Summary
A common feature of sex chromosomes is coordinated regulation of X-linked genes in one sex. Drosophila melanogaster males have one X chromosome, while females have two. The resulting imbalance in gene dosage is corrected by increased expression from the single X chromosome of males, a process known as dosage compensation. In flies, compensation involves recruitment of the Male Specific Lethal (MSL) complex to X-linked genes and modification of chromatin to increase expression. The extraordinary selectivity of the MSL complex for the X chromosome has never been explained. We previously demonstrated that the siRNA pathway, and siRNA from a family of X-linked satellite repeats (1.688X repeats), promote X-recognition. Now we test the ability of 1.688X DNA to attract compensation to genes nearby, and report that autosomal integration of 1.688X repeats increases MSL recruitment and gene expression in surrounding regions. Placement of 1.688X repeats opposite a lethal autosomal deletion achieves partial rescue of males, demonstrating functional compensation of autosomal chromatin. Females block formation of the MSL complex and are not rescued. The 1.688X repeats are therefore cis-acting elements that guide dosage compensation. Furthermore, 1.688X siRNA enhances rescue of males with a lethal deletion, but only when repeat DNA is present on the intact homolog. We propose that the siRNA pathway promotes X recognition by enhancing the ability of 1.688X DNA to attract compensation in cis. The dense and near-exclusive distribution of 1.688X sequences along the X chromosome suggests that they play a primary role in determining X identity during dosage compensation.
Keywords: Dosage compensation, sex chromosomes, X recognition, satellite repeats, 1.688X repeats, roX RNA, siRNA, Drosophila
Introduction
Males of many species carry a gene-rich X chromosome and a gene-poor, heterochromatic Y chromosome. Dosage compensation equalizes X-linked gene expression between XY males and XX females, thus maintaining a constant ratio of X:A gene products [1]. Strategies to accomplish this differ, but a unifying theme is coordinated regulation of an entire chromosome. Drosophila melanogaster males increase expression from most X-linked genes approximately twofold [2]. This is achieved in part by the Male Specific Lethal (MSL) complex, consisting of five proteins and one of two redundant, long non-coding RNAs called RNA on the X 1 and -2 (roX1 and roX2) [3]. The roX RNAs are essential for proper identification and compensation of X-linked genes [3, 4]. The MSL complex is recruited into the bodies of transcribed genes on the X, where it modifies chromatin through deposition of the activating H4K16ac mark [5]. How the MSL complex selectively recognizes X chromatin has never been explained.
Co-transcriptional assembly of the MSL complex is thought to occur at sites of roX RNA transcription [6, 7]. This is followed by binding of the MSL complex to X-linked loci called Chromatin Entry Sites (CESs) that are functionally defined by their ability to retain core MSL proteins in the absence of the complete complex [8, 9]. CESs contain the MSL Recognition Element (MRE), a 21 base pair, GA-rich motif required for MSL complex recruitment. The CLAMP protein is bound at MREs and may recruit the assembled MSL complex [10]. The MSL complex then spreads into nearby transcribed genes by recognition of active chromatin marks [11-13]. This elegant model suffers from major drawbacks. For example, X-identification cannot be attributed solely to MREs, since they are only ∼2 fold enriched on the X chromosome [8]. The discovery that the CLAMP protein enables MREs to recruit the MSL complex was a significant finding, but CLAMP-bound MRE motifs are present on all chromosomes. Modified MREs that also contact one of the MSL proteins are enriched on the X, but this motif is similarly found throughout the genome [14]. Although MREs play an important role in recruiting the MSL complex, their genome-wide distribution suggests that other factors uniquely specify X chromatin.
Both roX genes overlap CES and are X-linked, suggesting a role in marking the X for compensation. Indeed, when inserted on an autosome the roX genes recruit the MSL complex to the insertion site, and spreading to polytene bands hundreds of kb from the transgene is sometimes observed [15]. Subsequent studies confirmed up to two-fold activation of reporters and genes close to autosomal roX transgenes [16, 17]. The action of roX could be considered analogous to that of the long non-coding X-inactive specific transcript (Xist) that inactivates an X chromosome in mammalian dosage compensation. Xist is a part of the X inactivation center (Xic), a region that is necessary and sufficient to silence the entire chromosome [18]. Silencing is limited to chromatin in cis to the Xic, thus sparing the active X chromosome [19]. However, when roX genes are deleted from the X and placed on an autosome, roX RNA is incorporated into the MSL complex, which then travels to the X chromosome and rescues compensation [3]. Furthermore, MSL recruitment near autosomal roX insertions is considerably weaker than recruitment to the X chromosome. These observations indicate that the roX genes do not specify X identity by themselves. Exclusive X-recognition by the MSL complex must therefore involve additional mechanisms.
Our lab previously demonstrated a role for the siRNA pathway in X chromosome recognition [20]. This led to the discovery that siRNA from a repetitive element that is strikingly enriched on the X chromosome enhanced X-identification by the MSL complex and rescued the lethality of roX1 roX2 mutant males [21]. These repetitive sequences, the 1.688 g/cm3 satellite related repeats (hereafter 1.688X; superscript denotes cytological position) are ∼359 bp, AT-rich and found in short, tandem clusters in X euchromatin [22, 23]. Related repeats are found on other chromosomes, but clades that are near-exclusive to the X chromosome have been noted for 30 years [24, 25]. The 1.688X repeats are dissimilar in sequence to MREs within the CES, and display low recruitment of MSL proteins, suggesting that any role in recruitment of the MSL complex is indirect [26].
We postulated that the 1.688X repeats might act cooperatively with roX genes to identify X chromatin. The presence of hundreds of dispersed 1.688X repeats along the X chromosome makes evaluation of the effect of individual repeats impractical. Instead, we devised a functional approach that tested the ability of roX1, and three different 1.688X repeats, to recruit compensation to an autosome. A transgene carrying roX1 and 1.688X was integrated on an autosome. roX1 or 1.688X was then excised from the transgene, allowing testing of individual elements. As expected, the full transgene and roX1 alone recruit the MSL complex in cis. To our surprise, all 1.688X repeats tested were also able to recruit compensation to surrounding genes by themselves. All transgenes partially rescued a lethal autosomal deficiency of the homologous chromosome in males, but not in females, demonstrating functional compensation of autosomal chromatin. Finally, we demonstrate that ectopic production of siRNA from 1.6883F, previously shown to promote X recognition, enhances rescue of deficiency males if 1.688X sequences are present on the intact homolog. These findings suggest that recruitment of dosage compensation to nearby genes is a general property of 1.688X repeats. We postulate that the siRNA pathway normally acts at the dispersed, X-linked 1.688X sequences to promote identification of X chromatin.
Results
To test the idea that 1.688X act cooperatively with roX to identify X chromatin we generated transgenes with roX1 and 1.688X repeats [>roX1> w+mC>1.688X >]. LoxP and FRT sites (>) enable excision of roX1 or 1.688X to permit testing of individual elements (Fig. S1). Transgenes with 1.688X repeats from 1A, 3C and 3F (1.6881A, 1.6883C and 1.6883F) were integrated on 2L at cytological position 22A3, and the transgene carrying 1.6883F was also integrated at 24A2 and 25C7 (see STAR methods for details). Full transgenes are henceforth denoted as [roX1+1.688X] and reduced transgenes as [roX1] or [1.688X]. These repeats were selected for examination because ectopic 1.6883F siRNA production from a hairpin transgene ([hp 1.6883F]) achieved rescue of roX1 roX2 males, while similar transgenes producing hairpin RNA from 1.6881A (89 % identity to 1.6883F) and 1.6883C (68% identity to 1.6883F) afforded little or no rescue [21]. This suggested that 1.6883F, located on the X chromosome immediately distal to roX1, could have an unusual function, perhaps related to its situation near roX1. However, if 1.688X sequences generally identify X chromatin for compensation, we expect that many different repeat clusters will attract compensation to nearby genes.
1.688X and roX1 transgenes recruit MSL proteins to autosomal sites
Polytene chromosome preparations were made from a laboratory reference strain, and from males with full or reduced integrations at 22A3 ([roX1+1.6883F]22A3, [roX1]22A3, [1.6883F]22A3). These were immunostained for the core MSL complex component Male Specific Lethal 2 (MSL2). As expected, all preparations displayed strong recruitment to the X chromosome. No autosomal recruitment of MSL2 to 2L was detected in the laboratory reference strain (Fig. 1A, B). In contrast, males carrying complete or reduced transgenes displayed recruitment near the tip of 2L at cytological position 22A3 (yellow arrows, Fig. 1C-F). Surprisingly, recruitment of MSL2 by [1.6883F]22A3 was comparable to [roX1+1.6883F]22A3, and more robust than [roX1]22A3 (Table 1, Fig. S2). Both [1.6883F]22A3 and [roX1+1.6883F]22A3 support easily discernable spreading of MSL2 into the chromatin of 2L, most strikingly to a strong, subtelomeric band observed in most preparations with these transgenes (white arrows, Fig. 1D, F; Table 1). Interestingly, about half the preparations of [1.6883F]22A3 and [roX1+1.6883F]22A3 also showed MSL2 recruitment to the 3L telomere (white arrowheads, Fig. S2B, C). Although MSL2 did not spread into subtelomeric regions of [roX1]22A3 preparations, some proximal spreading was observed in about 20% of samples (Table 1). To determine if additional members of the MSL complex are similarly recruited, polytene preparations from [1.6883F]22A3 males were probed with antibodies to Male Specific Lethal 3 (MSL3) and Maleless (MLE). A partial complex capable of binding to the CES forms in the absence of MSL3, but spreading to nearby genes is reduced [12]. MLE colocalizes with MSL2, but is not considered part of the complex core [26]. Both antibodies detect minor signal at 22A3, as well as spreading into the subtelomeric region (Figure S3A-D). The 1.6883F repeats by themselves thus appear capable of recruiting intact MSL complexes.
Fig. 1. Autosomal insertions of roX1 or 1.6883F recruit MSL2.
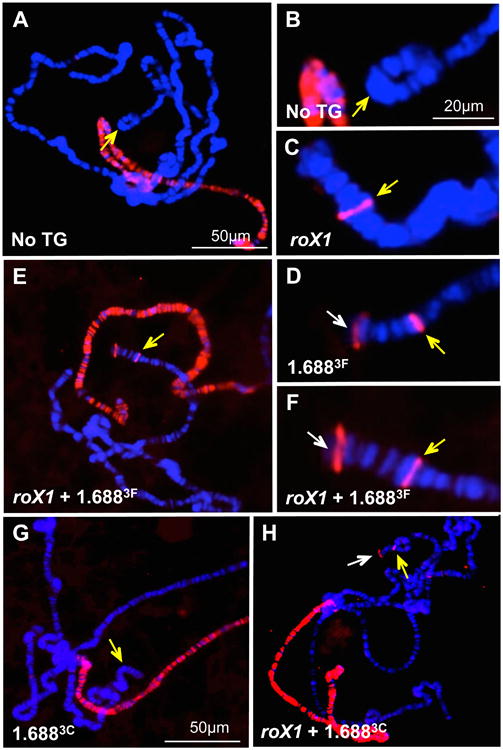
A) MSL2 recruitment (red) is limited to the X chromosome in a control male. B) Enlargement of cytological position 22A3 (yellow arrow) from A. C) [roX1]22A3 recruits MSL2 to a single band near 22A3. D) [1.6883F]22A3 recruits MSL2 robustly near the integration site (yellow arrow). Spreading to a sub-telomeric region is observed (white arrow). E) MSL2 recruitment by [roX1+1.6883F]22A3 is robust and similar to [1.6883F]22A3. Yellow arrow marks the 22A3 integration site. F) Detail from E. White arrow marks sub-telomeric spreading. Scoring of MSL2 recruitment and spreading is presented in Table 1. G) Visible MSL2 recruitment (red) in a [1.6883C]22A3 male larvae is limited to the X chromosome. Yellow arrow marks the insertion site at 22A3. H) Robust MSL2 recruitment (yellow arrow) and spreading to a subtelomeric region (white arrow) is observed in [roX1+1.6883C]22A3 males. Chromosome preparations were probed with anti-MSL2 antibody and detected by Texas Red. DNA is counterstained with DAPI (blue). See also Figures S1-S3 and Figure S5.
Table 1. Autosomal MSL2 recruitment by [roX1]22A3; [1.6883F]22A3 and [roX1+1.6883F]22A3.
Recruitment close to the integration site is ranked from none (-) to very strong (++++). Additional autosomal signals are categorized by cytological position. The percentage of nuclei in each category is followed by the number of nuclei (parentheses). Counts were derived from 3 individuals with no transgene (yw laboratory reference strain) and groups of 5 individuals with [roX1]22A3, [1.6883F]22A3 or [roX1+1.6883F]22A3. Scoring was done on coded samples to prevent bias. Representative images depicting weak (+) to very strong (++++) recruitment are presented in Figure S2A.
| Transgene | |||||
|---|---|---|---|---|---|
| None | roX1 | 1.6883F | roX1 + 1.6883F | ||
| 22A3 Recruitment | - | 100 (63) | 0 (0) | 0 (0) | 1.01 (1) |
| + | 0 (0) | 10.78 (11) | 0 (0) | 0 (0) | |
| ++ | 0 (0) | 65.68 (67) | 15.17 (17) | 30.3 (30) | |
| +++ | 0 (0) | 23.52 (24) | 55.35 (62) | 54.54 (54) | |
| ++++ | 0 (0) | 0 (0) | 29.46 (33) | 14.14 (14) | |
| Additional signal on 2L | None | 98.41 (62) | 75.49 (77) | 9.82 (11) | 15.15 (15) |
| Sub-telomeric | 0 (0) | 6.86 (7) | 89.3 (100) | 78.78 (78) | |
| Proximal to 22A3 | 1.5 (1) | 19.6 (20) | 8.03 (9) | 4.04 (4) | |
| 3L telomere | None | 98.41 (62) | 95.09 (97) | 49.1 (55) | 45.45 (45) |
| One Band | 1.58 (1) | 4.90 (5) | 50.89 (57) | 54.54 (54) | |
| Other | Ectopic bands on other chr | 3.17 (2) | 36.27 (37) | 7.14 (8) | 9.09 (9) |
| X-staining normal | 100 (63) | 100 (102) | 100 (112) | 100 (99) | |
| Total nuclei counted | 63 | 102 | 112 | 99 | |
To determine if recruitment is a general property of 1.688X repeats, we generated roX1 transgenes with 1.6881A and 1.6883C repeats, sharing 89% and 68% identity with 1.6883F, respectively. MSL2 was not detected on 2L in [1.6881A]22A3 or [1.6883C]22A3, and may be below the level detectable by antibody staining (Fig. 1G). However, [roX1+1.6881A]22A3 and [roX1+1.6883C]22A3 display recruitment of MSL2 to 2L and robust spreading into the sub-telomeric region (yellow and white arrows, Fig. 1 H). As recruitment is considerably stronger than that achieved by roX1 alone, 1.6883C and 1.6881A act synergistically with roX1 to elevate recruitment and spreading. We conclude that all three 1.688X repeats facilitate MSL2 recruitment and spreading, either independently or in cooperation with roX1.
We examined 1.688 X sequences for similarity to the CES consensus and found none (Fig. S4)[27]. To determine whether a group of closely related 359 bp repeats that comprise 10 Mb of pericentromeric X heterochromatin recruit MSL2, we performed immunostaining of mitotic embryo preparations. No convincing signal could be detected within X heterochromatin, largely composed of this repeat (Fig. S5). These findings are consistent with the fact that 1.688X repeats do not generally display MSL enrichment [26]. Taken together, these observations suggest that 1.688X repeats do not directly recruit MSL proteins, and likely act indirectly to promote MSL recognition of nearby chromatin.
Increased autosomal gene expression near transgene integrations
Recruitment of MSL2 by roX1 and 1.6883F integrations suggested a possible increase in expression of nearby genes in males. To test this, we generated chromosomes with three complete integrations on 2L, increasing the number of genes near integration sites ([roX1+1.6883F]22A3+24A2+25C7). Chromosomes with roX1 only ([roX1]22A3+24A2+25C7) or 1.6883F only ([1.6883F]22A3+24A2+25C7) were produced by Cre or FLP expression. Quantitative RT-PCR was used to measure mRNA from test genes situated near integration sites (Fig. 2A) and control genes on different chromosome arms. Control genes showed no expression differences between males of the laboratory reference strain and males with complete or reduced transgenes (white boxes, Fig. 2B). In contrast, genes near integration sites (gray boxes, Fig. 2B) displayed slightly elevated expression in [roX1]22A3+24A2+25C7 males. A greater, statistically significant increase of 30% was observed in [1.6883F]22A3+24A2+25C7 males, and [roX1+1.6883F]22A3+24A2+25C7 animals displayed a 50% increase in expression of nearby genes.
Fig. 2. Increased expression of autosomal genes near roX1 and 1.6883F transgenes.
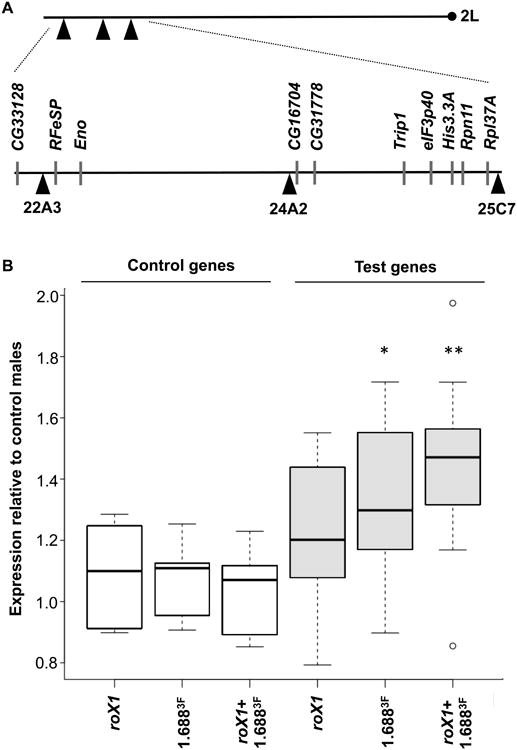
A) Three transgene integration sites on distal 2L are depicted by black triangles. The centromere is shown by a black circle. Positions of test genes and integration sites are depicted below. B) Relative expression of control genes (located on 2R or 3) and test genes was measured by quantitative RT-PCR. The ratio of expression in larval males with the indicated transgenes to control males (no transgenes) is depicted for 5 control genes (white) and 10 test genes (gray). Control genes show no changes in expression, but test genes display significant increases when [1.6883F]22A3+24A2+25C7 or [roX1+1.6883F]22A3+24A2+25C7 transgenes are present. * p<0.05, ** p<0.01. See also Figure S1 and Table S1.
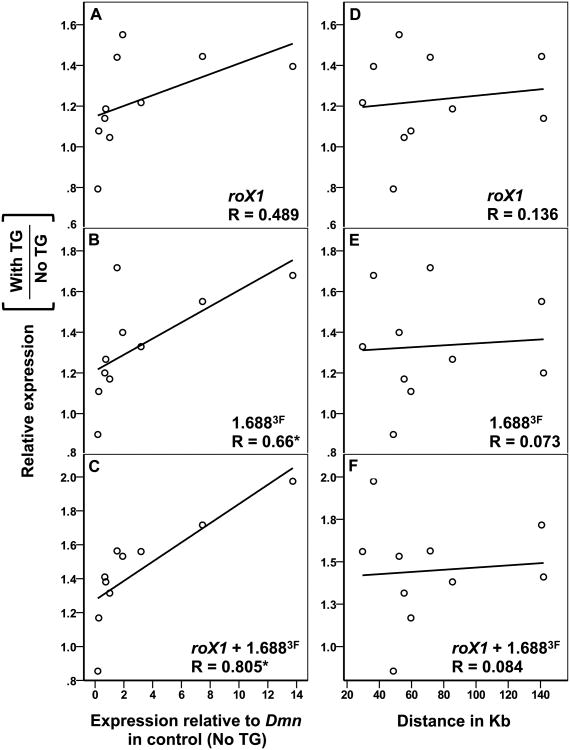
Since local MSL complex recruitment is cotranscriptional, highly expressed genes should be more strongly compensated [28, 29]. Indeed, the most highly expressed gene tested, RpL37A, displayed a 2-fold increase in expression, indicative of full compensation (Table S1). As a group, the test genes revealed a positive correlation between expression in the control and the relative increase achieved when roX1 and 1.6883F transgenes were present (Fig. 3A-C). The correlation was strongest when both roX1 and 1.6883F were present on the transgene, reflecting the presence of two recruiting elements. In contrast, no correlation between increased expression and distance to the nearest transgene was detected up to 142 kb (Fig. 3D-F). We conclude that local recruitment of the MSL complex by roX1 and 1.6883F transgenes displays the properties of dosage compensation as it favors active genes and achieves up to a two-fold increase in expression. The ability of roX1 and 1.6883F to attract compensation to genes over 100 kb away is consistent with the long-range effects of transgenes containing only roX [17].
Fig. 3. Increased expression near roX1 and 1.6883F correlates with gene activity. A-C).
The fold change in expression of individual genes near 2L transgenes (Y-axis) is plotted against expression in control males (no transgenes; X-axis). Each gene was internally normalized to Dmn before calculating fold change. Dots represent the average of two biological replicates. A significant correlation is observed for [1.6883F]22A3+24A2+25C7 (B) and [roX1+1.6883F]22A3+24A2+25C7 (C) but not [roX1]22A3+24A2+25C7 (A). D-F) The fold change in expression of individual genes near 2L transgenes (Y-axis) is plotted against the distance between each gene and the nearest transgene (X-axis). No significant correlation was detected in these analyses. Pairwise correlation was performed using SPSS, the Pearson Correlation Coefficient (R) is shown (* p < 0.05). See also Figure S1 and Table S1.
1.688X and roX1 transgenes functionally compensate a 2L deficiency
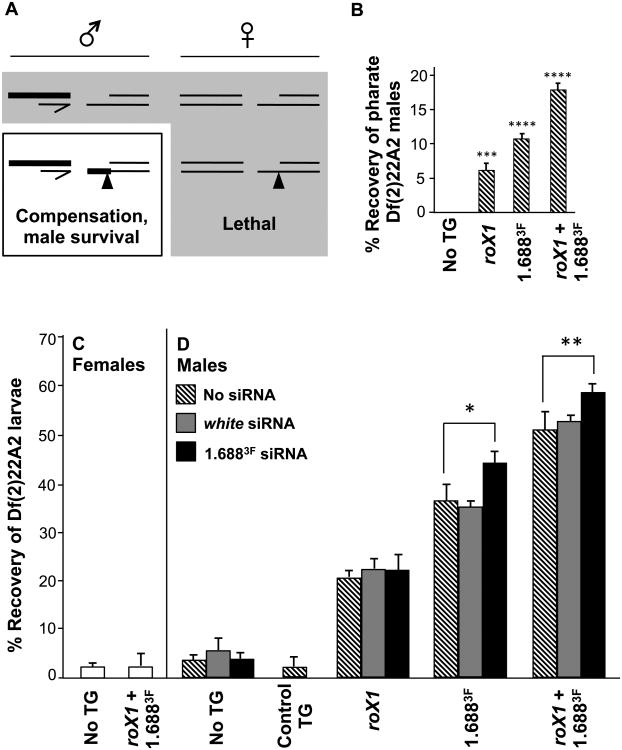
To determine if increased gene expression near roX1 and 1.688X transgenes constituted a functional dosage compensation system, we attempted rescue of the lethality produced by hemizygosity of distal 2L. Males were generated with a lethal 2nd chromosome deficiency and a single [roX1+1.6883F]22A3 transgene on the intact homolog, close to the deficiency break point. As the dosage compensation complex is only assembled in males, females served as a control (Fig. 4A). Significantly, this study was performed in flies that were completely wild type for the canonical dosage compensation machinery. Chromatin recognition and spreading will therefore occur in the context of a fully functional MSL complex.
Fig. 4. Insertions of roX1 and 1.6883F on 2L partially rescue males with a lethal 2L deficiency.
A) Df(2)22A2 is lethal in males (top left) and females (top right). If a transgene on the homolog recruits compensation, male-limited rescue will occur (bottom left). Thick lines indicate compensated chromatin. Females block formation of the MSL complex, preventing rescue (bottom right). B) Pharate Df(2)22A2 males are observed only when [roX1]22A3, [1.6883F]22A3 or [roX1+1.6883F]22A3 is present on the homolog. The recovery of pharate males is calculated from brothers that emerged with an intact 2nd chromosome (See Fig. S6A for mating strategy). C) Recovery of Df(2)22A2 female larvae is unaffected by [roX1+1.6883F]22A3 on the homolog (See Fig. S7B for mating strategy). D) Recovery of Df(2)22A2 male larvae increases when [roX1]22A3, [1.6883F]22A3 or [roX1+1.6883F]22A3 is present on the intact homolog (hatched bars), but not when a control P{EPgy2}hafEY08668 transgene marked with y+ and w+ is inserted at 22A3 (Control TG). Expression of siRNA to white (gray) has no effect on recovery of male larvae, but expression of 1.6883F siRNA (black) enhances recovery when transgenes containing 1.6883F DNA are present on the intact homolog (mating strategy in Fig. S7A, C). See Figure S1 for transgene composition and Figure S4 for 1.6883F sequence. Error bars represent SEM. * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
We generated translocation T(2;Y)22A2 by moving the distal 1.5 Mb of 2L onto the Y chromosome [30]. T(2;Y)22A2 can be separated into a terminal deficiency that is completely lethal (Df(2)22A2) and a reciprocal duplication on the Y chromosome (Dp(2:Y)22A2) by mating (Fig. S6A). Transgenes at the integration site closest to the deficiency break point, 22A3, were tested first. Neither [roX1]22A3, [1.6883F]22A3 or [roX1+1.6883F]22A3 rescued Df(2)22A2 males to adulthood. Df(2)22A2 adults could not be recovered when three copies of full or reduced transgenes were present at 22A3, 24A2 and 25C7. This was not surprising as the 24A2 and 25C7 integration sites are several Mb proximal to the deficiency. However, examination of vials producing Df(2)22A2 males with full or reduced transgenes at 22A3 revealed some adult Df(2)22A2 males within the pupal cuticle (pharate males), as well as a few dead but partially eclosed males (Fig. 4B). Dissection of pupal cases revealed adult males with eye color indicative of the Df(2)22A2 chromosome. The presence of Df(2)22A2 and lack of Dp(2:Y)22A2 was confirmed by PCR (Fig. S6B). In contrast, no pharate Df(2)22A2 males were observed from matings that lack transgene integrations, and no pharate females could be recovered with or without transgenes. roX1 alone enabled recovery of 6% pharate males, based on the survival of brothers with two intact 2nd chromosomes. [roX1+1.6883F]22A3 supported 18% pharate males. Intriguingly, the repeats alone, [1.6883F]22A3, supported 11% male pharate males, exceeding rescue with roX1 (Fig. 4B). While this suggests that roX1 and 1.6883F transgenes do achieve partial compensation of 2L, the difficulty in scoring pharate animals prompted a switch to examination of 3rd instar larvae.
Male larvae show functional compensation of autosomal chromatin
Late 3rd instar larvae from matings that produce Df(2)22A2 males and females were sexed and genotyped using visible markers, and a subset of Df(2)22A2 larvae were genotyped by PCR to confirm the deficiency (see mating strategies in Fig. S7A, B). Third instar Df(2)22A2 females were recovered at 3% the anticipated number in control matings in which no transgene is present on 2L, and female recovery is unchanged by the full [roX1+1.6883F]22A3 transgene (Fig. 4C). Df(2)22A2 males with no transgene are recovered at 4% (hatched bars, Fig. 4D). However, when [roX1]22A3 was present on the intact 2nd chromosome, 20% of Df(2)22A2 3rd instar males were recovered. This increased to 36.5% with [1.6883F]22A3, and to 51% when [roX1+1.6883F]22A3 was present (Fig. 4D). This confirms that 1.6883F repeats are more effective in recruiting compensation to nearby genes than roX1 itself, and suggests that roX1 and 1.6883F act in a cooperative manner.
The 22A3 landing site is marked with y+, and the intact [roX1+1.6883F]22A3 transgene also carries the w+mC marker. Both y and w are X-linked, and w+mC is partially compensated at many autosomal insertion sites. This raised the concern that the genetic markers might contribute to local recruitment. To address this, we tested the P{EPgy2}hafEY08668 insertion, 8 kb distal to the 22A3 landing site, marked with y+ and w+mC (Control TG, Fig. 4D). No rescue of 3rd instar Df(2)22A2 male larvae was detected, eliminating the possibility that the genetic markers alone are effective recruiting elements.
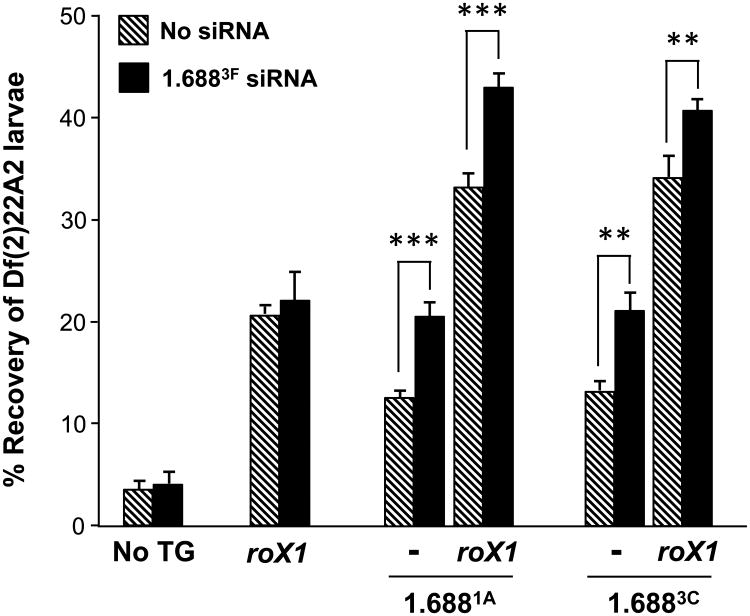
The absence of visible MSL2 recruitment by [1.6881A]22A3 and [1.6883C]22A3 suggested that these repeats might be ineffective by themselves. To test this we measured the survival of Df(2)22A2 male larvae with insertions of 1.6881A and 1.6883C repeats alone or with roX1 (hatched bars, Fig. 5). Surprisingly, [1.6881A]22A3 and [1.6883C]22A3 partially rescued Df(2)22A2 males, supporting 13% recovery of 3rd instar larvae. Recovery increased to 33-34% when roX1 was also present, exceeding rescue by roX1 alone. We conclude that even though [1.6881A]22A3 and [1.6883C]22A3 do not recruit visibly detectable levels of MSL2 on polytene preparations, they do in fact recruit dosage compensation and act cooperatively with roX1 to enhance compensation of nearby genes.
Fig. 5. Insertion of 1.6881A and 1.6883C on 2L partially rescue males with a lethal 2L deficiency.
Recovery of Df(2)22A2 male larvae increases when [1.6881A]22A3, [roX1+1.6881A]22A3, [1.6883C]22A3 or [roX1+1.6883C]22A3 is present on the intact homolog (hatched bars). The ratio of male larvae carrying Df(2)22A2 to brothers with an intact 2nd chromosome is presented. Recovery of males with no transgene or [roX1]22A3 is redrawn from Fig. 4D. Ectopic expression of 1.6883F siRNA (black) enhances recovery when 1.6881A or 1.6883C DNA is present on the homolog. The mating strategy is presented in Figure S7A, C. See Figure S1 for transgene composition and Figure S4 for comparison of 1.688X sequences used. Error bars represent SEM. ** p < 0.01; *** p < 0.001.
Ectopic expression of 1.6883F siRNA enhances rescue of male larvae when cognate sequence is present on 2L
Ectopic expression of hairpin RNA from 1.6883F produces abundant siRNA and partially rescues the lethality of roX1 roX2 males [21]. It is possible that the siRNA pathway acts upon chromatin at related repeats throughout the X, and that this facilitates recruitment of the MSL complex to nearby genes. If this is indeed the case, 1.6883F siRNA may also enhance recruitment of compensation by autosomal 1.688X transgenes. Ectopic expression of 1.6883F siRNA had no effect on the recovery of Df(2)22A2 male larvae with no transgene, or with [roX1]22A3 on the intact homolog (black bars, Fig. 4D; mating strategy presented Fig. S7C). However, expression of hp 1.6883F increased recovery of larval males carrying [1.6883F]22A3 or [roX1+1.6883F]22A3 by 8% (black bars, Fig. 4D). In contrast, expression of hp RNA to the non-essential white gene did not influence rescue by any transgene (gray bars, Fig. 4D). Enhanced rescue is therefore not the result of non-specific small RNA production.
To determine if 1.6883F siRNA also modulates the recruitment of compensation by 1.6881A and 1.6883C transgenes, we expressed hp 1.6883F in Df(2)22A2 males carrying [1.6881A]22A3, [roX1+1.6881A]22A3, [1.6883C]22A3 or [roX1+1.6883C]22A3 on the intact homolog. Expression of hp 1.6883F enhanced male survival when 1.6881A or 1.6883C was present on 2L, either alone or with roX1 (black bars, Fig. 5). The increase in survival, 7-9%, is comparable to that achieved in transgenes carrying 1.6883F. Taken together, these studies demonstrate that 1.6883F siRNA does not modulate the intrinsic activity of the MSL complex or influence recruitment by roX1, but acts through cognate sequence to elevate compensation at nearby genes. Complete sequence identity is not necessary, as the increase in survival achieved by production of 1.6883F siRNA in flies carrying [1.6883C]22A3 or [1.6883F]22A3 transgenes, containing repeats that share only 69% identity, was essentially identical. We propose that 1.6883F siRNA acts at numerous 1.688X repeats along the X chromosome, enabling these to more effectively recruit compensation to nearby genes.
Discussion
Previous studies have identified GA-rich MRE elements, as well as variations on this sequence, that directly recruit the CLAMP adapter protein and MSL complex [10, 14]. The enrichment of these motifs on the X chromosome is modest, suggesting that additional features contribute to X recognition [8]. Our prior studies led to the surprising conclusion that the siRNA pathway, and siRNA from a 1.688X repeat, participated in X recognition [20, 21, 31]. Many of the 1.688X repeats are transcribed and produce siRNA, making them attractive candidates for involvement in an siRNA-mediated process. The current study now demonstrates that 1.688X DNA itself is capable of attracting compensation to nearby transcribed genes, an effect that is enhanced by cognate siRNA. The mechanism by which 1.688X sequences on the X contribute to MSL recruitment is under investigation, but the repeats are not themselves sites of strong MSL recruitment, and thus anticipated to act indirectly. One hypothesis is that 1.688X sequences influence the architecture of the X chromosome to facilitate spreading of MSL complex along the chromosome [32]. Interestingly, the male X chromosome assumes a distinct interphase organization with compensated genes close together [33]. Parallel ideas have been proposed in mammalian dosage compensation, where the L1 (LINE-1) elements, and small RNA have been implicated in formation of a silencing domain [34].
Regardless of the mode of action, our study demonstrates that different 1.688X repeats share a remarkable ability to recruit compensation to nearby transcribed genes. Although visible recruitment of MSL2 by repeat-only transgenes was only observed for 1.6883F, 1.6881A and 1.6883C enhanced visible recruitment when roX1 was also present. More importantly, all three repeats alone supported detectable levels of autosomal compensation, as revealed by partial rescue of males with a lethal deficiency opposite a chromosome with a single transgene insertion. Functional compensation is thus achieved by MSL protein recruitment that is below the threshold for visual detection. It is important to note that, in all cases, partial rescue of Df(2)22A2 males was achieved by insertion of a single transgene with less than 2 kb of repeat DNA. In contrast, hundreds of 1.688X repeat clusters are broadly distributed across the X chromosome, an arrangement that provides redundancy of function.
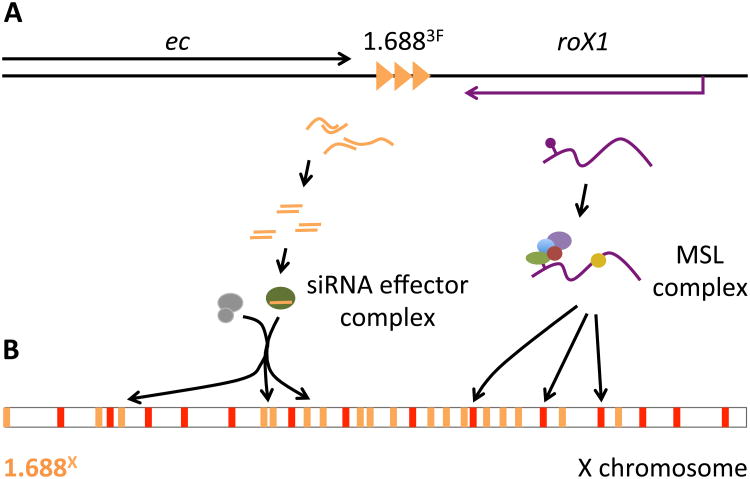
The finding that ectopic production of 1.6883F siRNA increased autosomal compensation when 1.6881A, 1.6883C or 1.6883F was present on the autosome links the compensation function of the siRNA pathway to the 1.688X repeats. Although the mechanism by which 1.6883F siRNA acts at disparate 1.688X sequences is unknown, siRNA has previously been found to regulate genes and mobile elements in fly somatic cells [35, 36]. It is plausible that 1.6883F siRNA enables recruitment of chromatin modifying activities to cognate loci, as in other organisms, and that these modifications alter properties of 1.688X chromatin (Fig. 6) [37, 38].
Fig. 6. Linkage of 1.6883F and roX1 could coordinate function.
A) 1.6883F is located between echinus (ec) and roX1. Bidirectional transcription of 1.6883F may generate siRNA that is loaded onto an effector complex (left). Ago2, and other genes in the siRNA pathway, participate in X recognition [20]. The roX1 transcript is assembled into the MSL complex (right). roX1 and 1.6883F produce different classes of noncoding RNA, and each element retains biological activity when separated from the other. It is possible that the proximity of 1.6883F and roX1 coordinates different pathways that cooperate to identify X chromatin. B) A siRNA-containing effector complex may recruit chromatin modifiers (gray) to 1.688X repeats (gold) across the euchromatic X chromosome. We hypothesize that this produces epigenetic or architectural changes that facilitate MSL recruitment, or spreading of the MSL complex along the X. The MSL complex is initially recruited to Chromatin Entry Sites (CES, red), and spreads into active genes nearby. No direct association between proteins of the siRNA pathway and the MSL complex has been reported, suggesting that the siRNA pathway influences MSL recruitment indirectly.
The unusual properties of 1.6883F siRNA, and the situation of this cluster immediately distal to roX1, raised suspicions that 1.6883F harbored a novel function related to roX1 [21]. For example, special properties of 1.6883F DNA could facilitate MSL complex spreading from roX1. In accord with this idea, 1.6883F does appear to recruit compensation more vigorously than 1.6881A and 1.6883C. One intriguing possibility is that the physical linkage of 1.6883F and roX1 coordinates two pathways that cooperate to identify X chromatin (Fig. 6). While the significance of this is unclear, it is interesting that roX1 is one of the earliest zygotic transcripts, and supports initial X recognition at 3 h of development [39, 40]. In contrast, roX2 is first expressed several hours later. It is possible that 1.6883F and roX1 collaborate in initial X recognition, and their close localization coordinates these activities.
Fly X chromosomes display the evolutionary signature of adaptation to compensation. Mutations, and expansions of GA on the fly X chromosome that contributed to the rise of MREs have been reported, and MREs have propagated across the D. miranda X chromosome by their inclusion in a mobile element [41-43]. More broadly, Drosophilid X chromosomes are strikingly enriched for chromosome-specific repeats, which account for 45 times more coverage on the D. melanogaster X chromosome than the autosomes [22]. This raises the possibility that additional families of repeats might function in a manner similar to the 1.688X repeats. Interestingly, a newly evolved X chromosome arm in D. pseudoobscura rapidly acquired satellite repeats that are present on the ancestral X [22]. How these repeats proliferate is unknown, but the current study suggests a mechanism that restricts 1.688X repeats to the X chromosome. The demonstration that autosomal insertions of 1.688X repeats induce misregulation of nearby genes in males reveals that autosomal 1.688X repeats would be targeted for elimination by natural selection. In contrast, 1.688X sequences on the X chromosome would enhance compensation of nearby genes, making them subject to positive natural selection.
STAR Methods
Contact For Reagent And Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Victoria H. Meller (vmeller@biology.biosci.wayne.edu).
Experimental Model And Subject Details
Fly culture and strains
Flies were maintained at 25°C on standard cornmeal agar diet in a humidified incubator. Strains are provided in the Star Methods section. Survival of male and female larvae to the end of pupation (pharate pupal stage) and to late 3rd instar (wandering stage) was measured. Animal sex was determined by gonad morphology (larvae) or adult genitalia (pharate pupae).
Method Details
Cloning and transgene integration
The [>roX1> w+mC>1.6883F >], [>roX1> w+mC>1.6881A >] and [>roX1> w+mC>1.6883C >] transgenes were assembled in the pUASTB vector by traditional cloning techniques[44]. Details of assembly and primer sequences are available upon request. 1.6883F repeats were contained in a 2 kb genomic fragment.1.6881A and 1.6883C repeats were introduced as 1.4 and 1.6 kb of amplified DNA, respectively. Construction was verified by restriction mapping, sequencing and PCR at each stage. Injections were performed by Genetic Services Inc. (Sudbury, MA) and Rainbow Transgenics (Camarillo, CA) in stocks containing 22A3 [VK00037], 24A2 [su(HW)attP6] and 25C7 [attP40] landing sites. In-situ hybridization to polytene preparations verified integration.
Scoring of larval rescue
Matings to generate Df(2)22A2 male and female larvae are presented in Fig. S7A, B. The intact 2nd chromosome was marked with p[Sqh-mCherry.M] to allow visual identification of Df(2)22A2 larvae. All third instar larvae were sexed and scored 3 times daily. PCR was performed on a subset of non-fluorescent larvae to confirm Df(2)22A2. Primers used for genotyping are available on request.
Immunodetection
Immunodetection of MSL2, MSL3 and MLE on polytene chromosomes was done as previously described [7, 45]. Briefly, transgenic larvae were grown at 18°C in uncrowded vials on standard cornmeal molasses food. Larvae were dissected in PBS, 4% formaldehyde, 1% Triton X100, and fixed for 45 sec. The fixative was replaced with 50% acetic acid, 4% formaldehyde for 2 min and then the glands were placed in lactoacetic acid (lactic acid:water:acetic acid, 1:2:3) and spread under a coverslip. The slides were blocked with 0.2% BSA and treated with affinity-purified primary antibodies (for antibody descriptions please refer the Key Resources Table). Visualization and photography were performed with an Olympus IX81 inverted microscope fitted with a Photometrics CoolSNAP EZ CCD camera.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-MSL3 | [45] | N/A |
| Rabbit polyclonal anti-MLE | [45] | N/A |
| Rabbit polyclonal anti-MSL2 | [45] | N/A |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster y1 w1118;PBac{y+-attP-3B}VK00037 | Bloomington Drosophila Stock Center |
BDSC: 9752, Flybase ID: FBst000972 |
| D. melanogaster y1 w*;P{CaryIP}su(Hw)attP6 | Bloomington Drosophila Stock Center |
BDSC: 34767, Flybase ID: FBst0034767 |
| D. melanogaster y1 v1; P{y+t7.7CaryP}attP40 | Bloomington Drosophila Stock Center |
BDSC: 36304; Flybase ID: FBst0036304 |
| D. melanogaster w*;P{sqh-mCherry.M}3 | Bloomington Drosophila Stock Center |
BDSC:59024, Flybase ID: FBst0059024 |
| D. melanogaster P{sqh-GAL4}2 | Bloomington Drosophila Stock Center |
Flybase ID: FBrf0191737 |
| D. melanogaster y1 w67c23; ;P {w+mC y+ mDint2 EPgy2}hafEY08668 | Bloomington Drosophila Stock Center |
BDSC: 17484 Flybase ID: FBst0017484 |
| D. melanogaster T(2;Y)22A2 | [30] | N/A |
| D. melanogaster y*w*; P{w+mC hp-1.6883F} | [21] | N/A |
| Recombinant DNA | ||
| pUASTB | [44] | N/A |
| Software and Algorithms | ||
| T-Coffee | [27] | N/A |
Mitotic chromosome preparations
Timed collections of early embryos were dechorionated, homogenized in PBST with 2% formaldehyde by two strokes with a tight pestle and filtered through Nytex (Millipore). Nuclei were collected by centrifugation at 2 K RPM for 5 min, fixative removed and nuclei suspended in 60 ml hexylene glycol fixative (1 mM HEPES pH 6.8, 1 mM CaCl2, 3.7% freshly made paraformaldehyde with 26% hexylene glycol). Five ml drops are squashed between cover slip and slide. Slides were plunged into liquid nitrogen, cover slips removed and slides stored at -20°C in 95% EtOH. After rehydration in PBST, prep arations were refixed in 4% formaldehyde in PBST, washed, blocked and incubated with antibody as described for polytene chromosome preparations.
Quantitative RT-PCR
Total RNA was prepared from two biological replicates of 50 third instar males using Trizol reagent (Invitrogen). One microgram of total RNA was reverse transcribed using ImProm-II reverse transcriptase following manufacturer recommendations (Promega). Duplicate reactions were amplified using iTaq Universal SYBR Green Supermix (Bio-Rad) with an Mx3000P Real-Time PCR system (Stratagene). The genes analyzed were stably expressed in late third instar larvae. Gene and primer information is available upon request. Values were normalized to Dmn and expression calculated using the efficiency corrected comparative quantification method [46].
Quantification And Statistical Analysis
After measuring gene expression using quantitative RT-PCR, box and whiskers plots of control and test genes were generated with SPSS. One-way ANOVA followed by Tukey's HSD post-hoc tests were used to determine the significance of gene expression. The Pearson correlation coefficients of relationships between change in gene expression and the distance from the transgene or basal expression were generated using SPSS.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. M. Kuroda, R. Findley and X-D. Zhang for generous contributions of reagents and equipment, Dr. S. K. Koya for assistance with data analysis, B. Patel for cloning, N. Deshpande for larval collection and J. Butts for technical support. We thank Drs. D. Barbash, and anonymous reviewers, for helpful comments. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. S. S. J. was supported in part by a T. C. Rumble University Graduate Fellowship. This work was supported by the Wayne State University Bridge Funding Program and NIH award GM 093110 to V. H. M.
Footnotes
Author Contributions: SSJ and VHM conducted experiments and SSJ analyzed the data. VHM designed this study and SSJ and VHM wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Disteche CM. Dosage compensation of the sex chromosomes. Annu Rev Genet. 2012;46:537–560. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucchesi J, Kelly W, Panning B. Chromatin remodeling in dosage compensation. Annual Review of Genetics. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 3.Meller V, Rattner B. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. The EMBO Journal. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng X, Meller VH. roX RNAs are required for increased expression of X-linked genes in Drosophila melanogaster males. Genetics. 2006;174:1859–1866. doi: 10.1534/genetics.106.064568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Molecular and Cellular Biology. 2000;20:312–318. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park Y, Kelley R, Oh H, Kuroda M, Meller V. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science. 2002;298:1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- 7.Kelley R, Meller VH, Gordadze P, Roman G, Davis R, Kuroda M. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- 8.Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soruco MM, Chery J, Bishop EP, Siggers T, Tolstorukov MY, Leydon AR, Sugden AU, Goebel K, Feng J, Xia P, et al. The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev. 2013;27:1551–1556. doi: 10.1101/gad.214585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Sural TH, Peng S, Li B, Workman JL, Park PJ, Kuroda MI. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat Struct Mol Biol. 2008;15:1318–1325. doi: 10.1038/nsmb.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramirez F, Lingg T, Toscano S, Lam KC, Georgiev P, Chung HR, Lajoie BR, de Wit E, Zhan Y, de Laat W, et al. High-Affinity Sites Form an Interaction Network to Facilitate Spreading of the MSL Complex across the X Chromosome in Drosophila. Mol Cell. 2015;60:146–162. doi: 10.1016/j.molcel.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villa R, Schauer T, Smialowski P, Straub T, Becker PB. PionX sites mark the X chromosome for dosage compensation. Nature. 2016;537:244–248. doi: 10.1038/nature19338. [DOI] [PubMed] [Google Scholar]
- 15.Kelley R, Meller V, Gordadze P, Roman G, Davis R, Kuroda M. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- 16.Henry RA, Tews B, Li X, Scott MJ. Recruitment of the male-specific lethal (MSL) dosage compensation complex to an autosomally integrated roX chromatin entry site correlates with an increased expression of an adjacent reporter gene in male Drosophila. J Biol Chem. 2001;276:31953–31958. doi: 10.1074/jbc.M103008200. [DOI] [PubMed] [Google Scholar]
- 17.Park SW, Oh H, Lin YR, Park Y. MSL cis-spreading from roX gene up-regulates the neighboring genes. Biochem Biophys Res Commun. 2010;399:227–231. doi: 10.1016/j.bbrc.2010.07.059. [DOI] [PubMed] [Google Scholar]
- 18.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 19.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 20.Menon DU, Meller VH. A role for siRNA in X-chromosome dosage compensation in Drosophila melanogaster. Genetics. 2012;191:1023–1028. doi: 10.1534/genetics.112.140236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menon DU, Coarfa C, Xiao W, Gunaratne PH, Meller VH. siRNAs from an X-linked satellite repeat promote X-chromosome recognition in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2014;111:16460–16465. doi: 10.1073/pnas.1410534111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallach M. Recurrent turnover of chromosome-specific satellites in Drosophila. Genome Biol Evol. 2014;6:1279–1286. doi: 10.1093/gbe/evu104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn G, Kuttler H, Moreira-Filho O, Heslop-Harrison J. The 1.688 repetitive DNA of Drosophila: concerted evolution at different genomic scales and association with genes. Molecular Biology and Evolution. 2011. [DOI] [PubMed]
- 24.Di Bartolomeis S, Tartof K, Jackson FR. A superfamily of Drosophila satellite related (SR) DNA repeats restricted to X chromosome euchromatin. Nucleic Acids Research. 1992;20:1113–1116. doi: 10.1093/nar/20.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waring GL, Pollack JC. Cloning and characterization of a dispersed, multicopy X chromosome sequence in Drosophila melanogaster. PNAS. 1987;84:2843–2847. doi: 10.1073/pnas.84.9.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Straub T, Zabel A, Gilfillan GD, Feller C, Becker PB. Different chromatin interfaces of the Drosophila dosage compensation complex revealed by high-shear ChIP-seq. Genome Res. 2013;23:473–485. doi: 10.1101/gr.146407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kind J, Akhtar A. Cotranscriptional recruitment of the dosage compensation complex to X-linked target genes. Genes Dev. 2007;21:2030–2040. doi: 10.1101/gad.430807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joshi SS, Cheong H, Meller VH. A strategy for generation and balancing of autosome: Y chromosome translocations. Fly (Austin) 2014;8:58–62. doi: 10.4161/fly.27814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menon DU, Meller VH. Imprinting of the Y chromosome influences dosage compensation in roX1 roX2 Drosophila melanogaster. Genetics. 2009;183:811–820. doi: 10.1534/genetics.109.107219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menon DU, Meller VH. Identification of the Drosophila X chromosome: The long and short of it. RNA Biol. 2015;12:1088–1093. doi: 10.1080/15476286.2015.1086864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimaud C, Becker PB. The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. Genes Dev. 2009;23:2490–2495. doi: 10.1101/gad.539509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 35.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meller VH, Joshi SS, Deshpande N. Modulation of Chromatin by Noncoding RNA. Annu Rev Genet. 2015;49:673–695. doi: 10.1146/annurev-genet-112414-055205. [DOI] [PubMed] [Google Scholar]
- 38.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meller VH. Initiation of dosage compensation in Drosophila embryos depends on expression of the roX RNAs. Mechanisms of Development. 2003;120:759–767. doi: 10.1016/s0925-4773(03)00157-6. [DOI] [PubMed] [Google Scholar]
- 40.Chen K, Johnston J, Shao W, Meier S, Staber C, Zeitlinger J. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. Elife. 2013;2:e00861. doi: 10.7554/eLife.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellison CE, Bachtrog D. Dosage compensation via transposable element mediated rewiring of a regulatory network. Science. 2013;342:846–850. doi: 10.1126/science.1239552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuzu G, Kaye EG, Chery J, Siggers T, Yang L, Dobson JR, Boor S, Bliss J, Liu W, Jogl G, et al. Expansion of GA Dinucleotide Repeats Increases the Density of CLAMP Binding Sites on the X-Chromosome to Promote Drosophila Dosage Compensation. PLoS Genet. 2016;12:e1006120. doi: 10.1371/journal.pgen.1006120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alekseyenko AA, Ellison CE, Gorchakov AA, Zhou Q, Kaiser VB, Toda N, Walton Z, Peng S, Park PJ, Bachtrog D, et al. Conservation and de novo acquisition of dosage compensation on newly evolved sex chromosomes in Drosophila. Genes Dev. 2013;27:853–858. doi: 10.1101/gad.215426.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgeneic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Copps K, Richman R, Lyman L, Chang K, Rampersand-Ammons J, Kuroda M. Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. The EMBO Journal. 1998;17:5409–5417. doi: 10.1093/emboj/17.18.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfaffle M. A new mathamatical model for relative quantification of real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.