Abstract
The platinum-based drugs cisplatin, carboplatin and oxaliplatin are often used for chemotherapy, but drug resistance is common. The prediction of resistance to these drugs via genomics is a challenging problem since hundreds of genes are involved. A possible alternative is to use mass spectrometry to determine the propensity for cells to form drug-DNA adducts—the pharmacodynamic drug-target complex for this class of drugs. The feasibility of predictive diagnostic microdosing was assessed in non-small cell lung cancer (NSCLC) cell culture and a pilot clinical trial. Accelerator mass spectrometry (AMS) was used to quantify [14C]carboplatin-DNA monoadduct levels in the cell lines induced by microdoses and therapeutic doses of carboplatin, followed by correlation with carboplatin IC50 values for each cell line. The adduct levels in cell culture experiments were linearly proportional to dose (R2=0.95, p<0.0001) and correlated with IC50 across all cell lines for microdose and therapeutically relevant carboplatin concentrations (p=0.02 and p=0.01, respectively). A pilot microdosing clinical trial was conducted to define protocols and gather preliminary data. Plasma pharmacokinetics (PK), and [14C]carboplatin-DNA adducts in white blood cells and tumor tissues from six NSCLC patients were quantified via AMS. The blood plasma half-life of [14C]carboplatin administered as a microdose was consistent with the known PK of therapeutic dosing. The optimal [14C]carboplatin formulation for the microdose was 107 dpm/kg of body weight and 1% of the therapeutic dose for the total mass of carboplatin. No microdose-associated toxicity was observed in the patients. Additional accruals are required to significantly correlate adduct levels with response.
INTRODUCTION
Lung cancer is the worldwide leading cause of cancer related deaths, with the majority of cases comprising of non-small cell lung cancer (NSCLC).1, 2 Most NSCLC patients present at advanced stages and are treated with platinum (Pt)-based chemotherapy as a first line or maintenance therapy. Despite the wide-spread use, the response rate is less than 30%.3 These statistics imply that the majority of patients will not benefit from this chemotherapy regimen while still incurring associated toxicities and costs, and also delaying the use of other potentially useful chemotherapy regimens for several months. Therefore, assays for prospective identification of chemosensitivity prior to initiation of therapy are much needed. Numerous groups have used gene expression analysis and other genomic approaches to gain insights into the mechanisms of Pt-based drug resistance. This led to the discovery of hundreds of genes that are involved in cellular response toward these drugs.4–6 However, translating these findings into a clinically useful test for predicting tumor resistance prior to the initiation of chemotherapy has proven to be difficult.
Cisplatin and carboplatin commonly used in the treatment of NSCLC and many other cancer types.7 Both drugs kill cells mainly through formation of intra- and interstrand DNA crosslinks (called adducts), which induce cell death via apoptosis or necrosis (Figure 1A).8–11 The adduct levels are governed by a variety of factors including genetics, tumor microenvironment, kidney function, overall patient health and others.12, 13 The relationship between therapy-induced drug-DNA adduct levels and clinical response has been reported for a variety of cancers, including NSCLC.10, 14–16 In some studies, a positive correlation of adduct level in normal surrogate cells (e.g. PBMC) and good clinical outcome have been observed16–18, whereas other reports failed to show such a correlation, possibly due to different study designs, drug regimes, analytical methods employed and small numbers of patients.19, 20 Despite the previous contradictory reports, drug-DNA adduct levels as pharmacodynamic (PD) endpoints remain potentially more informative and useful than genomic analysis of drug response, particularly for cytotoxic chemotherapy agents.
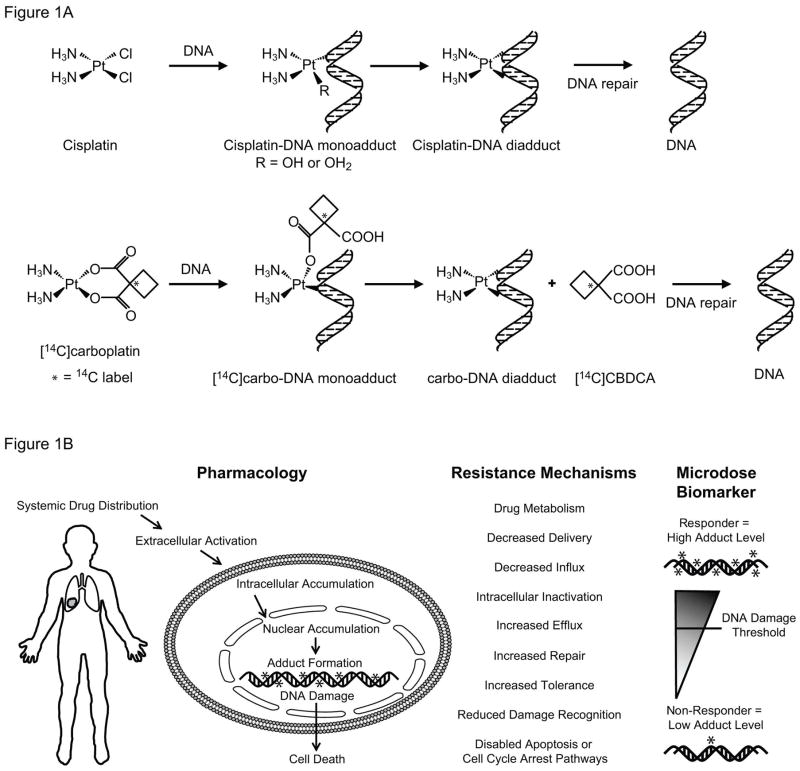
Figure 1. Illustrations of cisplatin and carboplatin chemical structure and biological mechanisms of action.
A. Simplified schematic of cisplatin and carboplatin structure and DNA adduct formation (R=OH or H2O, * = represents 14C label, which can be detected by AMS). B. Diagram of the major mechanistic steps of platinum (Pt) based chemotherapy. Formation of Pt-induced DNA damage (adducts) is the most critical step of Pt-induced cell death. Other major steps involved in damage accumulation, such as drug metabolism, cell accumulation (uptake/efflux), intracellular inactivation and DNA can also affect the levels of cytotoxicity.
Accelerator mass spectrometry (AMS) is an ultrasensitive technique for measuring rare isotopes such as 14C, which relies on dissociation of the sample into CO2 or graphite followed by isotope ratio analysis in a small particle accelerator. The resulting ratio data allow calculation of the labeled drug concentration in tissues, blood, protein or nucleic acids at concentrations that are difficult or impossible to measure with other techniques. AMS therefore enables human studies with radiolabeled drug while circumventing the need for toxic drug or radiation exposures.4, 21, 22 Carboplatin is easily labeled with a 14C atom, and therefore detectable by AMS, whereas cisplatin is unable to be detected by AMS since it cannot be labeled with a 14C atom. However, both drugs form the same final drug-DNA diadduct crosslink structures and are sometimes used interchangeably in clinical practice because clinical cross-resistance is common.23 AMS specifically detects carboplatin-DNA monoadducts, since the 14C-label in the cyclobutane dicarboxylate (CBDCA) group is released once the diadduct is formed (Figure 1A). Since drug-DNA monoadducts are the precursors of all other types of platinum-based drug-DNA adducts, we hypothesize that carboplatin-DNA monoadduct levels are predictive of carboplatin or cisplatin cytotoxicity, and that microdose-induced adduct levels are biomarkers of drug response to cytotoxic chemotherapy agents.24 We have previously reported that relatively high levels of microdose-induced monoadducts correlated with high cytotoxicity of carboplatin in cancer cell lines from different cancer types.24 We report herein investigation of carboplatin microdosing with six NSCLC cell lines and six NSCLC patients.
MATERIALS AND METHODS
Chemicals
Unlabeled carboplatin (CARBOplatin®, 10 mg/mL) was obtained from Hospira (Lake Forest, IL, USA). [14C]carboplatin (specific activity 53 mCi/mmol with the 14C-label in the cyclobutane dicarboxcylic group) was obtained from GE Healthcare (Waukesha, WI, USA). [14C]carboplatin for injection was prepared under good manufacturing practices (GMP) at the GMP facility at UC Davis. The [14C]carboplatin drug substance was dissolved with sterile water for injection (WFI). The resulting solution was filter sterilized with 0.2 μm PES syringe filter into sterile glass vials and sealed with a rubber septum. Specific activity was determined by liquid scintillation counting (LSC). Mixtures of 14C-labeled and unlabeled drug were used to minimize the usage of radiocarbon and achieve the different specific activities required for microdoses and therapeutic doses. Drug solutions for the indicated experiments were prepared immediately before use.
Cell lines and cytotoxicity assay
Six human NSCLC cell lines (H23, H460, H727, HCC827, H1975 and A549) were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured with the recommended medium unless otherwise specified. Their key characteristics are summarized in SI Table 1. Carboplatin and cisplatin IC50 values were determined in triplicate after incubating cells for 72 hours with indicated drug concentrations. The MTT assay was performed as previously reported to determine the drug concentration of cisplatin or carboplatin required to inhibit cell growth by 50% (IC50). 25
Drug treatment and AMS analysis
Cells were cultured to >90% confluence, dosed with [14C]carboplatin, and subjected to DNA isolation and AMS analysis as previously described.24 Briefly, cells were seeded at 1 × 106 cells/60 mm dish and allowed to attach overnight at 37°C and 5% CO2 in a humidified atmosphere. At hour 0, cells were dosed with 1 μM (microdose) or 100 μM carboplatin (therapeutic dose), each supplemented with 0.3 μM [14C]carboplatin (50,000 dpm/mL). Cultures were incubated with [14C]carboplatin for 4 h then washed twice with phosphate-buffered saline (PBS) and maintained thereafter with drug-free culture media for indicated periods of time to mimic the in vivo carboplatin half-life in patients (1.1–5.9 hours).26 Cells were harvested at 0, 2, 4, 8 and 24 hours after initiation of dosing. DNA was extracted from collected cells with the Wizard Genomic DNA Purification Kit according to the manufacturer’s instruction (Promega, Madison, WI, USA). DNA quantity and quality was determined with a NanoDrop 1000 spectrophotometer, the purity was ensured by obtaining a 260/280 nm OD ratio of approximately 1.9. All DNA samples were submitted to Lawrence Livermore National Laboratory (LLNL) for AMS analysis of radiocarbon content using an established protocol.27 Ten micrograms of DNA per sample was converted to graphite and measured by AMS for 14C quantification as previously described. Triplicate sets of AMS experiments were performed for each cell line and time point. The data was plotted as carboplatin-DNA monoadducts per 108 nucleotides (nt) over time.
Molecular analysis
Total RNA was extracted from cells using the RNeasy mini kit (Qiagen, Valencia, Ca) following manufacturer’s instructions. cDNA was synthesized from 500 ng of total RNA using the SuperScript® III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, Ca). Real-time PCR was conducted using the SYBR® GreenER™ qPCR SuperMix (Invitrogen) and an iQ5 Real-Time PCR detection system (BioRad, Hercules, Ca) for ERCC1 with β-actin as a reference gene.28 Previously reported primers and optimized PCR conditions were used for each gene. The software package Q-Gene was used to determine the mean normalized expression (MNE) for each gene for three independent experiments and the MNE, along with the standard error, was plotted using GraphPad Prism 5 version 5.03 for Windows (GraphPad Software, San Diego Ca).29
A pilot feasibility diagnostics clinical trial
A study titled “A phase 0 clinical trial of microdosing carboplatin and molecular profiling for chemoresistance” (ClinicalTrials.gov identifier NCT01261299) is an ongoing, multisite feasibility study of the diagnostic microdosing approach. This clinical trial was approved by the UC Davis Institutional Review Board and conducted under an IND from the FDA. The patient population consisted of non-small cell lung cancer patients (NSCLC), stage IV with measurable lesions, and bladder transitional cell carcinoma (TCC) patients, stage II disease and above for neoadjuvant treatment, or stage III and IV metastatic disease (only data for NSCLC are presented herein). Microdoses of [14C]carboplatin were administered to patients as a diagnostic reagent, followed by standard of care full dose platinum-based chemotherapy and evaluation of response. PBMC and tumor tissue were collected from the patients for analysis of carboplatin-DNA monoadduct frequencies. Toxicity of the [14C]carboplatin administered as a microdose was assessed using Common Terminology Criteria for Adverse Events (CTCAE). Patient response to chemotherapy was evaluated using RECIST criteria for correlation to carboplatin-DNA monoadduct frequency. The carboplatin dose for human chemotherapy was calculated using the Calvert formula with an AUC of 6. Therefore, individual patients were given a microdose of [14C]carboplatin (1.0 × 107 dpm/kg of body weight) containing a total carboplatin dose at 1% of their therapeutic dose by a 2-minute bolus intravenous infusion (IV). Unlabeled carboplatin and [14C]carboplatin were mixed immediately before dosing and injected through the peripheral vein at one arm. Peripheral blood specimens were drawn into BD Vacutainer CPTTM tubes with sodium citrate (Becton Dickinson) from the other arm at specified time points before and after the administration of the microdose. Tubes were immediately placed on ice and PBMC were isolated within 2 hours of collection by centrifugation according to manufacturer’s instruction. A proportion of total plasma was used for liquid scintillation counting (LSC) PK determination. Outcomes related to chemotherapy (including response and adverse events) were collected and correlated with carboplatin-DNA monoadduct data.
Statistical analysis
All statistical analyses were performed using GraphPad Prism™ software (GraphPad Software Inc., CA, USA) using a two-tailed Student’s t-test or when appropriate a 1-way ANOVA with Bonferroni’s post hoc test. A p-value below 0.05 was considered statistically significant. All experiments were carried out at least in triplicate in order to enable statistically significant comparisons of the results. All results are expressed as the mean ± SD unless otherwise noted. A simple correlation of the adduct levels and response is reported.
RESULTS
Comparison of NSCLC cell line sensitivity to carboplatin and cisplatin
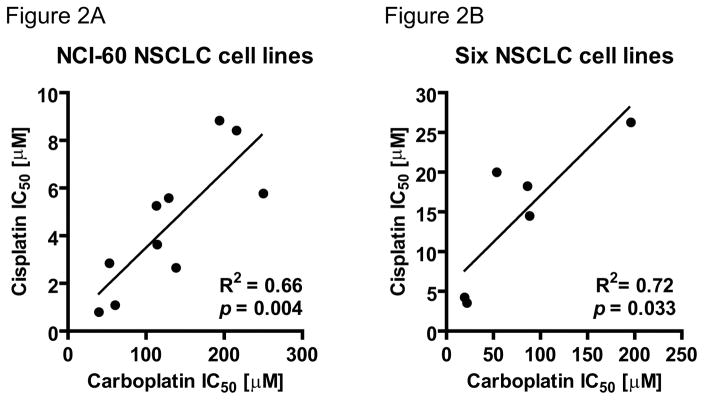
Cisplatin and carboplatin form the same DNA crosslinks or diadducts (Figure 1A) and clinical cross resistance is common. We proposed that the sensitivity of cell cultures to these two drugs might be similar. If so, the carboplatin microdosing approach described in this project could potentially be applied to the study in cellular sensitivity to cisplatin, since only carboplatin can be labeled with 14C. Therefore we compared the publicly available cytotoxicity data from the NIH Developmental Therapeutics Program (http://dtp.nci.nih.gov/docs/compare/compare.html) (IC50 or growth inhibition of 50%) of cisplatin and carboplatin in the NCI-60 NSCLC cell lines (Figure 2A, A549, EKVX, HOP-62, HOP-92, NCI-H226, NCI-H23, NCI-H322M, NCI-H460, NCI-H522 and LXFL 529) and six NSCLC cell lines (Figure 2B, H23, H460, H727, HCC827, H1975 and A549) that are used in this study for AMS analysis.30, 31 For the NCI-60 panel, Rixe et al. found that cisplatin and carboplatin show a similar sensitivity profile with a high Pearson correlation coefficient of 0.798. This indicates that both drugs share similar resistance mechanisms. Concordantly, we observed statistically significant linear correlation of the cytotoxicity of these two drugs in the ten NSCLC cell lines of the NCI-60 panel (R2 = 0.66, p = 0.004) and in the 6 NSCLC cell lines used in this study (including two NCI-60 cell lines, R2 = 0.72, p = 0.033).
Figure 2. Linear correlation of IC50 for cisplatin and carboplatin in NSCLC cell lines.
Linear correlation of cisplatin IC50 with carboplatin IC50 in A. ten NSCLC cell lines from the NCI-60 panel (R2 = 0.66, p = 0.004) and B. six additional NSCLC cell lines used in this manuscript (R2 = 0.72, p = 0.033). Only mean values are shown.
DNA monoadduct levels induced by microdose and therapeutic concentrations of carboplatin in cell culture are linearly proportional
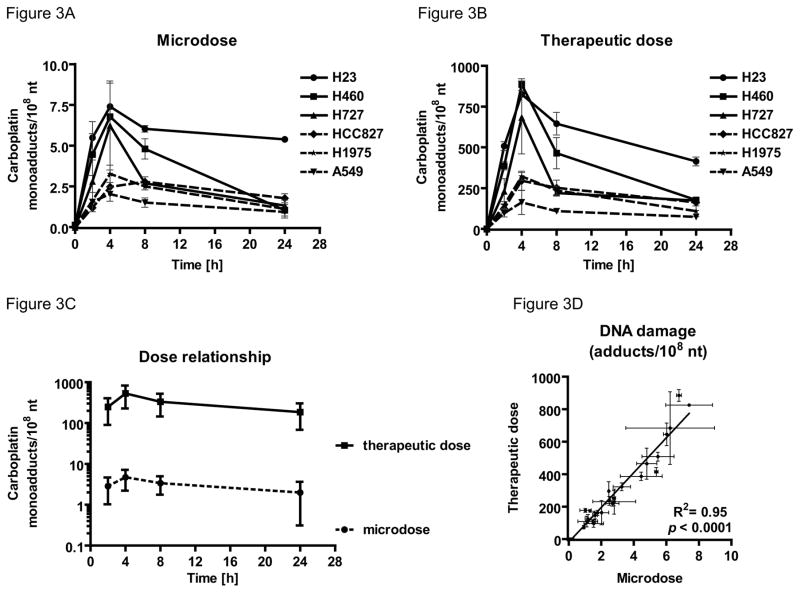
DNA damage induced by therapeutically relevant doses (100 μM) or microdoses (1 μM) of carboplatin was determined through the AMS analysis of purified genomic DNA. To mimic the in vivo carboplatin half-life of 1.1–5.9 hours26 the six NSCLC cell lines were treated for 4 hours and drug-DNA monoadduct levels were measured over a period of 24 hours (Figure 3). There was a time-dependent increase in carboplatin-DNA monoadduct levels during the first 4 hours of incubation with microdose (Figure 3A) or therapeutic (Figure 3B) concentrations of carboplatin followed by a gradual decrease over the subsequent 20 hours of incubation in drug-free medium. Peak adduct levels varied considerably between different cell lines even though all cell lines showed the same overall trend with respect to reaching a maximum adduct level and a gradual decrease after the drug was removed. Carboplatin-DNA monoadduct level at each time point in the microdosing group ranged from ~1 to 10 monoadducts per 108 nucleotides (nt) and are approximate 100-fold lower adduct levels then the therapeutic group which ranged from ~100–1,000 monoadducts per 108 nt (Figure 3C). Linear regression analysis showed the monoadduct levels induced by the two carboplatin doses were highly linear and statistically significantly correlated (R2 = 0.95, p < 0.0001, Figure 3D), suggesting that the nontoxic microdosing approach can be used to predict the DNA monoadduct levels induced by therapeutic carboplatin.
Figure 3. Microdose-induced carboplatin–DNA monoadduct levels correlate therapeutic adduct levels.
Indicated six NSCLC cells lines were dosed for 4 h followed by washing and incubation in drug-free medium as described. A. Monoadduct formation over time induced by microdoses (1 μM). B. Monoadduct formation over time induced by therapeutic doses (100 μM). C. Dose proportionality of microdose and therapeutic dose induced carboplatin-DNA monoadduct level onto log scale. D. Linear correlation of carboplatin-DNA monoadduct level induced by microdosing and therapeutic carboplatin, showing that the monoadduct levels induced by therapeutic carboplatin was highly linear to the monoadduct levels induced by microdosing carboplatin (R2 = 0.95, p < 0.0001). Mean values and standard error are shown.
Correlation of monoadduct levels and resistance to Pt analogs
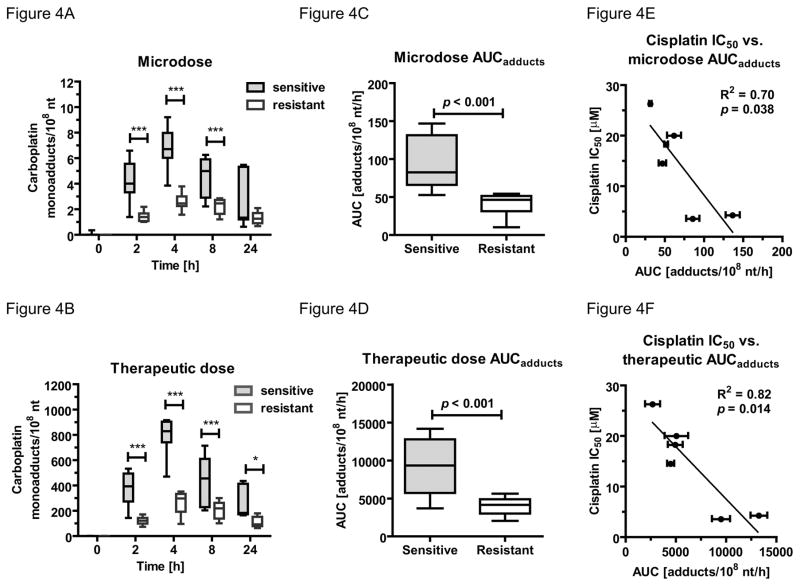
We correlated carboplatin-DNA monoadduct levels from both doses over 24 h to the IC50 data for each cell line (Figure 4). At each time point the three most resistant cell lines A549, H1975 and HCC827 had lower carboplatin-DNA monoadduct levels than the three most sensitive cell lines H23, H460 and H727 (Figure 4A+B and SI Table 1). The average area under curve (AUC) of the three resistant cell lines were significant lower than the mean AUC of the sensitive cell lines (40.98 ± 14.3 versus 94.61 ± 33.99 monoadducts per 108 nt per hour, p = 0.005 (Figure 4C) and 4034 ± 1172 versus 9265 ± 3656 monoadducts per 108 nt per hour, p = 0.0009 (Figure 4D) for microdose and therapeutic dose, respectively). Correlation analysis was performed to determine the relation of AUC and IC50 values of the six cell lines (Figure 4E+F). The AUC of monoadducts induced by microdosing (R2 = 0.70, p = 0.038) and therapeutic (R2 = 0.82, p = 0.014) carboplatin are demonstrating an inverse relationship to the cisplatin IC50 values. Similar correlations were also observed regarding cellular sensitivity to carboplatin (Supplemental Figure 1A and B).
Figure 4. Correlation between NSCLC cell line sensitivity and carboplatin-DNA monoadduct levels.
Box plot (whiskers min to max) comparing monoadduct levels in carboplatin-sensitive (H23, H460, H727) and –resistant (HCC827, H1975, A549) NSCLC cell lines over 24 h. Three most resistant cell lines (white box) had significant (*** = p < 0.001, * = p < 0.05, one-way ANOVA with Bonferroni’s multiple comparison test) lower adduct level A. after microdose or B. therapeutic dose. Comparison of area under the adduct curve (AUCadduct) between sensitive (grey box) and resistant (white box) in NSCLC cell lines after 4h of treatment with C. 1 μM (microdose) or D. 100 μM (therapeutic dose) of carboplatin. The sensitive cell lines had significant higher AUCadduct (p < 0.001, student t-test) the resistant NSCLC cell lines. Linear correlation of AUCadduct E. microdose (R2 = 0.70, p = 0.038) or F. therapeutic dose (R2 = 0.82, p = 0.014) with cell sensitivity (IC50) towards cisplatin.
ERCC1 expression does not correlate with cellular sensitivity towards carboplatin or cisplatin
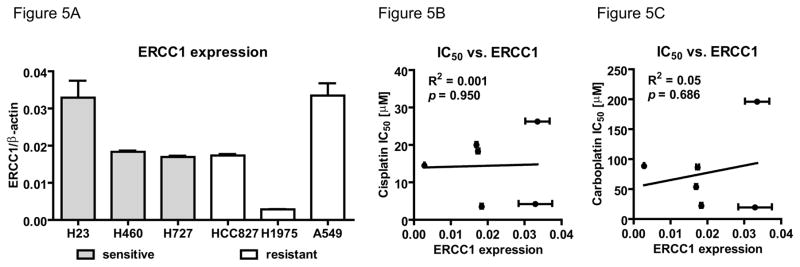
Quantitative analysis of ERCC1 expression in NSCLC tissue has been evaluated as a predictive biomarker for Pt-based chemotherapy in the clinic.32 We determined ERCC1 expression by quantitative RT-PCR (Figure 5A) and correlated cisplatin (Figure 5B) and carboplatin (Figure 5C) IC50 values of six NSCLC cell lines levels to the ERCC1 transcripts. Contrary to the inverse correlation of AUC with IC50 values, there was no such an association between the ERCC1 expression in these six cell lines (p = 0.950 and 0.686 for cisplatin and carboplatin, respectively).
Figure 5. No correlation between cisplatin or carboplatin sensitivity and ERCC1 mRNA levels.
A. Bar graph of relative ERCC1 (ERCC1/β-actin ratio) mRNA expression level of indicated NSCLC cell lines (grey bar = sensitive, white bar = resistant) shown as mean and SD. Linear regression analysis of B. cisplatin (R2 = 0.001, p = 0.950) or C. carboplatin (R2 = 0.05, p = 0.686) IC50 with relative ERCC1 expression.
A pilot diagnostic feasibility trial in human NSCLC patients
To assess the possibility to use microdosing to predict clinical outcome in NSCLC patients a clinical feasibility trial was initiated at UC Davis. A total of 21 patients were accrued, including 14 bladder cancers, 6 NSCLC and one mediastinal mass that was subsequently identified as Hodgkin’s lymphoma. This paper only discusses the data for NSCLC patients. The bladder cancer data was published separately, along with a supporting mouse study.33
Of the 6 NSCLC patients that completed the microdosing and blood sampling, only three received subsequent full dose platinum based chemotherapy and could be evaluated for clinical response. The enrolled NSCLC patients in this pilot study included two males and four females with the age ranging from 51 to 72 years. The patients’ characteristics are summarized in SI Table 2.
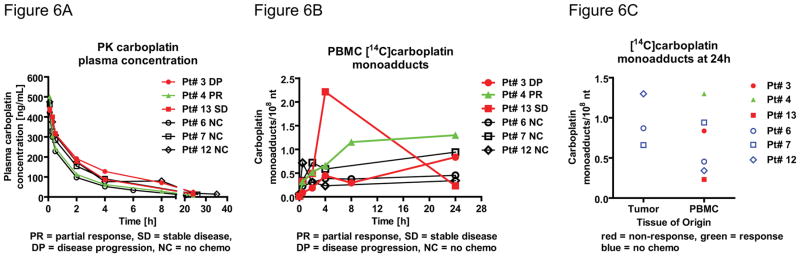
With the microdose composition of [14C]carboplatin of 107 dpm/kg and a total carboplatin dose of 1% of the therapeutic dose (target AUC of 6), the 14C-signal in DNA isolated from PBMC was approximately 10–100 times the background, which allowed accurate adduct measurement by AMS (Figure 6). As expected, no microdose-associated adverse events were observed in any of the patients. The administered diagnostic [14C]carboplatin microdose appears to be safe in this patient population. In comparison to the annual effective radiation dose equivalent from natural internal sources of 1.6 mSv per person,34 and a radiation exposure for an abdominal CT scan of 10 mSv, 35 the average administration of the diagnostic microdose was not greater than 0.1 mSv.
Figure 6. Carboplatin microdose elimination kinetics during a feasibility clinical trial.
A. Carboplatin plasma concentration over 24 hours after microdosing as determined from whole plasma by LSC (N = 6 patients). Initial elimination T1/2α: 38–57 minutes, second phase T1/2β: 6.7–11.2 hours. B. Pharmacodynamic analysis of carboplatin-DNA monoadducts in PBMC over 24 hours. C. Carboplatin-DNA monoadducts in PBMC (N = 6) and in tumor (N = 3) biopsy specimens at 24h. (red = non-responder, green = responder, black line = no response correlation possible, PR = partial response, SD = stable disease, DP disease progression, NC = no chemotherapy).
Immediately following the blood draws, whole plasma was isolated for PK analysis via liquid scintillation counting (LSC). The PK of the diagnostic microdoses (Figure 6A) showed biphasic elimination kinetics, with an initial T1/2α (absorption and distribution) of 0.63–0.95 hours and T1/2β (elimination) of 6.7–11.2 hours, which are similar to the published values.26, 36–39 At 24 hours after dosing, over 99% of 14C label was cleared from the blood, suggesting this is the optimal time for biopsy with little concern for 14C contamination.
Carboplatin-DNA monoadduct levels in genomic DNA from PBMC was chosen as a surrogate biomarker for analysis in tumor since several studies showed that platinum-based drug-DNA adducts in PBMC correlate with tumor response to chemotherapy, and because blood samples are easy to obtain.16–18, 40–44 Radiocarbon content in the DNA was measured by AMS according to published protocols.27 The levels of carboplatin-DNA monoadducts were readily detectable and ranged from 0.23 to 1.30 monoadducts per 108 nucleotides (Figure 6B and SI table 2). Three patients received subsequent platinum-based chemotherapy within four weeks after the microdosing procedure and clinical response could be correlated to adduct level (Figure 6B). One patient responded to chemotherapy (green symbols and line) whereas two patients showed stable disease or progression (red symbols and lines). At 24h the responding patient exhibited higher PBMC monoadduct level (1.30 monoadducts/108 nt) than the two non-responders (0.837 and 0.232 monoadducts/108 nt, respectively). However, the non-responder with stable disease (red square symbols) showed the highest AUC (27.9 monoadducts/108 nt per hour) compared to the other non-responder (11.4 monoadducts/108 nt per hour) and the responder (25.2 monoadducts/108 nt per hour). Adduct correlation to clinical outcome was not possible for three patients that underwent the microdosing procedure but that did not receive subsequent chemotherapy. Although the adduct level difference between response groups are not yet statistically significant, the early trend associating DNA monoadduct levels with patient response in encouraging.
Even though most of the [14C]carboplatin was cleared from blood over 24h, monoadduct levels were readily measurable in DNA isolated from PBMC and tumor tissue. However, more patients need to be accrued to determine if monoadduct levels are dependent upon tissue type and are predictive of clinical response in NSCLC.
There was no significant correlation between the plasma half-life values (T1/2α and T1/2β) and 24h PBMC drug-DNA monoadduct levels (SI figure 2), suggesting that the intracellular mechanisms predominate in the formation and repair of DNA damage rather than serum PK.
Discussion
We determined the feasibility of a diagnostic microdosing approach for the study of cellular sensitivity to cisplatin and carboplatin. Our data support the idea that microdose-induced carboplatin-DNA monoadduct levels are predictive of response to either cisplatin or carboplatin. There was a significant linear correlation between cisplatin and carboplatin cell IC50 values in a panel of NSCLC cell lines. The IC50 values for both drugs also significantly correlated to carboplatin-DNA monoadduct levels for each cell line. We also observed that carboplatin-DNA monoadduct levels induced by therapeutically relevant concentrations of carboplatin significantly correlated to those formed by microdoses.
The positive correlation of adduct levels to IC50 was superior compared to ERCC1 mRNA levels (Figure 4 and 5). ERCC1 was previously shown to correlate with resistance to platinum-based chemotherapy in NSCLC.32 However, this protein is only one of many that participate in the major DNA repair pathways that interact with platinum-DNA adducts such as nucleotide excision repair, mismatch repair and recombinational repair. High levels of ERCC1 indicate the capability of fast DNA repair and probably confer resistance to Pt therapy in some patients. However, enhancement of nucleotide excision repair by ERCC1 may not be relevant if other cellular drug resistance mechanisms predominate. Although other DNA repair mechanisms certainly act upon platinum-DNA adducts, the use of the related gene expression or protein levels as biomarkers of response have not proven to be clinically useful (reviewed in 45). For example, ERCC1 antibodies are known to have lot-to-lot variation in substrate binding. A prospective phase III trial including 275 patients with stage IV lung cancer failed to validate RRM1 tumor expression as a biomarker of platinum-based treatment response. BRCA1 mRNA levels in tumor tissue also failed to predict platinum resistance in 287 stage IV NSCLC patients. Of the other multivariate panels for NSCLC, few have been replicated in independent cohorts for prognostic value, and none were externally validation for predictive value. Clearly, such tests still need substantial additional validation before inclusion into routine practice for cytotoxic chemotherapy.45
ERCC2 single nucleotide polymorphisms (SNPs) and clinical outcome in NSCLC have been intensively studied. However, inconsistent and in some cases contradictory results have been published. For example, the results of one study, where the rs50872 locus variation was linked to poorer response and outcomes,46 could not be duplicated by another group, where it was found to be a favorable marker.47 In another prospective study no correlation of ERCC2 SMP with chemotherapy response was found, only a significant association with radiotherapy-derived toxicity was established.48
Other mechanisms include, but are not limited to, decreased drug uptake, increased efflux, drug inactivation by cellular antioxidants such as glutathione and others (Figure 1B).4 In the six cell line panel we examined, carboplatin-DNA monoadduct levels were superior predictors of cellular resistance compared to ERCC1 expression levels (Figure 4).
Diagnostic microdosing is an attractive concept due to the simplicity of measuring a single marker that is influenced by a large array resistance factors. Furthermore, measurement of drug-DNA adduct levels may be useful as a tool for better understanding DNA repair pathways of clinical relevance. For example, the combination of carboplatin with DNA repair inhibitors such as a poly ADP ribose polymerase (PARP) may be most efficacious in patients with a propensity for forming high levels to carboplatin-DNA adducts or those patients with rapid DNA repair. Mouse and clinical studies have already shown that ABT-888, an oral PARP inhibitor, potentiates the anti-tumor activity of carboplatin.49, 50
Our preclinical cell line data form the foundation of a currently active feasibility microdosing trial aimed at determining whether carboplatin-DNA monoadduct levels in PBMC and tumor tissue correlate with the response to carboplatin-based chemotherapy in NSCLC and bladder cancer. Our pilot study showed that a diagnostics feasibility study can be performed in patients without any detectable toxicity associated with the microdose. This pilot study was not designed to demonstrate statistical significance of carboplatin-DNA adduct frequency as a biomarker, but still yielded some encouraging results (Figure 6). First, we developed protocols for conducting a clinical trial with a 14C-labeled drug and processing samples for AMS analysis. Based on this effort, we decided to perform the biopsy at 24 hours after dosing. At this time point, there were large inter-patient variations in drug-DNA adduct levels and the [14C]carboplatin concentration in plasma was low, minimizing the possibility of contamination. Second, we defined a clinically useful of carboplatin microdose formulation consisting of 1% of the therapeutic dose of carboplatin and 107 dpm/kg of body weight of labeled drug—enough to detect carboplatin-DNA adducts but with minimal clinically acceptable radiation exposure to the patient. We were able to correlate carboplatin-DNA monoadduct levels with clinical response in three patients. One patient with the highest adduct levels after 24h had a positive clinical response, whereas the two patients with lower adduct levels did not respond. Patient 13 was potentially an outlier with respect to a very high initial binding of carboplatin to DNA followed by a rapid decrease in signal over time. Additional accruals are needed to better understand this observation.
In conclusion, we developed a highly sensitive AMS-based assay that can possibly identify cellular sensitivity to platinum-based drugs prior to toxic treatment. Based on this pilot study, a diagnostics feasibility clinical trial is currently in progress with enough power for statistical analysis to determine if carboplatin-DNA monoadduct levels correlate with cancer resistance.
Supplementary Material
Novelty and Impact.
This study assessed the feasibility of using microdoses (approximately 1/100th the therapeutic dose) of [14C]carboplatin to predict response to cisplatin or carboplatin in non-small cell lung cancer (NSCLC). The 14C label enabled detection of [14C]carboplatin-DNA adducts by accelerator mass spectrometry. The adduct levels correlated with drug cytotoxicity in six NSCLC cell lines. A diagnostic microdosing clinical trial was initiated to establish a useful microdose [14C]carboplatin formulation and obtain preliminary clinical data.
Acknowledgments
We are grateful to Laurel Beckett, Judy Li and Primo Lara for helpful discussions and advice. Work was funded by NIH grants CA93373, SBIR contracts to AMD Phase I HHSN261201000133C, Phase II HHSN261201200048C, LLNL grants LDRD 08-LW-100, NIH/NIGMS P41 RR13461, American Cancer Society, the Knapp Family Fund, and VA Career Development Award-2. Work performed (partially) at the Research Resource for Biomedical AMS, which is operated at LLNL under the auspices of the U.S. Department of Energy under contract DE-AC52-07NA27344. The Research Resource is supported by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program grant P41 RR13461. Drs. Pan, Henderson, Zimmermann and Cimino are shareholders of Accelerated Medical Diagnostics, Inc.
Abbreviations
- NSCLC
non-small cell lung cancer
- Pt
platinum
- AMS
accelerator mass spectrometry
- PBMC
peripheral blood mononuclear cells
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cytotoxicity assay
- PK
pharmacokinetics
- PD
pharmacodynamics
- LSC
liquid scintillation counting
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Mao Y, Yang D, He J, Krasna MJ. Epidemiology of Lung Cancer. Surgical Oncology Clinics of North America. 2016;25:439–45. doi: 10.1016/j.soc.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. Comparison of Four Chemotherapy Regimens for Advanced Non–Small-Cell Lung Cancer. New England Journal of Medicine. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 4.Cimino GD, Pan CX, Henderson PT. Personalized medicine for targeted and platinum-based chemotherapy of lung and bladder cancer. Bioanalysis. 2013;5:369–91. doi: 10.4155/bio.12.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dasari S, Bernard Tchounwou P. Cisplatin in cancer therapy: Molecular mechanisms of action. European Journal of Pharmacology. 2014;740:364–78. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen D-W, Pouliot LM, Hall MD, Gottesman MM. Cisplatin Resistance: A Cellular Self-Defense Mechanism Resulting from Multiple Epigenetic and Genetic Changes. Pharmacological Reviews. 2012;64:706–21. doi: 10.1124/pr.111.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nature reviews Cancer. 2007;7:573–84. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 8.Chaney SG, Campbell SL, Temple B, Bassett E, Wu Y, Faldu M. Protein interactions with platinum–DNA adducts: from structure to function. Journal of inorganic biochemistry. 2004;98:1551–59. doi: 10.1016/j.jinorgbio.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 9.Goodisman J, Hagrman D, Tacka KA, Souid A-K. Analysis of cytotoxicities of platinum compounds. Cancer chemotherapy and pharmacology. 2006;57:257–67. doi: 10.1007/s00280-005-0041-4. [DOI] [PubMed] [Google Scholar]
- 10.Unger FT, Klasen HA, Tchartchian G, de Wilde RL, Witte I. DNA damage induced by cis- and carboplatin as indicator for in vitro sensitivity of ovarian carcinoma cells. BMC cancer. 2009;9:1–9. doi: 10.1186/1471-2407-9-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nature reviews Drug discovery. 2005;4:307–20. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 12.Wheate NJ, Walker S, Craig GE, Oun R. The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 2010;39:8113–27. doi: 10.1039/c0dt00292e. [DOI] [PubMed] [Google Scholar]
- 13.Massari F, Santoni M, Ciccarese C, Brunelli M, Conti A, Santini D, Montironi R, Cascinu S, Tortora G. Emerging concepts on drug resistance in bladder cancer: Implications for future strategies. Critical reviews in oncology/hematology. 2015;96:81–90. doi: 10.1016/j.critrevonc.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Veal GJ, Dias C, Price L, Parry A, Errington J, Hale J, Pearson ADJ, Boddy AV, Newell DR, Tilby MJ. Influence of Cellular Factors and Pharmacokinetics on the Formation of Platinum-DNA Adducts in Leukocytes of Children Receiving Cisplatin Therapy. American Association for Cancer Research. 2001;7:2205–12. [PubMed] [Google Scholar]
- 15.Kim ES, Lee JJ, He G, Chow C-W, Fujimoto J, Kalhor N, Swisher SG, Wistuba II, Stewart DJ, Siddik ZH. Tissue Platinum Concentration and Tumor Response in Non–Small-Cell Lung Cancer. Journal of Clinical Oncology. 2012;30:3345–52. doi: 10.1200/JCO.2011.40.8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schellens JH, Ma J, Planting AS, van der Burg ME, van Meerten E, de Boer-Dennert M, Schmitz PI, Stoter G, Verweij J. Relationship between the exposure to cisplatin, DNA-adduct formation in leucocytes and tumour response in patients with solid tumours. British journal of cancer. 1996;73:1569–75. doi: 10.1038/bjc.1996.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reed E, Parker RJ, Gill I, Bicher A, Dabholkar M, Vionnet JA, Bostick-Bruton F, Tarone R, Muggia FM. Platinum-DNA Adduct in Leukocyte DNA of a Cohort of 49 Patients with 24 Different Types of Malignancies. Cancer research. 1993;53:3694–99. [PubMed] [Google Scholar]
- 18.Parker RJ, Gill I, Tarone R, Vionnet JA, Grunberg S, Muggia FM, Reed E. Platinum—DNA damage in leukocyte DNA of patients receiving carboplatin and cisplatin chemotherapy, measured by atomic absorption spectrometry. Carcinogenesis. 1991;12:1253–58. doi: 10.1093/carcin/12.7.1253. [DOI] [PubMed] [Google Scholar]
- 19.Bonetti A, Apostoli P, Zaninelli M, Pavanel F, Colombatti M, Cetto GL, Franceschi T, Sperotto L, Leone R. Inductively coupled plasma mass spectroscopy quantitation of platinum-DNA adducts in peripheral blood leukocytes of patients receiving cisplatin- or carboplatin-based chemotherapy. Clinical Cancer Research. 1996;2:1829–35. [PubMed] [Google Scholar]
- 20.Motzer RJ, Reed E, Perera F, Tang D, Shamkhani H, Poirier MC, Tsai W-Y, Parker RJ, Bosl GJ. Platinum-DNA adducts assayed in leukocytes of patients with germ cell tumors measured by atomic absorbance spectrometry and enzyme-linked immunosorbent assay. Cancer. 1994;73:2843–52. doi: 10.1002/1097-0142(19940601)73:11<2843::aid-cncr2820731130>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 21.FDA. U.S. Food and Drug Administration website. [Accessed March 3, 2015]. Guidance for industry, investigators, and reviewers: exploratory IND studies. Published January 2006. [Google Scholar]
- 22.Garner RC. Practical experience of using human microdosing with AMS analysis to obtain early human drug metabolism and PK data. Bioanalysis. 2010;2:429–40. doi: 10.4155/bio.10.6. [DOI] [PubMed] [Google Scholar]
- 23.Ho GY, Woodward N, Coward JIG. Cisplatin versus carboplatin: comparative review of therapeutic management in solid malignancies. Critical reviews in oncology/hematology. 2016;102:37–46. doi: 10.1016/j.critrevonc.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Henderson PT, Li T, He M, Zhang H, Malfatti M, Gandara D, Grimminger PP, Danenberg KD, Beckett L, de Vere White RW, Turteltaub KW, Pan CX. A microdosing approach for characterizing formation and repair of carboplatin-DNA monoadducts and chemoresistance. International journal of cancer Journal international du cancer. 2011;129:1425–34. doi: 10.1002/ijc.25814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. Journal of Immunological Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 26.Hospira I. Carboplatin Injection Product Insert. 2015. Revised: 8/2015. [Google Scholar]
- 27.Ognibene TJ, Bench G, Vogel JS, Peaslee GF, Murov S. A high-throughput method for the conversion of CO2 obtained from biochemical samples to graphite in septa-sealed vials for quantification of 14C via accelerator mass spectrometry. Analytical chemistry. 2003;75:2192–6. doi: 10.1021/ac026334j. [DOI] [PubMed] [Google Scholar]
- 28.Vallböhmer D, Iqbal S, Yang DY, Rhodes KE, Zhang W, Gordon M, Fazzone W, Schultheis AM, Sherrod AE, Danenberg KD, Lenz H-J. Molecular determinants of irinotecan efficacy. International Journal of Cancer. 2006;119:2435–42. doi: 10.1002/ijc.22129. [DOI] [PubMed] [Google Scholar]
- 29.Simon P. Q-Gene: processing quantitative real-time RT–PCR data. Bioinformatics. 2003;19:1439–40. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- 30.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nature reviews Cancer. 2006;6:813–23. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 31.Rixe O, Ortuzar W, Alvarez M, Parker R, Reed E, Paull K, Fojo T. Oxaliplatin, tetraplatin, cisplatin, and carboplatin: Spectrum of activity in drug-resistant cell lines and in the cell lines of the national cancer institute’s anticancer drug screen panel. Biochemical pharmacology. 1996;52:1855–65. doi: 10.1016/s0006-2952(97)81490-6. [DOI] [PubMed] [Google Scholar]
- 32.Lord RVN, Brabender J, Gandara D, Alberola V, Camps C, Domine M, Cardenal F, Sánchez JM, Gumerlock PH, Tarón M, Sánchez JJ, Danenberg KD, et al. Low ERCC1 Expression Correlates with Prolonged Survival after Cisplatin plus Gemcitabine Chemotherapy in Non-Small Cell Lung Cancer. American Association for Cancer Research. 2002;8:2286–91. [PubMed] [Google Scholar]
- 33.Zimmermann M, Wang SS, Zhang H, Lin TY, Malfatti M, Haack K, Ognibene T, Yang H, Airhart S, Turteltaub KW, Cimino GD, Tepper CG, et al. Microdose-induced Drug-DNA Adducts as Biomarkers of Chemotherapy Resistance in Humans and Mice. Molecular cancer therapeutics. 2016 doi: 10.1158/1535-7163.MCT-16-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.103 IP. The 2007 Recommendations of the International Commission on Radiological Protection. Annals of the ICRP. 2007;37:2–4. doi: 10.1016/j.icrp.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Brenner DJ, Hall EJ. Computed Tomography — An Increasing Source of Radiation Exposure. New England Journal of Medicine. 2007;357:2277–84. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- 36.van der Vijgh WF. Clinical Pharmacokinetics of Carboplatin. Clinical pharmacokinetics. 1991;21:242–61. doi: 10.2165/00003088-199121040-00002. [DOI] [PubMed] [Google Scholar]
- 37.Duffull S, Robinson B. Clinical Pharmacokinetics and Dose Optimisation of Carboplatin. Clinical pharmacokinetics. 1997;33:161–83. doi: 10.2165/00003088-199733030-00002. [DOI] [PubMed] [Google Scholar]
- 38.Sharma H, Thatcher N, Baer J, Zaki A, Smith A, McAucliffe CA, Crowther D, Owens S, Fox BW. Blood clearance of radioactively labelled cis-diammine 1,1-cyclobutane dicarboxylate platinum (II) (CBDCA) in cancer patients. Cancer chemotherapy and pharmacology. 1983;11:5–7. doi: 10.1007/BF00257407. [DOI] [PubMed] [Google Scholar]
- 39.Harland SJ, Newell DR, Siddik ZH, Chadwick R, Calvert AH, Harrap KR. Pharmacokinetics of cis-diammine-1,1-cyclobutane dicarboxylate platinum(II) in patients with normal and impaired renal function. Cancer research. 1984;44:1693–7. [PubMed] [Google Scholar]
- 40.Fichtinger-Schepman AMJ, van der Velde-Visser SD, van Dijk-Knijnenburg HCM, van Oosterom AT, Baan RA, Berends F. Kinetics of the Formation and Removal of Cisplatin-DNA Adducts in Blood Cells and Tumor Tissue of Cancer Patients Receiving Chemotherapy: Comparison with in Vitro Adduct Formation. Cancer research. 1990;50:7887–94. [PubMed] [Google Scholar]
- 41.Reed E, Ozols RF, Tarone R, Yuspa SH, Poirier MC. Platinum-DNA adducts in leukocyte DNA correlate with disease response in ovarian cancer patients receiving platinum-based chemotherapy. Proc Natl Acad Sci USA. 1987:84. doi: 10.1073/pnas.84.14.5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed E, Ozols RF, Tarone R, Yuspa SH, Poirier MC. The measurement of cisplatin-DNA adduct levels in testicular cancer patients. Carcinogenesis. 1988;9:1909–11. doi: 10.1093/carcin/9.10.1909. [DOI] [PubMed] [Google Scholar]
- 43.van de Vaart PJM, Belderbos J, de Jong D, Sneeuw KCA, Majoor D, Bartelink H, Begg AC. DNA-adduct levels as a predictor of outcome for NSCLC patients receiving daily cisplatin and radiotherapy. International Journal of Cancer. 2000;89:160–66. [PubMed] [Google Scholar]
- 44.Poirier MC, Reed E, Shamkhani H, Tarone RE, Gupta-Burt S. Platinum drug-DNA interactions in human tissues measured by cisplatin-DNA enzyme-linked immunosorbent assay and atomic absorbance spectroscopy. Environmental health perspectives. 1993;99:149–54. doi: 10.1289/ehp.9399149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergot E, Levallet G, Campbell K, Dubois F, Lechapt E, Zalcman G. Predictive biomarkers in patients with resected non-small cell lung cancer treated with perioperative chemotherapy. European Respiratory Review. 2013;22:565–76. doi: 10.1183/09059180.00007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiyohara C, Takayama K, Nakanishi Y. Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: A meta-analysis. Lung Cancer. 2006;54:267–83. doi: 10.1016/j.lungcan.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Wang S, Wang J, Bai Y, Wang Q, Liu L, Zhang K, Hong X, Deng Q, Zhang X, He M, Wu T, Xu P, et al. The genetic variations in DNA repair genes ERCC2 and XRCC1 were associated with the overall survival of advanced non-small-cell lung cancer patients. Cancer medicine. 2016;5:2332–42. doi: 10.1002/cam4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan I, Salazar J, Majem M, Pallares C, Del Rio E, Paez D, Baiget M, Barnadas A. Pharmacogenetics of the DNA repair pathways in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer letters. 2014;353:160–6. doi: 10.1016/j.canlet.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 49.Rugo HS, Olopade OI, DeMichele A, Yau C, van ‘t Veer LJ, Buxton MB, Hogarth M, Hylton NM, Paoloni M, Perlmutter J, Symmans WF, Yee D, et al. Adaptive Randomization of Veliparib–Carboplatin Treatment in Breast Cancer. New England Journal of Medicine. 2016;375:23–34. doi: 10.1056/NEJMoa1513749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karginova O, Siegel MB, Van Swearingen AED, Deal AM, Adamo B, Sambade MJ, Bazyar S, Nikolaishvili-Feinberg N, Bash R, O’Neal S, Sandison K, Parker JS, et al. <div xmlns=“http://www.w3.org/1999/xhtml”>Efficacy of Carboplatin Alone and in Combination with ABT888 in Intracranial Murine Models of BRCA-Mutated and BRCA–Wild-Type Triple-Negative Breast Cancer</div>. Molecular cancer therapeutics. 2015;14:920–30. doi: 10.1158/1535-7163.MCT-14-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.