs. (A) Proliferation of MSCs transduced to express the indicated shRNAs was estimated using an MTT assay (measuring optic density of formazan formation). Expression of both shRNAs (shHMGA2-1 and shHMGA2-2) significantly reduced the proliferation of MSCs (N = 5). (B) Similarly, an MTT assay was used to quantify proliferation of MSCs transduced with either a control lentiviral vector (Control) or MSCs over-expressing HMGA2, in either the presence or absence of FGF-2 (10 ng/ml). FGF-2 only induced a significant increase in proliferation of MSCs transduced with the control lentiviral vector, but not in MSCs over-expressing HMGA2 (N = 8). (C) FGFR1 mRNA levels were decreased upon over-expression of HMGA2, as measured by RT-PCR. Transduced cells (with either control lentiviral vector or to over-express HMGA2) were exposed to 24 h 10 ng/ml FGF-2 (N = 5). *P <0.05, **P <0.005.
HMGA2 REGULATES ADIPOGENESIS BUT NOT OSTEOGENESIS OF MSCs
To study a potential effect of HMGA2 on osteogenesis of MSCs, we performed differentiation assays and measured alkaline phosphatase activity (ALP) at day 8 and calcium precipitation (using Alizarin Red S [ARS] staining) at day 14. In agreement with previous reports [Tsutsumi et al., 2001; Ng et al., 2008], addition of FGF-2 inhibited both ALP and ARS. However, either over-expression or silencing HMGA2 did not affect these parameters (Fig. S3). Therefore, the inhibitory effect of FGF-2 in osteogenesis appears to be independent of HMGA2. Our results also suggest that osteogenesis of MSCs is not affected by HMGA2 expression.
To measure adipogenesis in MSCs, we used Oil-Red O staining to quantify triglyceride accumulation and measured gene expression of adipogenic markers adipsin and fatty acid binding protein 4 (fabp4). As previously described [Tsutsumi et al., 2001; Ng et al., 2008], supplementation with FGF-2 inhibited adipogenesis. Remarkably, over-expression of HMGA2 also inhibited adipogenesis, while the combination of FGF-2 and HMGA2 showed an additive effect, at least at gene expression level (Fig. 5A–D). However, silencing HMGA2 had an inhibitory effect too (Fig. 5E–H), suggesting that, although excess HMGA2 inhibits adipogenesis, basal levels of HMGA2 are necessary during adipogenic differentiation of MSCs.
Fig. 5.
Both over-expressing and silencing HMGA2 inhibit adipogenesis of MSCs. (A–D) MSCs were transduced with Control or HMGA2 lentiviral vectors and cultured for 14 days in adipogenic media, in the presence or absence of FGF-2 (10 ng/ml). (E–H) Similarly, MSCs were transduced with lentivirus for the indicated shRNAs and cultured for 14 days in adipogenic medium. (D) and (E) show representative images of triglyceride accumulation, after staining with Oil Red O. (A) and (F) show the quantification of Oil Red O staining, which was found to be significantly lower after over-expression or silencing of HMGA2 (for both, N = 7). (B), (C), (G), and (H) show expression of adipogenic genes adipsin and fabp4, measured at day 14 by RT-PCR (for B and C, N = 4 and for G and H, N = 3). *P <0.05, **P <0.005.
DISCUSSION
HMGA2 plays a crucial role promoting self-renewal in many types of stem cells, whereas aberrant expression of HMGA2 associates with malignancy in multiple cancers [Mayr et al., 2007]. Although in vitro expanded MSCs clearly show proliferation and differentiation potential, their self-renewal capacity seems limited, rising the question whether expanded MSCs are truly stem cells [Bianco et al., 2008]. In fact, most of clinical applications of MSCs do not rely on their differentiation potential, but rather focus on their paracrine activity; MSCs secrete factors and interact with host cells to promote wound healing and modulate the immune system, among other effects [Uccelli et al., 2008; Egana et al., 2009; Caplan and Correa, 2011].
Our results support the notion that MSCs in culture may not completely self-renew, but rather proliferate and differentiate similarly to many lineage-restricted progenitor cells. In line with this notion, HMGA2 expression (which is necessary for self-renewal of other stem cells [Melton et al., 2010; Cimadamore et al., 2013; Copley et al., 2013; Worringer et al., 2014]) was strongly reduced during early expansion of the cells. Our observations encourage the search for novel culture conditions that may promote true self-renewal of MSCs in culture. Previous attempts include genetic engineering of the cells [Jun et al., 2004; Boker et al., 2008], growing cells in suspension [Baksh et al., 2005], culturing MSCs under low oxygen levels [Basciano et al., 2011], and/or supplementing the culture media with FGF-2 [Tsutsumi et al., 2001]. Despite the fact that HMGA2 alone was insufficient to promote self-renewal, we propose that HMGA2 could be used as a molecular marker for self-renewal in MSCs.
We also propose that self-renewal of MSCs requires the orchestration of HMGA2 with additional factors, which remain unknown. Of note, we observed that over-expression of LIN28 (hence, repression of let-7) increased the proliferation rate of MSCs expanded for 9–12 days, but did not affect MSCs in later stages (expanded for 20 days or more; not shown). Since MSCs expanded for 9–12 days proliferated faster than further expanded MSCs (Fig. 1B) and showed the highest levels of HMGA2 (Fig. 1C), we propose that, in MSCs, the effect of LIN28/let-7 on proliferation depends on HMGA2, as previously described for other stem cells.
In order to study possible mechanisms involved in the rapid repression of HMGA2, we examined let-7 levels. In fact, the reduction of HMGA2 coincided with an increase in let-7 during early expansion. However, on later stages let-7 decreased while HMGA2 levels remained low, suggesting that additional mechanisms are involved in the repression of HMGA2. Since the methylation status of HMGA2 promoter region was overall low, we propose that other mechanisms are possibly involved, such as histone modifications [Ferguson et al., 2003].
During early expansion of MSCs, expression levels of all let-7 measured (let-7a through let-7f) showed very similar dynamics. That is a sharp increase after 16 days in culture and a decrease after 30 days. Of note, the small nucleolar RNA U6 used as loading control for let-7 measurements showed no changes in expression over time (not shown), suggesting that in general the canonical miRNA machinery might be differentially expressed or active during early expansion of MSCs. A similar effect has been described for cells under hypoxia, affecting the expression of all canonical miRNAs by inhibiting the miRNA processing factor Dicer [Ho et al., 2012; van den Beucken et al., 2014].
The effect of FGF-2 on HMGA2 showed a positive correlation between mRNA and protein levels. This is important to highlight, since during early expansion of MSCs, HMGA2 was only measured at mRNA level, due to the technical challenge of obtaining sufficient MSCs in early days for protein extraction. Of note, repression of protein synthesis by miRNAs such as let-7 may or may not imply the degradation of the target mRNA [Bartel, 2004; Zamore and Haley, 2005]. In our experiments, LIN28 induced a repression of let-7 and increased HMGA2 at both protein and mRNA levels, suggesting that repression of HMGA2 by let-7 implies the degradation of the target mRNA (Fig. 6).
Fig. 6.
Schematic representation of the role of HMGA2 in FGF-2 signaling. The effect of FGF-2 on proliferation and adipogenesis of MSCs is dependent on HMGA2 expression. Of note, silencing HMGA2 represses both, proliferation and differentiation, suggesting that HMGA2 is necessary for these processes. Over-expression of HMGA2 abrogates adipogenesis and inhibits FGFR1 expression, blocking the mitogenic effect of FGF-2.
We show that FGF-2 regulates HMGA2 at mRNA level through a pathway involving FGFR1 and downstream ERK1/2, independent from let-7 (Fig. 6). However, the exact regulatory mechanism remains unknown. Of note, FGF-2 significantly increased both the endogenous and the transgene HMGA2 mRNA levels (Fig. S2B). Since in MSCs over-expressing HMGA2 the transgene is under a constitutive promoter and does not contain the 3′-untranslated region (3′UTR) of HMGA2, we propose that FGF-2 may increase HMGA2 mRNA levels by mechanisms such as modulating miRNAs targeting the coding region or by affecting the methylation of HMGA2 mRNA [Meyer et al., 2012; Fustin et al., 2013; Grabole et al., 2013].
Over-expression of HMGA2 suppressed the mitogenic effect of FGF-2 (Fig. 4B), suggesting that HMGA2 acts as a negative feedback for FGF-2 signaling. A possible mechanism is the significant inhibition of FGFR1 observed when over-expressing HMGA2 (Fig. 6). Human MSCs also express FGFR2-4, however at much lower levels than FGFR1 [Delorme et al., 2008; Lai et al., 2011]. Alternatively, it is possible that HMGA2 is required in a cell cycle-dependent manner [Pfannkuche et al., 2009; Yu et al., 2013], where HMGA2 is up-regulated during certain stages but reduced during others, in order to allow FGF-2-mediated cell cycle progression. This concept could be tested with future studies on cell cycle-related target genes, common to HMGA2 and FGF-2.
In line with previous work on human umbilical cord blood-derived stromal cells (UC-MSCs) [Yu et al., 2013], silencing HMGA2 inhibited proliferation and adipogenesis of MSCs, suggesting that HMGA2 is necessary for these processes. However, Yu and collaborators showed a mitogenic effect when over-expressing HMGA2 in UC-MSCs, which we did not observe. Our results, in bone marrow-derived MSCs resemble what had been described for fibroblasts [Narita et al., 2006]. In addition, in contrast to a previous report [Wei et al., 2014] in adipose tissue-derived MSCs (AT-MSCs), we did not observe an effect of HMGA2 on osteogenesis. These discrepancies might be due to the fact that bone marrow-derived MSCs are initially obtained in very low frequencies and have to undergo additional population doublings as compared to UC-MSCs or AT-MSCs. Another factor is that shRNAs from different studies vary in their silencing efficiency and specificity. Our results are strengthened by the use of two different shRNAs against HMGA2.
Finally, we show that both FGF-2 and over-expression of HMGA2 reduce adipogenesis of MSCs, suggesting that FGF-2 inhibits adipogenesis of MSCs via HMGA2 (Fig. 6). Nonetheless, both adipsin and fabp4 mRNA levels were further reduced upon combination of FGF-2 and HMGA2, suggesting that both factors act in an additive way.
In conclusion, we propose that HMGA2 is rapidly reduced during expansion of MSCs in culture, but can be restored by supplementation with FGF-2. Our results also suggest that HMGA2 is necessary for normal proliferation and differentiation of MSCs. Therefore, we propose that maintenance of endogenous HMGA2 levels may promote self-renewal of MSCs, allowing further expansion of the cells without hampering their therapeutic efficacy.
Supplementary Material
Acknowledgments
NIH Transformative Grant; Grant number: 1R01GM099688; Grant sponsor: Council on International Educational Exchange/Hungarian-American Enterprise Scholarship Fund.
We would like to thank Dr. Steven Frese for his critical review of this manuscript. This project was funded by the NIH Transformative Grant 1R01GM099688 (awarded to JN). KL was funded by the Council on International Educational Exchange/Hungarian-American Enterprise Scholarship Fund.
Footnotes
Conflict of interest: All authors declare no conflicts of interest regarding this publication or any information related to it.
Additional supporting information may be found in the online version of this article at the publisher’s website.
References
- Ankrum JA, Ong JF, Karp JM. Mesenchymal stem cells: Immune evasive, not immune privileged. Nat Biotechnol. 2014;32:252–260. doi: 10.1038/nbt.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoubi TA, Jansen E, Meulemans SM, Van de Ven WJ. Regulation of HMGIC expression: An architectural transcription factor involved in growth control and development. Oncogene. 1999;18:5076–5087. doi: 10.1038/sj.onc.1202881. [DOI] [PubMed] [Google Scholar]
- Baksh D, Davies JE, Zandstra PW. Soluble factor cross-talk between human bone marrow-derived hematopoietic and mesenchymal cells enhances in vitro CFU-F and CFU-O growth and reveals heterogeneity in the mesenchymal progenitor cell compartment. Blood. 2005;106:3012–3019. doi: 10.1182/blood-2005-01-0433. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Basciano L, Nemos C, Foliguet B, de Isla N, de Carvalho M, Tran N, Dalloul A. Long term culture of mesenchymal stem cells in hypoxia promotes a genetic program maintaining their undifferentiated and multipotent status. BMC Cell Biol. 2011;12:12. doi: 10.1186/1471-2121-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beegle J, Lakatos K, Kalomoiris S, Stewart H, Isseroff RR, Nolta JA, Fierro FA. Hypoxic preconditioning of mesenchymal stromal cells induces metabolic changes, enhances survival and promotes cell retention in vivo. Stem Cells. 2015;33:1818–1828. doi: 10.1002/stem.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: Revisiting history, concepts, and assays. Cell Stem Cell. 2008;2:313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boker W, Yin Z, Drosse I, Haasters F, Rossmann O, Wierer M, Popov C, Locher M, Mutschler W, Docheva D, Schieker M. Introducing a single-cell-derived human mesenchymal stem cell line expressing hTERT after lentiviral gene transfer. J Cell Mol Med. 2008;12:1347–1359. doi: 10.1111/j.1582-4934.2008.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AI, Correa D. The MSC: An injury drugstore. Cell Stem Cell. 2011;9:11–15. doi: 10.1016/j.stem.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimadamore F, Amador-Arjona A, Chen C, Huang CT, Terskikh AV. SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc Natl Acad Sci USA. 2013;110:E3017–E3026. doi: 10.1073/pnas.1220176110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EA, Kalomoiris S, Nolta JA, Fierro FA. Concise review: MicroRNA function in multipotent mesenchymal stromal cells. Stem Cells. 2014;32:1074–1082. doi: 10.1002/stem.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley MR, Babovic S, Benz C, Knapp DJ, Beer PA, Kent DG, Wohrer S, Treloar DQ, Day C, Rowe K, Mader H, Kuchenbauer F, Humphries RK, Eaves CJ. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15:916–925. doi: 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- Delorme B, Ringe J, Gallay N, Le Vern Y, Kerboeuf D, Jorgensen C, Rosset P, Sensebe L, Layrolle P, Haupl T, Charbord P. Specific plasma membrane protein phenotype of culture-amplified and native human bone marrow mesenchymal stem cells. Blood. 2008;111:2631–2635. doi: 10.1182/blood-2007-07-099622. [DOI] [PubMed] [Google Scholar]
- Egana JT, Fierro FA, Kruger S, Bornhauser M, Huss R, Lavandero S, Machens HG. Use of human mesenchymal cells to improve vascularization in a mouse model for scaffold-based dermal regeneration. Tissue Eng Part A. 2009;15:1191–1200. doi: 10.1089/ten.tea.2008.0097. [DOI] [PubMed] [Google Scholar]
- Ferguson M, Henry PA, Currie RA. Histone deacetylase inhibition is associated with transcriptional repression of the Hmga2 gene. Nucleic Acids Res. 2003;31:3123–3133. doi: 10.1093/nar/gkg403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro FA, Kalomoiris S, Sondergaard CS, Nolta JA. Effects on proliferation and differentiation of multipotent bone marrow stromal cells engineered to express growth factors for combined cell and gene therapy. Stem Cells. 2011;29:1727–1737. doi: 10.1002/stem.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, Okamura H. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Grabole N, Tischler J, Hackett JA, Kim S, Tang F, Leitch HG, Magnusdottir E, Surani MA. Prdmpromotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 2013;14:629–637. doi: 10.1038/embor.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JJ, Metcalf JL, Yan MS, Turgeon PJ, Wang JJ, Chalsev M, Petruzziello-Pellegrini TN, Tsui AK, He JZ, Dhamko H, Man HS, Robb GB, Teh BT, Ohh M, Marsden PA. Functional importance of Dicer protein in the adaptive cellular response to hypoxia. J Biol Chem. 2012;287:29003–29020. doi: 10.1074/jbc.M112.373365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenmann S, Arthur A, Zannettino AC, Turner JL, Shi S, Glackin CA, Gronthos S. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27:2457–2468. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]
- Isern J, Martin-Antonio B, Ghazanfari R, Martin AM, Lopez JA, Del Toro R, Sanchez-Aguilera A, Arranz L, Martin-Perez D, Suarez-Lledo M, Marin P, Van Pel M, Fibbe WE, Vazquez J, Scheding S, Urbano-Ispizua A, Mendez-Ferrer S. Self-renewing human bone marrow mesenspheres promote hematopoietic stem cell expansion. Cell Rep. 2013;3:1714–1724. doi: 10.1016/j.celrep.2013.03.041. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, Du J, Aldrich S, Lisberg A, Low WC, Largaespada DA, Verfaillie CM. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- Jun ES, Lee TH, Cho HH, Suh SY, Jung JS. Expression of telomerase extends longevity and enhances differentiation in human adipose tissue-derived stromal cells. Cell Physiol Biochem. 2004;14:261–268. doi: 10.1159/000080335. [DOI] [PubMed] [Google Scholar]
- Lai WT, Krishnappa V, Phinney DG. Fibroblast growth factor 2 (Fgf2) inhibits differentiation of mesenchymal stem cells by inducing Twist2 and Spry4, blocking extracellular regulated kinase activation, and altering Fgf receptor expression levels. Stem Cells. 2011;29:1102–1111. doi: 10.1002/stem.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos K, Kalomoiris S, Merkely B, Nolta JA, Fierro FA. Mesenchymal stem cells respond to hypoxia by increasing diacylglycerols. J Cell Biochem. 2015;117:300–307. doi: 10.1002/jcb.25292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendicino M, Bailey AM, Wonnacott K, Puri RK, Bauer SR. MSC-based product characterization for clinical trials: An FDA perspective. Cell Stem Cell. 2014;14:141–145. doi: 10.1016/j.stem.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, Lowe SW. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- Ng F, Boucher S, Koh S, Sastry KS, Chase L, Lakshmipathy U, Choong C, Yang Z, Vemuri MC, Rao MS, Tanavde V. PDGF, TGF-beta, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): Transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood. 2008;112:295–307. doi: 10.1182/blood-2007-07-103697. [DOI] [PubMed] [Google Scholar]
- Pfannkuche K, Summer H, Li O, Hescheler J, Droge P. The high mobility group protein HMGA2: A co-regulator of chromatin structure and pluripotency in stem cells? Stem Cell Rev. 2009;5:224–230. doi: 10.1007/s12015-009-9078-9. [DOI] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Schopman NC, Liu YP, Konstantinova P, ter Brake O, Berkhout B. Optimization of shRNA inhibitors by variation of the terminal loop sequence. Antiviral Res. 2010;86:204–211. doi: 10.1016/j.antiviral.2010.02.320. [DOI] [PubMed] [Google Scholar]
- Shibata KR, Aoyama T, Shima Y, Fukiage K, Otsuka S, Furu M, Kohno Y, Ito K, Fujibayashi S, Neo M, Nakayama T, Nakamura T, Toguchida J. Expression of the p16INK4A gene is associated closely with senescence of human mesenchymal stem cells and is potentially silenced by DNA methylation during in vitro expansion. Stem Cells. 2007;25:2371–2382. doi: 10.1634/stemcells.2007-0225. [DOI] [PubMed] [Google Scholar]
- Solchaga LA, Penick K, Porter JD, Goldberg VM, Caplan AI, Welter JF. FGF-2 enhances the mitotic and chondrogenic potentials of human adult bone marrow-derived mesenchymal stem cells. J Cell Physiol. 2005;203:398–409. doi: 10.1002/jcp.20238. [DOI] [PubMed] [Google Scholar]
- Trounson A, McDonald C. Stem cell therapies in clinical trials: Progress and challenges. Cell Stem Cell. 2015;17:11–22. doi: 10.1016/j.stem.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Tsutsumi S, Shimazu A, Miyazaki K, Pan H, Koike C, Yoshida E, Takagishi K, Kato Y. Retention of multilineage differentiation potential of mesenchymal cells during proliferation in response to FGF. Biochem Biophys Res Commun. 2001;288:413–419. doi: 10.1006/bbrc.2001.5777. [DOI] [PubMed] [Google Scholar]
- Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- van den Beucken T, Koch E, Chu K, Rupaimoole R, Prickaerts P, Adriaens M, Voncken JW, Harris AL, Buffa FM, Haider S, Starmans MH, Yao CQ, Ivan M, Ivan C, Pecot CV, Boutros PC, Sood AK, Koritzinsky M, Wouters BG. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun. 2014;5:5203. doi: 10.1038/ncomms6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bahr L, Sundberg B, Lonnies L, Sander B, Karbach H, Hagglund H, Ljungman P, Gustafsson B, Karlsson H, Le Blanc K, Ringden O. Long-term complications, immunologic effects, and role of passage for outcome in mesenchymal stromal cell therapy. Biol Blood Marrow Transplant. 2012;18:557–564. doi: 10.1016/j.bbmt.2011.07.023. [DOI] [PubMed] [Google Scholar]
- Wei J, Li H, Wang S, Li T, Fan J, Liang X, Li J, Han Q, Zhu L, Fan L, Zhao RC. Let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cells Dev. 2014;23:1452–1463. doi: 10.1089/scd.2013.0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worringer KA, Rand TA, Hayashi Y, Sami S, Takahashi K, Tanabe K, Narita M, Srivastava D, Yamanaka S. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell. 2014;14:40–52. doi: 10.1016/j.stem.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchter P, Bieback K, Schrezenmeier H, Bornhauser M, Muller LP, Bonig H, Wagner W, Meisel R, Pavel P, Tonn T, Lang P, Muller I, Renner M, Malcherek G, Saffrich R, Buss EC, Horn P, Rojewski M, Schmitt A, Ho AD, Sanzenbacher R, Schmitt M. Standardization of Good Manufacturing Practice-compliant production of bone marrow-derived human mesenchymal stromal cells for immunotherapeutic applications. Cytotherapy. 2015;17:128–139. doi: 10.1016/j.jcyt.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu KR, Park SB, Jung JW, Seo MS, Hong IS, Kim HS, Seo Y, Kang TW, Lee JY, Kurtz A, Kang KS. HMGA2 regulates the in vitro aging and proliferation of human umbilical cord blood-derived stromal cells through the mTOR/p70S6K signaling pathway. Stem Cell Res. 2013;10:156–165. doi: 10.1016/j.scr.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.