Abstract
Alterations to the expression and activity of androgen receptor (AR) co-regulators in prostate cancer is an important mechanism driving disease progression and therapy resistance. Using a novel proteomic technique, we identified a new AR co-regulator, the transcription factor Grainyhead-like 2 (GRHL2), and demonstrated its essential role in the oncogenic AR signaling axis. GRHL2 colocalized with AR in prostate tumors and was frequently amplified and upregulated in prostate cancer. Importantly, GRHL2 maintained AR expression in multiple prostate cancer model systems, was required for cell proliferation, enhanced AR's transcriptional activity, and co-located with AR at specific sites on chromatin to regulate genes relevant to disease progression. GRHL2 is itself an AR-regulated gene, creating a positive feedback loop between the two factors. The link between GRHL2 and AR also applied to constitutively active truncated AR variants (ARVs), as GRHL2 interacted with and regulated ARVs and vice versa. These oncogenic functions of GRHL2 were counterbalanced by its ability to suppress epithelial-mesenchymal transition and cell invasion. Mechanistic evidence suggested that AR assisted GRHL2 in maintaining the epithelial phenotype. In summary, this study has identified a new AR co-regulator with a multifaceted role in prostate cancer, functioning as an enhancer of the oncogenic AR signaling pathway but also a suppressor of metastasis-related phenotypes.
Keywords: Prostate cancer, androgen receptor, transcription factor, Grainyhead-like, castration-resistant prostate cancer
Introduction
Prostate cancer (PCa) is the most frequent male non-skin cancer and a major cause of cancer-associated death. PCa growth and survival is driven by androgen receptor (AR) activation in response to ligand (androgen) binding. Accordingly, androgen deprivation therapy (ADT) is the first line treatment for locally-advanced and metastatic disease. However, resistance to ADT and disease progression, termed castrate resistant prostate cancer (CRPC), inevitably occurs within 18-24 months. The recent success of second-generation ADT agents (i.e. the AR antagonist enzalutamide and the androgen synthesis inhibitor abiraterone acetate) in achieving a survival benefit in men with CRPC highlights an ongoing dependence on AR signaling in this disease setting (1). Unfortunately, even these new, more potent, drugs are only palliative, with a mean average survival benefit measured in months, and the resultant disease generally remains AR-driven (1).
Mechanisms underlying continued AR activity in CRPC include deregulation of androgen biosynthesis and direct changes to the AR, including gene amplification, gain-of-function mutations and the emergence of constitutively active truncated AR variants (ARVs) that lack a functional LBD (2). Another adaptive response that can mediate AR signaling in the castrate environment is altered expression and activity of AR co-regulator proteins. Over 130 AR co-regulators have been identified, comprising a highly complex system for shaping AR function in the normal and malignant prostate (3). In general, CRPC is characterized by gain of co-activators and loss of co-repressors, which collectively enhances AR signalling in the castrate environment.
Here, we utilized an unbiased proteomics technique, termed rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) (4), to elucidate the chromatin-associated AR interactome. We identified a novel AR-associated factor, Grainyhead-like 2 (GRHL2), and demonstrated that it plays a major role in maintaining AR expression and signaling in PCa while simultaneously suppressing metastasis-associated phenotypes.
Materials and Methods
Cell line culture and transfection
R1-AD1 and R1-D567 have been described previously (5). C4-2B, LNCaP, VCaP, 22Rv1, PC3 and DU145 human prostate carcinoma cells were obtained from ATCC. Cells were cultured as described previously (6). The number of passages between thawing cell lines and their use in the described experiments were as follows: R1-AD1, R1-D567 and 22Rv1 (passages 2-30); C4-2B and PC3 (2-15); LNCaP (2-20); VCaP (2-8); DU145 (2-10). All cell lines underwent: i) verification by short-tandem repeat profiling in July or August 2016 by CellBank Australia; and ii) regular (approximately every 4 months) testing using a custom PCR-based assay to ensure lack of contamination with the most common Mycoplasma and Acholeplasma strains.
Cells were transfected with 20 nM siRNA using RNAiMAX transfection reagent (Life Technologies), according to the manufacturer's instructions. The following siRNAs were used in this study: GRHL2 (sc77606a, sc77606b and sc77606c (Santa Cruz Biotechnology)); AR (AR Silencer 51539, #4390826 (Ambion)). For transfecting cells with the GRHL2 over-expression plasmid (pCMV-NEO-GRHL2, #SC305189 (Origene Technologies)), we used Lipofectamine 2000 (Life Technologies).
Stable LNCaP-shGRHL2 cell lines were generated using GRHL2-specific shRNA plasmids (TRCN0000015808 and TRCN0000015810) and an shRNA control plasmid (Shc002) from Sigma as described previously (7).
Rapid Immunoprecipitation Mass spectrometry of Endogenous proteins (RIME)
RIME was carried out essentially as described (4). 3 × 107 R1-AD1 cells or 6 × 107 R1-D567 cells were subjected to immunoprecipitation with 10 μg of AR antibody (Santa Cruz AR N20; sc-816). Only co-precipitating proteins that occurred in 3/3 independent biological replicates were considered, and further filtering was achieved by excluding proteins that appeared in >1 of the 3 matching IgG controls.
Co-immunoprecipitation
R1-AD1 and R1-D567 cells were seeded at 2.5 ×106 cells/plate and LNCaP cells were seeded at 3.0 × 106 cells/plate in RMPI 1640 +10% CSS. After 72 hours, media was replaced with fresh RPMI 1640 + 10% CSS containing 10 nM DHT. After an additional 4 hours, nuclear pellets were made according to the RIME protocol. Nuclear pellets were sonicated for 10 cycles (30sec on, 30 sec off) using a Diagenode Bioruptor. After centrifugation, lysates were immunoprecipitated overnight with 5 μg AR antibody (Santa Cruz #sc-816×) or 5 μg of GRHL2 antibody (Sigma HPA004820) pre-coupled to Protein G Dynabeads (Life Technologies, 10004D). The following day, the beads were washed 3 times with PBS and then boiled in SDS-PAGE sample buffer, prior to analysis by Western blotting.
Analysis of GRHL2 expression in published datasets
Transcriptomic data was downloaded from GEO (GSE35988 and GSE21032), The Cancer Genome Atlas (TCGA) data portal and cBioPortal (8). GRHL2 copy number data was obtained via cBioPortal (8).
Western blotting and antibodies
Protein extraction from cells, using RIPA buffer, and Western blotting was done as described previously (9). Antibodies used in Western blotting were: AKT 1/2 (Santa Cruz N-19; sc-1619); pAKT (Cell Signalling Thr308, #9275S); AR (Santa Cruz N20; sc-816); E-Cadherin (BD Biosciences, clone 36/E); ERK1 (Santa Cruz K23; sc-94); pERK (Cell Signalling Thr202/Tyr204, #9101); GRHL2 (Sigma; HPA004820); GAPDH (EMD Millipore; MAB374); N-cadherin (Santa Cruz H-63; sc-7939); Tubulin (EMD Millipore; 05-829); Vimentin (Abcam EPR3776); ZEB1 (Santa Cruz H102; Sc-25388); and ZO-1 (Cell Signalling, D7D12).
Quantitative real-time PCR (RT-qPCR)
RNA extraction from cells, using Trizol reagent, and RT-qPCR was done as described previously (9). GAPDH was used for normalization of RT-qPCR data. Fold changes in mRNA expression levels were calculated using the comparative Ct method as described. Primer sequences are available on request.
Immunofluorescence
4μm sections of paraffin-embedded human transurethral resection of the prostate (TURP) tumor samples were cut and adhered to Superfrost Ultra Plus coated slides (Menzel-Glaser) overnight at 45°C, then de-waxed and immunolabelled as previously described (10). Primary antibodies (1:400 rabbit anti-GRHL2 (Sigma HPA004820) or 1:60 mouse anti-AR (Dako M3562)) were incubated overnight at 4°C and secondary antibodies (goat anti-mouse AlexaFluor 488 (Thermo A-11029) or goat anti-rabbit Alexa 594 (Thermo A-11037)) incubated for 30 minutes at room temperature. Images were acquired on a Zeiss 700 confocal microscope. Two-dimension expression intensity histograms and Spearman's correlation coefficients were generated using ImageJ software and the Coloc2 plugin.
Immunohistochemistry
Immunohistochemistry for GRHL2 was carried out as described (11) using anti-GRHL2 antibody (Sigma HPA004820) at a 1:8000 dilution. Slide digitization, annotation, and immunohistochemical quantification was done as described (12).
Transactivation assays
Transactivation assays with the probasin luciferase reporter construct were carried out essentially as described previously (13), with the exception that cells were co-transfected with 1 ng of pCMV-NEO-GRHL2 plasmid, 1 ng of pcDNA-AR, 1 ng of pRL-CMV and 100 ng of the probasin reporter construct.
Chromatin immunoprecipitation (ChIP) and ChIP-seq
LNCaP cells were seeded at 3×106 cells/plate in 15c m plates in RPMI 1640 + 10% FBS and allowed to grow for 3 days prior to fixation. ChIP was then performed as described previously (14). For ChIP-qPCR, 2 μl of DNA was used in 10 μl PCR reactions. For ChIP-seq, 5 ng of DNA (ChIP-enriched or input) was used for library creation with a Illumina TruSeq ChIP Library Prep Kit (Illumina) and sequenced (75 bp single-end reads) at the South Australian Health and Medical Research Institute Genomics Facility using an Illumina NextSeq 500. Mapping of sequencing data and peak calling (using input DNA as a control) were performed as described previously (6). Consensus peaks were determined by including only regions identified by in two biological replicates. ChIP-seq data are available through NCBI's Gene Expression Omnibus (GSE80256).
To identify AR/GRHL2 shared binding regions, ChIPpeakAnno (15) (implemented in R 2.12.0) was used with a maxgap value of 1000. Cistrome (16) was used to generate heatmaps. HOMER (17) was used to generate histograms of tag density around peaks. Regions of AR/GRHL2 binding were annotated with respect to neighbouring genes (≤50 kb from the transcription start site) using CisGenome (18). Identification of enriched sequence motifs (known and de novo) were identified as described previously (19). Visualization of ChIP-seq data was performed using Integrated Genomics Viewer (IGV 2.3, Broad Institute, (20)).
Our AR/GRHL2 ChIPSeq data was also compared to other published datasets: H3K4me2, H3K4me3, H3K27ac, P300, MED12, FoxA1 from Wang and colleagues (21); H3K9me3 and H3K27me3 from Yu and colleagues (22); and RNAPII from He and colleagues (23). Read data from these studies were downloaded and mapped around AR and GRHL2 peaks sets using HOMER.
RNA sequencing (RNA-seq)
RNA from LNCaP cells was prepared using Trizol 2 days after transfection with siGRHL2. Libraries were generated using 10 ng of RNA and NEXTflex Rapid Illumina Directional RNA-Seq Library Prep Kits (Bioo Scientific), according to the manufacturer's instructions. Sequencing was carried out at the South Australian Health and Medical Research Institute Genomics Facility using an Illumina NextSeq 500 (75 bp single-end reads). Reads were mapped using TopHat v2.0.9 (24) in Galaxy (25,26), using default parameters and hg19 as the reference genome. Raw counts per exon (mapped to hg19_genes_2012-03-09.gtf) were estimated using htseq-count (27). Subsequently, DESeq2 (28) (implemented in Bioconductor v2.12, R v3.0.1) was used to identify differentially expressed genes. RNA-seq data are available through NCBI's Gene Expression Omnibus (GSE80452).
For gene set enrichment analysis (GSEA) (29), read counts were normalized using the DESeq2 rlog transformation algorithm, genes were ranked using a signal to noise metric, and the GSEAPreranked tool was run on the GenePattern server (http://www.broad.mit.edu/tools/software.html). A signature of GRHL2 activity was made by considering the top 100 differentially expressed genes in response to siGRHL2 knockdown (comprising 21 upregulated genes and 79 downregulated genes): GRHL2 activity scores for individual tumors from two large clinical cohorts were determined as described previously (23).
Chick chorioallantoic membrane (CAM) assays
CAM assays were approved by the University of Adelaide Animal Ethics Committee (approval number M-2014_079) and done using the in ovo method as described previously (7).
Statistical analyses
All statistical analyses were carried out using GraphPad Prism (version 5; GraphPad Software). Details of statistical tests used are provided in the figure legends.
Results
GRHL2 is a novel AR-interacting protein
To identify novel proteins that interact with AR and ARVs, we conducted RIME in R1-AD1 and R1-D567 cells. R1-D567 has been engineered such that AR exons 5-7 are deleted from the genome, leading to exclusive expression of the constitutively active ARv567es variant, whereas the R1-AD1 cell line expresses full-length AR only (5). RIME identified 54 and 75 proteins associated with AR and ARv567es, respectively (Supplementary Table 1). Analysis of the interactomes at the individual protein (Supplementary Table 1) and pathway (Figure 1A) level revealed high concordance between ligand-activated AR and ARv567es. AR itself was isolated with high confidence, and many known AR co-regulators, including FoxA1, P300, AP-1 and members of the SWI/SNF chromatin remodelling complex, were also identified in the interactomes, highlighting the robustness of the data.
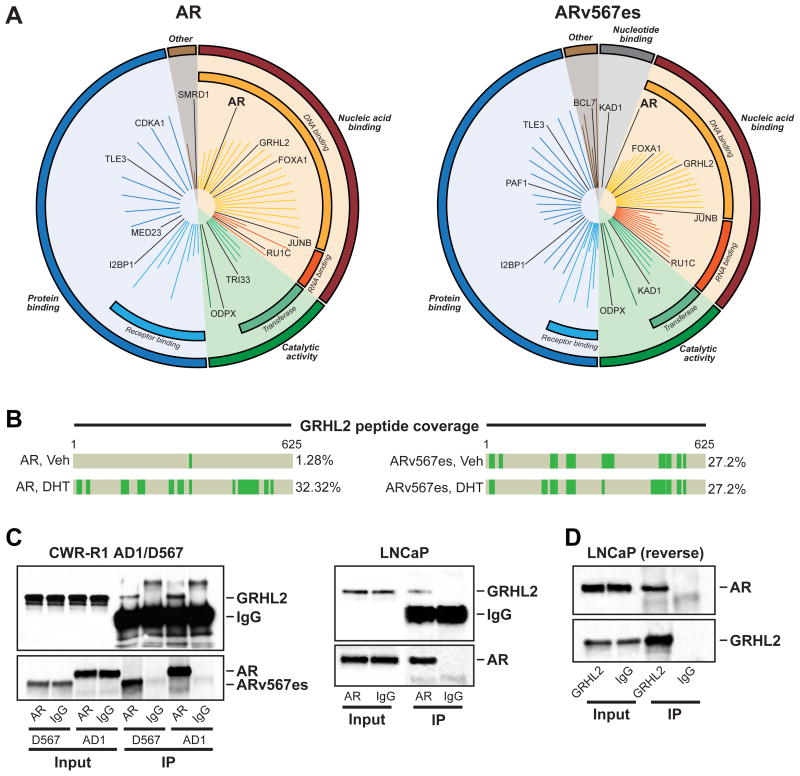
Figure 1. GRHL2 is a novel AR-interacting protein.
(A) Graphical plots (generated by MS-ARC) showing AR- and ARv567es-associated proteins, clustered according to molecular function. The length of the line represents the Mascot score. (B) Peptide coverage of GRHL2 from RIME experiments. (C-D) Validation of AR:GRHL2 interaction by co-immunoprecipitation. AR protein in R1-AD1 and R1-D567 cells (left) and LNCaP cells (right) was immunoprecipitated and GRHL2 was detected by immunoblotting (C). GRHL2 protein in LNCaP cells was immunoprecipitated and AR was detected by immunoblotting (D). For all co-immunoprecipitations, IgG served as a negative control.
Of the novel co-precipitating factors, the transcription factor GRHL2 was one of the most confident hits from both cell line models (Figure 1B). AR only bound GRHL2 robustly in androgen-replete conditions whereas, as expected, the constitutively active ARv567es variant interacted with GRHL2 in an androgen-independent manner (Figure 1B). The interaction was confirmed by co-immunoprecipitation of GRHL2 using an AR antibody in R1-AD1, R1-D567 and LNCaP cells (Figure 1C). A robust association between AR and GRHL2 in LNCaP cells was also evident by “reverse” co-immunoprecipitation (i.e. co-immunoprecipitation of AR with a GRHL2 antibody) (Figure 1D).
GRHL2 is commonly amplified and over-expressed in prostate cancer
To assess the clinical relevance of GRHL2 in PCa, we examined a series of published cohorts for GRHL2 alterations (30-32). GRHL2 resides at 8q.22.3, a genomic locus that is frequently amplified in PCa: indeed, GRHL2 gene copy number was gained/amplified in 54-71% of metastatic samples and 14-32% of primary tumors (Figure 2A), and this alteration was associated with a trend towards increased GRHL2 mRNA expression in CRPC (Figure 2B). Moreover, GRHL2 mRNA was elevated in malignant compared to non-malignant prostate tissues (Figure 2C) and was higher in Gleason score 8-10 versus Gleason 6 tumors (Figure 2D).
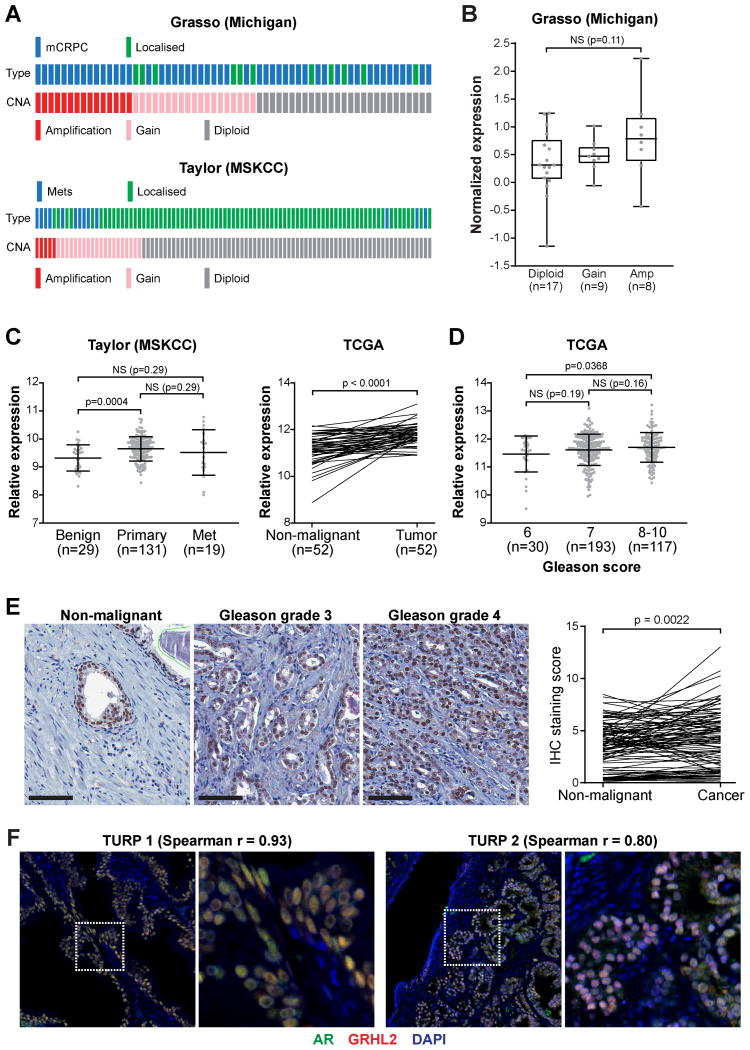
Figure 2. GRHL2 is commonly amplified and over-expressed in prostate cancer.
(A) Oncoprints representing GRHL2 copy-number alterations from the Grasso and Taylor datasets were generated using cBioPortal. mCRPC, metastatic castration-resistant prostate cancer (mCRPC); Mets, metastases. (B) GRHL2 gene amplification in the Grasso cohort is associated with increased mRNA expression. Boxes show minimum and maximum (bottom and top lines, respectively) and mean (line within the boxes) values. P value was determined an unpaired t test. (C) GRHL2 expression is elevated in primary prostate cancer. In the Taylor graph, lines represent the mean ± standard deviation (SD). P values were determined using unpaired (Taylor) or paired (TCGA) t tests. (D) GRHL2 expression is associated with Gleason grade. Lines in the graph represent the mean ± SD. P values were determined using unpaired t tests. (E) Representative staining of non-malignant and cancer tissues are shown on the left (bar = 100 microns). A graph of staining scores is shown on the right. P value was determined using an paired t test. (F) Cellular co-expression of GRHL2 and AR in prostate tissues, as demonstrated by dual immunofluorescence. Shown are two representative prostate tumors. Nuclei were stained with DAPI. Spearman r values represent the correlation between AR and GRHL2 staining (see Materials and Methods). Scale bars are 50 microns.
Extending upon the in silico analyses, we used immunohistochemistry to evaluate GRHL2 protein expression in 101 matched non-malignant prostate tissues and tumors. GRHL2 was detectable in all samples: staining was primarily nuclear and present in luminal epithelial cells, with minimal signal in the stroma. Representative images from strongly positive samples are shown in Figure 2E (left). Although staining was strong in benign prostate samples (median staining index = 3.66), levels of GRHL2 were significantly higher in tumors (4.26) (Figure 2E, right). However, GRHL2 expression was not associated with other clinicopathological parameters such as Gleason grade/score or serum prostate specific antigen levels (data not shown).
To determine the potential for functional interaction between AR and GRHL2 in clinical specimens, we examined their tissue localization using dual immunofluorescence in human prostate tumors obtained from transurethral resections. Two representative sections are shown in Figure 2F. AR and GRHL2 were primarily expressed in luminal epithelial cells. While most cells exhibited positivity for both factors, cells with high GRHL2 staining and low/no AR staining were evident. Using software to assess co-localization (see Materials and Methods), a high positive correlation between the expression of AR and GRHL2 was observed at the cellular level (Figure 2F and Supplementary Figure 1).
GRHL2 is an AR target gene
To identify appropriate model systems to characterize GRHL2 function, its expression was assessed in a panel of PCa cell lines. GRHL2 was most highly expressed in VCaP cells and not detectable in AR-negative PC3 or DU145 cells (Figure 3A, left). This expression pattern closely mirrored that of AR (Figure 3A, right), leading us to postulate that GRHL2 is an androgen-regulated factor. Supporting this idea, GRHL2 mRNA levels were significantly induced by DHT treatment in multiple cell lines (Figure 3B). This regulation was dependent on the AR, since siRNA-mediated AR knockdown cells led to a dramatic loss of GRHL2 protein in both the presence and absence of androgen (Figure 3C, left). Using the R1-D567 cell line model, we demonstrated that GRHL2 is also regulated by the constitutively-active ARV, ARv567es. In support of the in vitro data, GRHL2 expression decreased following ADT and was re-gained in CRPC in clinical specimens (Figure 3D) (33,34).
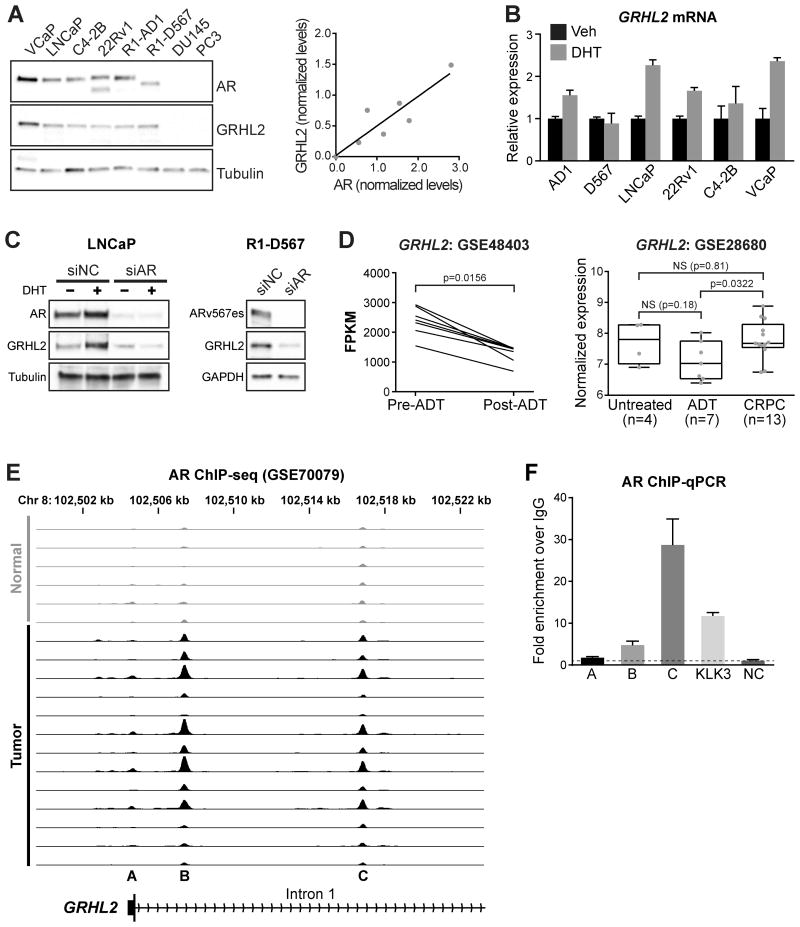
Figure 3. Regulation of GRHL2 expression by AR in prostate cancer.
(A) GRHL2 protein levels in a panel of prostate cancer cell lines (tubulin is the loading control). The graph on the right shows the correlation between tubulin-normalized AR and GRHL2 in all cell lines. (B) Cell lines were treated with 1 nM DHT for 4 hours and GRHL2 levels were assessed by RT-qPCR. Error bars represent ± SD. (C) LNCaP (left) or R1-D567 (right) cells were transfected with siRNA targeting AR (siAR) or control siRNA (siNC) for 48 hours. LNCaP cells were additional treated with 1 nM DHT (or ethanol as a control) for 24 hours. Tubulin or GAPDH are loading controls. (D) GRHL2 mRNA expression in two clinical cohorts. Boxes in the right graph show minimum and maximum (bottom and top lines, respectively) and mean (line within the boxes) values. P values were determined using a Wilcoxon matched-pairs signed rank test (GSE48403) or unpaired t tests (GSE28680). FPKM, fragments per kilobase of exon per million mapped reads; NS, not significant. (E) AR binding sites (from ChIP-seq) proximal to the GRHL2 gene in non-malignant and prostate tumor samples (35). Each track depicts ChIP-seq AR binding intensity for a given sample. (F) Validation of 3 putative AR binding sites (A-C; shown below the ChIP-seq tracks in E) by ChIP-qPCR. Error bars represent ± standard error of the mean (SEM).
To evaluate whether AR-mediated regulation of GRHL2 was direct, we examined ChIP-seq data for potential AR binding sites proximal to the GRHL2 gene. In multiple AR cistromes derived from distinct cell line models of PCa, we found evidence for AR (and ARV) binding near at two sites proximal to the GRHL2 promoter (Supplementary Table 2). Both of these binding sites were also evident in a more recent study (35) examining the AR cistrome in clinical specimens (sites B and C in Figure 3E); moreover, in this in vivo dataset, it was apparent that AR binding proximal to the GRHL2 locus was notably increased in malignant compared to normal tissues (Figure 3E). We validated AR binding at these sites by ChIP-qPCR in LNCaP cells (Figure 3F). The tissue ChIP-seq data also suggested a weak AR binding event at the GRHL2 promoter (Figure 3E, site A), but this was not evident by ChIP-qPCR (Figure 3F). Collectively, these data indicate that AR binds to multiple sites proximal to the GRHL2 transcriptional start site, supporting a direct mode of transcriptional regulation.
GRHL2 promotes prostate cancer growth and is critical for AR expression
The functional role of GRHL2 in PCa was assessed using siRNA-mediated knockdown. Three distinct GRHL2-targeted siRNAs, all of which were highly effective at silencing GRHL2 (Supplementary Figure 2), caused inhibition of proliferation in multiple PCa cell lines (Figure 4A). Verifying the specificity of knockdown, none of the siRNAs inhibited the growth of GRHL2-negative PC-3 cells (Supplementary Figure 3).
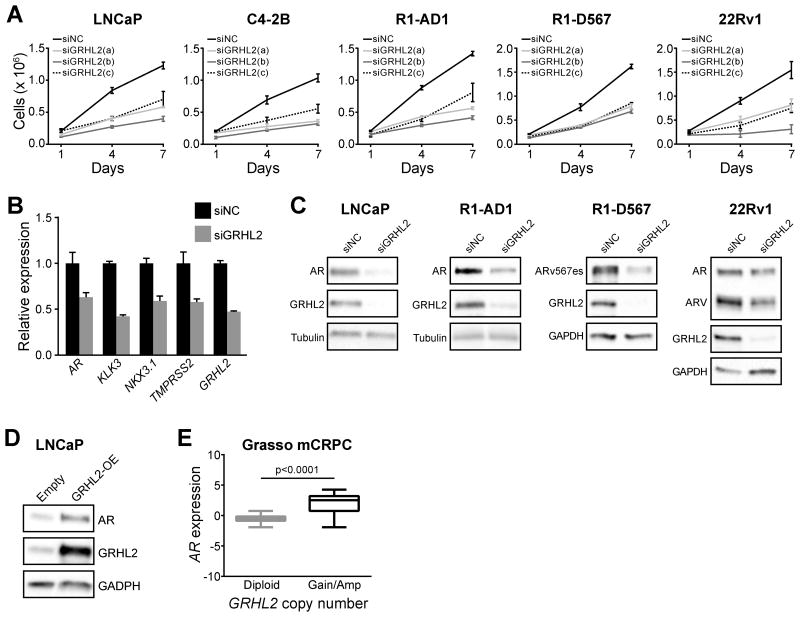
Figure 4. Regulation of prostate cancer cell growth and AR expression and signaling by GRHL2.
(A) Prostate cancer cell lines were transfected with 3 distinct siRNAs and Trypan blue growth assays were performed. Error bars represent ± SD. (B) LNCaP cells were transfected with GRHL2 siRNA (siGRHL2) or a control (siNC) and gene expression was measured by RT-qPCR after 48 hours. Error bars represent ± SEM of 3 independent experiments. (C) Prostate cancer cell lines were transfected with GRHL2 siRNA (siGRHL2) or a control (siNC) and, 2 days later, protein expression was assessed by Western blotting. Tubulin or GAPDH are shown as loading controls. (D) LNCaP cells were transfected with a GRHL2 over-expression vector (GRHL2-OE) or a control (Empty) and protein expression was assessed by Western blotting after 48 hours. GAPDH is shown as a loading control. (E) AR expression is higher in metastatic CRPC (mCRPC) tumors with GRHL2 copy number gain or amplification compared to tumors with no change in GRHL2 copy number (diploid). Data is from the Grasso cohort and was obtained via cBioPortal. P value was determined using an unpaired t test.
We subsequently tested the effect of a pool of the siRNAs (termed siGRHL2) on AR signaling. Knockdown of GRHL2 caused marked suppression of AR and AR target gene expression (Figure 4B) and decreased the levels of AR and ARV protein (Figure 4C). Conversely, GRHL2 over-expression resulted in accumulation of AR protein (Figure 4D). This latter finding is likely to be clinically relevant, since tumors with gain or amplification of GRHL2 exhibited higher AR expression than diploid tumors (Figure 4E).
We next assessed whether the physical association between GRHL2 and AR/ARV was a mechanism underlying GRHL2-mediated maintenance of AR/ARV expression. Following treatment with the protein biosynthesis inhibitor, cycloheximide, AR and ARV protein half-life was equivalent in siGRHL2 and control cells (Supplementary Figure 4). These data suggest that maintenance of AR/ARV protein levels by GRHL2 is not primarily mediated by post-translational mechanisms. By inference, we propose that the primary mechanism by which GRHL2 maintains AR/ARV protein levels is via induction of the AR gene.
Interplay between the transcriptional activities of AR and GRHL2
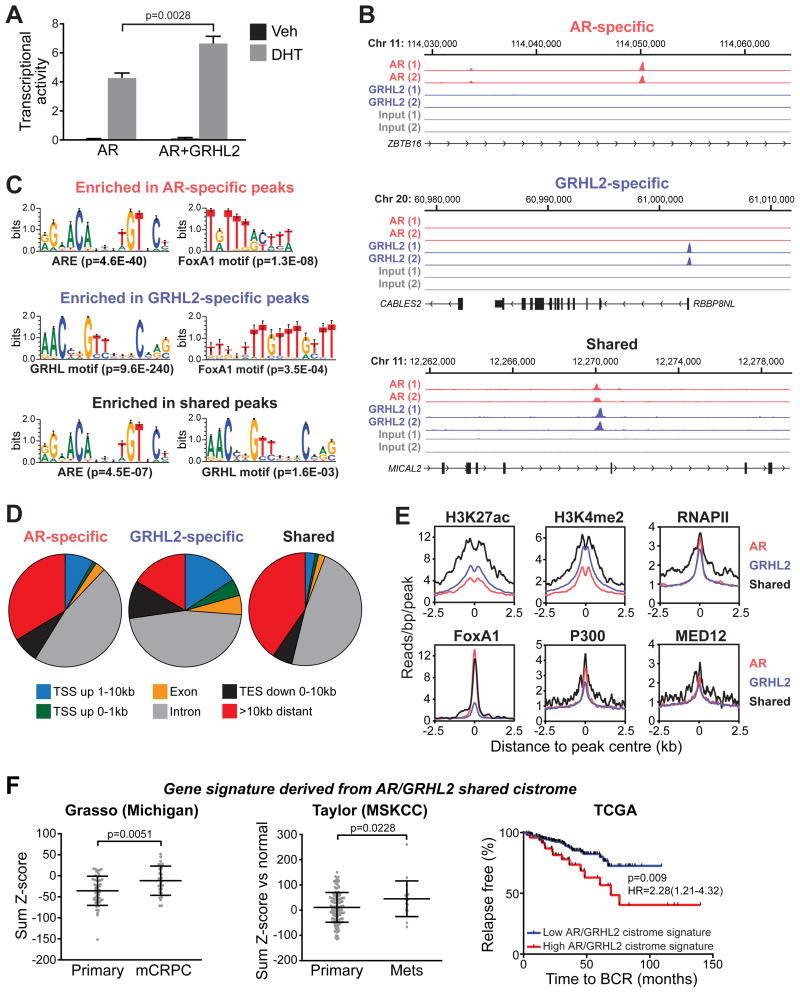
Given that RIME was designed to identify chromatin-associated protein:protein interactions, we speculated that GRHL2 could influence the transcriptional activity of AR. To test this hypothesis, we first evaluated the influence of GRHL2 on a well-characterized probasin reporter construct (13). For these experiments, we transiently expressed GRHL2 and AR in PC3 cells, which are negative for both factors: the rationale for this approach was that modulation of endogenous GRHL2 in AR-positive model systems resulted in down-regulation of AR expression, which complicates assessment of transcriptional outcomes. GRHL2 modestly but reproducibly enhanced AR's transcriptional activity (Figure 5A), indicating that it can act as a co-activator.
Figure 5. GRHL2 enhances AR's transcriptional activity and associates with AR on chromatin.
(A) PC3 cells were transfected with plasmids expressing AR, GRHL2 and a probasin-derived AR-responsive reporter and subsequently treated with 1 nM DHT or vehicle (Veh) control. Transcriptional activity values as assessed by luciferase assays represent the mean (±SEM) of 6 biological replicates; results are a representative of three independent experiments. An unpaired t test was used to assess the affect of GRHL2 on AR activity. (B) Examples of AR-specific, GRHL2-specific and shared binding sites (left). (C) Select motifs enriched in AR-specific, GRHL2-specific and shared peaksets. Motifs were identified using a de novo Gibbs motif sampling approach. P values for enrichment over genomic background are shown. (D) Genomic location summary of AR-specific, GRHL2-specific and shared binding events. (E) Distribution of normalized sequence tag density for H3K27ac, H3K4me2, RNAPII, FoxA1, P300 and Med12 in AR-specific, GRHL2-specific and shared binding events. (F) A gene signature based on shared GRHL2/AR binding events is upregulated in metastatic CRPC (two left panels) and associated with recurrence following radical prostatectomy (right panel).
We subsequently used ChIP-seq to elucidate genome-wide AR and GRHL2 chromatin binding patterns in LNCaP cells with the aim of identifying potential sites of coordinate transcriptional regulation. The reads obtained across two ChIP-seq replicates for each protein were highly concordant, highlighting the robustness of the assay (Supplementary Figure 5). Only peaks identified in both replicates were considered, yielding 5,378 GRHL2 and 1,202 AR high stringency binding sites. Comparison of the two cistromes revealed that 140 (11.6%) of AR binding sites were shared with GRHL2, highlighting potential for cooperative transcriptional regulation. Although not as extensive as the overlap between AR and the pioneer factors FoxA1 and GATA2 (21,23,36), the AR/GRHL2 overlap was highly significant compared to what would be expected by chance (Fisher's Exact Test p value = 3.40 E-24). An example of a peak from each of the “AR-specific”, “GRHL2-specific” and “shared” cistromes is shown in Figure 4A. Quantitative analysis of tag density demonstrated the accuracy of these peak subsets (Supplementary Figure 6).
Motif enrichment analysis of the cistromic data revealed strong enrichment for androgen response elements (AREs) and GRHL2 and FoxA1 motifs in the specific and shared peak subsets (Supplementary Table 3). Indeed, de novo motif generation identified a direct repeat of the FoxA1 motif as one of the most enriched sequences in the GRHL2-specific peak set (Figure 5C). Supporting this finding, the GRHL2 cistrome overlapped substantially (25-41%) with published LNCaP FoxA1 cistromes (Supplementary Figure 7), an interaction that was experimentally validated in a recent study (37).
The AR and GRHL2 cistromes were assessed with respect to genomic features. The bulk of binding events in all three peak sets (AR-specific, GRHL2-specific and shared) were located within introns and intergenic regions (Figure 5D), although GRHL2-specific peaks also exhibited enrichment in gene promoters (2.78-fold over genomic average). Importantly, the shared GRHL2/AR binding events were highly enriched for epigenetic marks (K27-specific acetylation of histone 3 (H3K27ac) and K4-specific methylation of histone 3 (H3K4me2)), general transcription machinery (RNAPII, Mediator) and transcription factors (FoxA1 and P300) that collectively demarcate enhancer sites and/or active transcription (Figure 5E), indicating that co-location of these factors on chromatin promotes high transcriptional activity. Marks of transcription inhibition (H3K9me3 and H3K27me3) showed no enrichment at shared or factor-specific binding sites (Supplementary Figure 8).
To further evaluate the relevance of shared AR/GRHL2 binding events in relation to transcriptional outputs, we identified genes with transcriptional start sites proximal to these sites to yield an “AR/GRHL2 cistrome-based signature” comprised of 194 genes. Importantly, this signature was elevated in clinical CRPC specimens and associated with a poor outcome following surgery, indicating that these genes are linked to PCa progression (Figure 5F). Moreover, by using gene set enrichment analysis (GSEA) and a published AR-responsive gene signature (23), we demonstrated that genes upregulated by AR were enriched near the shared AR/GRHL2 binding events (Supplementary Figure 9), suggesting that co-localization of these factors on chromatin influences the androgenic transcriptional program. Collectively, these data indicate that GRHL2 and AR co-occupy a set of genomic loci associated with genes that are relevant in PCa.
Elucidation of the GRHL2-regulated transcriptome in prostate cancer
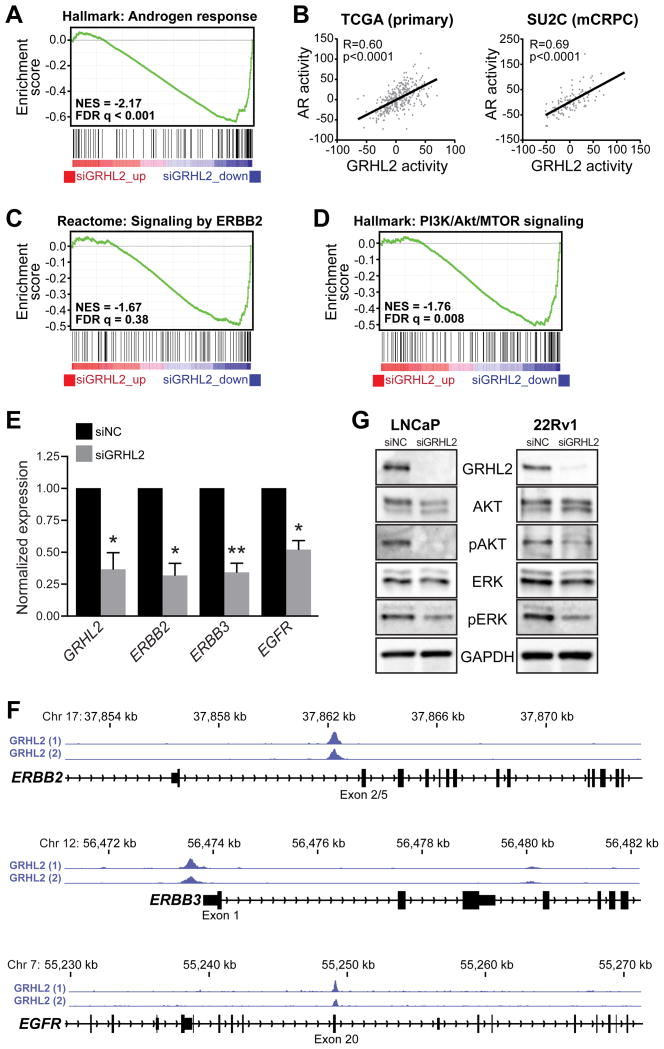
To further explore the function of GRHL2 in PCa, we used RNA-seq to measure transcriptional alterations in response to siGRHL2 in LNCaP cells. This analysis identified a GRHL2-regulated transcriptome comprised of 3,510 genes (Supplementary Table 4). GSEA was used to identify associations between the GRHL2 transcriptome and the “Hallmarks”, “Reactome” and “Kegg” gene set collections of the Molecular Signature Database (29) (Supplementary Table 5). Not unexpectedly, the hallmark most strongly enriched in genes down-regulated by siGRHL2 was “androgen response” (Figure 6A and Supplementary Table 5), further validating the critical requirement of GRHL2 for the AR signaling axis. Indeed, a signature of GRHL2 activity derived from the RNA-seq transcriptome was highly correlated with AR activity in both primary cancer and metastatic CRPC (Figure 6B).
Figure 6. GRHL2 regulates AR, ERBB and PI3K/Akt signaling pathways.
(A) GRHL2 knockdown is associated with decreased expression of an AR-regulated gene set (“Hallmark: Androgen reponse”), as demonstrated by GSEA. (B) Correlation between GRHL2- and AR-regulated gene signatures in primary prostate cancer (left, TCGA cohort) and metastatic CRPC (right, SU2C cohort). (C) GRHL2 knockdown is associated with decreased expression of the PI3K/Akt pathway (“Hallmark: PI3K/Akt/MTOR signaling”), as demonstrated by GSEA. (D) GRHL2 knockdown results in decreased expression of the ERBB2-regulated gene set (“Reactome: Signaling by ERBB2”), as demonstrated by GSEA. (E) GRHL2 knockdown (siGRHL2) leads to reduced expression of ERBB2, ERBB3 and EGFR. Values for the negative control (siNC) were set to 1, and error bars are ± SEM. P values were determined using one-sample t tests (*, P < 0.05; **, P < 0.01). (F) GRHL2 binding events in LNCaP cells, identified by ChIP-seq, proximal to ERBB2, ERBB3 and EGFR. (G) GRHL2 knockdown results in decreased phosphorylation of Akt (pAkt) and ERK (pERK) in LNCaP and 22Rv1 cells, as assessed by Western blotting.
The finding that the majority of GRHL2 DNA binding events were independent of AR suggested that this transcription factor would have functions in PCa beyond its critical role in the androgen signaling axis. To identify such functions, we considered other gene signatures enriched in the GRHL2-regulated transcriptome. Several consistent findings in response to loss of GRHL2 were observed (Supplementary Table 5), including: i) down-regulation of key pro-growth pathways, such as those regulated by ERBB2 (Figure 6C), PI3K/Akt (Figure 6D) and Hippo; ii) down-regulation of numerous biosynthetic pathways, most notably lipids, cholesterol and steroids; iii) up-regulation of ribosomal genes; and iv) down-regulation of epithelial signatures and up-regulation of epithelial-mesenchymal transition (EMT) signatures (Supplementary Figure 10). We validated a subset of these biological associations. First, we demonstrated that loss of GRHL2 caused down-regulation of ERBB2 and other ERBB family members ERBB3 and EGFR (Figure 6E), likely due to the capacity of GRHL2 to directly regulate these genes (Figure 6F and (38)). Second, we observed a significant decrease in active components of the PI3K/Akt pathway (i.e. phosphorylated Akt and ERK) following GRHL2 knockdown (Figure 6G). Finally, we validated the potent anti-EMT and anti-invasive activity of GRHL2 both in vitro and in vivo (below).
Loss of GRHL2 promotes EMT and invasion
EMT has been demonstrated to play a key role in certain phases of metastasis, including escape from the primary tumor, migration and invasion into the stroma, entry/exit from the bloodstream and suppression of senescence, apoptosis and anoikis (39). With this in mind, we hypothesized that the negative association between GRHL2 and EMT revealed by our RNA-seq data (Supplementary Figure 10) could have important biological ramifications.
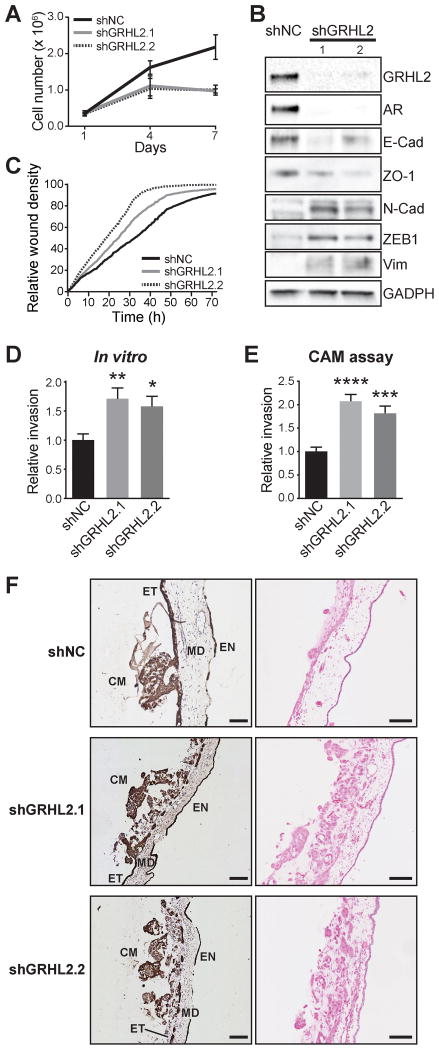
To address this hypothesis, we generated LNCaP cells with stable knockdown of GRHL2 using two distinct lentiviral shRNA constructs. Recapitulating the short-term siRNA experiments, GRHL2 silencing resulted in a reduced growth rate and a dramatic decrease in AR expression (Figures 7A,B). To examine how loss of GRHL2 influences the epithelial phenotype, the expression of epithelial and mesenchymal markers was measured by Western blotting in the LNCaP-shGRHL2 cells. As expected, knockdown of GRHL2 caused loss of epithelial and gain of mesenchymal factors, indicative of EMT (Figure 7B).
Figure 7. GRHL2 suppresses EMT and invasion of prostate cancer.
(A) The growth of two distinct clones with stable knockdown of GRHL2 (shGRHL2.1 and shGRHL2.2) were compared to a control (shNC) in Trypan blue growth assays. Error bars represent ± SD. (B) Stable knockdown of GRHL2 decreases epithelial marker (E-cad, ZO-1) and increases mesenchymal marker (N-cad, ZEB1, Vim) expression at the protein level in LNCaP cells. E-cad, E-cadherin; N-cad, N-cadherin; Vim, Vimentin. (C) Stable knockdown of GRHL2 increases migration of LNCaP cells in a scratch-wound assay. (D) Stable knockdown of GRHL2 increases invasion of LNCaP cells. Values for the negative control (NC) were set to 1, and error bars are ± SEM. P values were determined using unpaired t tests (**, P < 0.01). (E) Stable knockdown of GRHL2 promotes cancer cell invasion in CAM invasion assays. Data represents the mean percentage of images with invasion into the mesoderm ± SEM. P value was determined using an unpaired two-sided t test (***, P < 0.001; ****, P < 0.0001). (F) Representative images from the CAM assays. Cancer cell:matrigel grafts (CM) were placed on top of the ectoderm (ET) layer and cancer cell invasion into the CAM mesoderm (MD) was assessed in day 14 chick embryos. Endoderm, EN. Shown are cytokeratin (CK) IHC (left) and haematoxylin and eosin (right) images. Scale bars = 100 microns.
Recent studies have elucidated transcriptional targets via which GRHL2 suppresses EMT in breast and ovarian cancer (38,40). For example, GRHL2 binds to promoter and/or enhancer elements to directly activate CDH1 (E-cadherin), TJP1 (ZO-1), CLDN4, ELF3, EPCAM and RAB25, thereby promoting an epithelial phenotype. Analysis of our ChIP-seq data revealed that GRHL2 also associates with these cis-regulatory elements in LNCaP cells (Supplementary Figure 11), suggesting that the transcriptional targets via which GRHL2 fosters epithelial identity are concordant among distinct cancer types.
To investigate the phenotypic consequences of GRHL2-regulated EMT, we evaluated motility and invasion of the LNCaP-shGRHL2 cells using three distinct assays. We first used a scratch wound assay, and found that stable knockdown of GRHL2 significantly enhanced cell migration (Figure 7C). Subsequently, an in vitro transwell assay was employed to measure cell invasion through Matrigel, which mimics basement membrane. As expected, LNCaP-shGRHL2 cells were markedly more invasive than control cells (Figure 7D). Finally, the invasive capacity of the modified cell lines were evaluated in a more physiologically relevant setting using a chick chorioallantoic membrane (CAM) assay (41), which allows visualization of cell invasion through an ectoderm into mesoderm. Cancer cell/Matrigel grafts were implanted onto the CAM and, after 3 days, invasion was assessed by pan-cytokeratin immunohistochemistry. Control LNCaP cells invaded through the ectoderm of the CAM very poorly (Figure 7E,F). By contrast, knockdown of GRHL2 greatly enhanced the capacity of LNCaP cells to disrupt the ectodermal layer and invade into the mesoderm (Figure 7E,F).
To evaluate the generalizability of these findings, we conducted experiments in another model system, 22Rv1. In these experiments, cells were transfected with siGRHL2 to determine whether acute loss of GRHL2 was sufficient to promote EMT. As expected, transient knockdown of GRHL2 resulted in EMT within 2 days (Supplementary Figure 12). This experiment suggests that maintenance of epithelial status by GRHL2 is a general phenomenon in PCa, and that loss of GRHL2 could rapidly enhance mesenchymal, pro-invasive characteristics of tumor cells.
Discussion
Using a powerful and unbiased proteomic technique, we have identified GRHL2 as a new binding partner of AR, the key driver of PCa. Our data indicate that GRHL2 has a dichotomous oncogenic/protective role in this disease: it can act as an oncogene by enhancing AR signaling and promoting cancer cell proliferation; alternatively, it can potently suppress cancer cell EMT and invasion, phenotypes that are associated with disease progression and metastasis.
GRHL2 is a member of the Grainyhead-like family of transcription factors, which are expressed primarily in epithelia in a tissue- and developmental-specific manner and are critical for organogenesis, epidermal development and wound healing (42). More recently, a role for GRHL2 in various solid cancers has been proposed, although its precise functions are largely unknown. Increased GRHL2 expression is associated with poor outcomes in liver, kidney, gastric and colorectal cancers, and experimental studies have revealed that it can promote growth of certain cancer models in vitro and in vivo (42). By contrast, the action of GRHL2 in breast cancer is somewhat controversial, with studies purporting to demonstrate both oncogenic and tumor suppressive activities (43,44). A putative explanation for this apparent discrepancy was provided by Werner and colleagues, who presented evidence for dual functionality of GRHL2 in breast cancer: namely, it can both promote tumor growth but suppress EMT (38). Herein, we have shown that GRHL2 plays an analogous dual role in PCa growth and progression. The spatiotemporality of GRHL2 expression and activity in prostate (and breast) tumors is likely to be critical in dictating which of these roles dominate. More specifically, in early, localized tumors GRHL2 would act to promote cancer growth; however, as disease progressed, GRHL2 could act to suppress EMT and thereby curb stromal invasion, intravasation and survival of circulating tumor cells, with the collective outcome being inhibition of metastasis; finally, in micrometastases, GRHL2 could revert to an oncoprotein by reversing mesenchymal phenotypes, re-activating proliferation and facilitating establishment of clinically-significant metastases. Recent work suggests that dual functionality of GRHL2 (i.e. growth-promoting but EMT/invasion-suppressing) could be a generalizable phenomenon in carcinomas (40,45).
The data presented herein demonstrate that the relationship between GRHL2 and the AR signaling axis is multifaceted and complex. More specifically, GRHL2 is not only essential for the maintenance of AR expression, it also acts as an AR transcriptional co-activator. With regards to the latter function, is must be noted that GRHL2 and AR share only a small proportion of their respective cistromes. However, these shared binding events occur proximal to a set of genes that are relevant in PCa. These critical roles for GRHL2 in the AR signaling axis are particularly relevant in light of our finding that GRHL2 is a direct AR target gene. Reciprocal regulation creates a potent positive feed-forward loop between AR and GRHL2, which may be amplified further in the CRPC setting where the genes encoding both factors are commonly amplified and/or upregulated.
A key function of the AR signaling axis in both the normal and malignant prostate is to regulate epithelial differentiation (46,47). The work reported herein provides new insight into molecular mechanisms underlying this function: specifically, by directly promoting GRHL2 expression, AR indirectly activates a transcriptional program comprising key epithelial identity factors such as CDH1 (E-cadherin), TJP1 (ZO-1), ELF3, CLDN4 and EPCAM. Interestingly, by interrogating AR chromatin binding events in prostate tumors (35), we found evidence that it could directly regulate a subset of these genes in concert with GRHL2 (Supplementary Figure 11). Further investigation of the cistromic interplay between these two factors, including in the normal prostate, is required to definitively identify their shared target genes.
There is an urgent requirement for novel strategies that inhibit growth of CRPC and improve patient outcomes; one such strategy that has been proposed is targeting transcriptional co-regulators rather than AR itself. In this context, our study is significant because it identifies a critical new co-regulator that not only enhances AR activity but is also essential for the maintenance of AR expression. However, while targeting GRHL2 could disable the AR signaling axis at multiple levels, it may concomitantly enhance the metastatic capacity of the tumor. As such, if strategies to inhibit GRHL2 were developed, their application in PCa would require careful consideration.
Supplementary Material
Acknowledgments
The authors thank Drs Colm Morrisey, Larry True and Tony Rizzardi (University of Washington) for assistance with the prostate cancer tissue microarray, Mark Van der Hoek from the South Australian Health and Medical Research Institute Genomics Facility for assistance with ChIP-seq and RNA-seq, and Dr Nicholas Mitsiades (Baylor College of Medicine) for sharing unpublished data. The results published here are in part based on data generated by The Cancer Genome Atlas, established by the National Cancer Institute and the National Human Genome Research Institute, and we are grateful to the specimen donors and relevant research groups associated with this project.
Financial support: This work was supported by funding from the National Health and Medical Research Council of Australia (ID 1008349 to W. D. Tilley; ID 1083961 to L. A. Selth and W. D. Tilley), a Prostate Cancer Research Programs Transformative Impact Award from the US Department of Defense (W81XWH-13-2-0093 to S. R. Plymate, W. D. Tilley, G. V. Raj, S. M. Dehm and L. A. Selth) and NIH grant R01CA174777 (to S. M. Dehm). L. A. Selth was supported by a Young Investigator Award from the Prostate Cancer Foundation (Foundation 14 award).
Abbreviations list
- ADT
androgen deprivation therapy
- AR
androgen receptor
- ARV
androgen receptor variant
- ChIP
chromatin immunoprecipitation
- ChIP-seq
chromatin immunoprecipitation sequencing
- CRPC
castrate resistant prostate cancer
- EMT
epithelial-mesenchymal transition
- GRHL2
Grainyhead-like 2
- PCa
prostate cancer
- GSEA
gene set enrichment analysis
- RIME
rapid immunoprecipitation mass spectrometry of endogenous proteins
- RNA-seq
RNA sequencing
Reference List
- 1.Mitsiades N. A road map to comprehensive androgen receptor axis targeting for castration-resistant prostate cancer. Cancer research. 2013;73(15):4599–605. doi: 10.1158/0008-5472.CAN-12-4414. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt AW, Gleave ME. Targeting the adaptive molecular landscape of castration-resistant prostate cancer. EMBO Mol Med. 2015;7(7):878–94. doi: 10.15252/emmm.201303701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chmelar R, Buchanan G, Need EF, Tilley W, Greenberg NM. Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. Int J Cancer. 2007;120(4):719–33. doi: 10.1002/ijc.22365. [DOI] [PubMed] [Google Scholar]
- 4.Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, D'Santos CS. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11(2):316–26. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- 5.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KA, et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(43):17492–7. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan SC, Selth LA, Li Y, Nyquist MD, Miao L, Bradner JE, et al. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das R, Gregory PA, Fernandes RC, Denis I, Wang Q, Townley SL, et al. MicroRNA-194 Promotes Prostate Cancer Metastasis by Inhibiting SOCS2. Cancer research. 2017;77(4):1021–34. doi: 10.1158/0008-5472.CAN-16-2529. [DOI] [PubMed] [Google Scholar]
- 8.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selth LA, Das R, Townley SL, Coutinho I, Hanson AR, Centenera MM, et al. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene. 2016 doi: 10.1038/onc.2016.185. [DOI] [PubMed] [Google Scholar]
- 10.Tarulli GA, De Silva D, Ho V, Kunasegaran K, Ghosh K, Tan BC, et al. Hormone-sensing cells require Wip1 for paracrine stimulation in normal and premalignant mammary epithelium. Breast Cancer Res. 2013;15(1):R10. doi: 10.1186/bcr3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hickey TE, Irvine CM, Dvinge H, Tarulli GA, Hanson AR, Ryan NK, et al. Expression of androgen receptor splice variants in clinical breast cancers. Oncotarget. 2015;6(42):44728–44. doi: 10.18632/oncotarget.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizzardi AE, Rosener NK, Koopmeiners JS, Isaksson Vogel R, Metzger GJ, Forster CL, et al. Evaluation of protein biomarkers of prostate cancer aggressiveness. BMC Cancer. 2014;14:244. doi: 10.1186/1471-2407-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillis JL, Selth LA, Centenera MM, Townley SL, Sun S, Plymate SR, et al. Constitutively-active androgen receptor variants function independently of the HSP90 chaperone but do not confer resistance to HSP90 inhibitors. Oncotarget. 2013;4(5):691–704. doi: 10.18632/oncotarget.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48(3):240–8. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu LJ, Gazin C, Lawson ND, Pages H, Lin SM, Lapointe DS, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome biology. 2011;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38(4):576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26(11):1293–300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Need EF, Selth LA, Trotta AP, Leach DA, Giorgio L, O'Loughlin MA, et al. The unique transcriptional response produced by concurrent estrogen and progesterone treatment in breast cancer cells results in upregulation of growth factor pathways and switching from a Luminal A to a Basal-like subtype. BMC Cancer. 2015;15:791. doi: 10.1186/s12885-015-1819-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–4. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer cell. 2010;17(5):443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He B, Lanz RB, Fiskus W, Geng C, Yi P, Hartig SM, et al. GATA2 facilitates steroid receptor coactivator recruitment to the androgen receptor complex. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(51):18261–6. doi: 10.1073/pnas.1421415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010:1–21. doi: 10.1002/0471142727.mb1910s89. Chapter 19:Unit 19 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15(10):1451–5. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research Network. Electronic address scmo, Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer Cell. 2015;163(4):1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajan P, Sudbery IM, Villasevil ME, Mui E, Fleming J, Davis M, et al. Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. European urology. 2014;66(1):32–9. doi: 10.1016/j.eururo.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma NL, Massie CE, Ramos-Montoya A, Zecchini V, Scott HE, Lamb AD, et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer cell. 2013;23(1):35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nature genetics. 2015;47(11):1346–51. doi: 10.1038/ng.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahu B, Laakso M, Ovaska K, Mirtti T, Lundin J, Rannikko A, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. The EMBO journal. 2011;30(19):3962–76. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jozwik KM, Chernukhin I, Serandour AA, Nagarajan S, Carroll JS. FOXA1 Directs H3K4 Monomethylation at Enhancers via Recruitment of the Methyltransferase MLL3. Cell reports. 2016;17(10):2715–23. doi: 10.1016/j.celrep.2016.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werner S, Frey S, Riethdorf S, Schulze C, Alawi M, Kling L, et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. The Journal of biological chemistry. 2013;288(32):22993–3008. doi: 10.1074/jbc.M113.456293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nature reviews Cancer. 2013;13(2):97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 40.Chung VY, Tan TZ, Tan M, Wong MK, Kuay KT, Yang Z, et al. GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci Rep. 2016;6:19943. doi: 10.1038/srep19943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lokman NA, Elder AS, Ricciardelli C, Oehler MK. Chick chorioallantoic membrane (CAM) assay as an in vivo model to study the effect of newly identified molecules on ovarian cancer invasion and metastasis. Int J Mol Sci. 2012;13(8):9959–70. doi: 10.3390/ijms13089959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mlacki M, Kikulska A, Krzywinska E, Pawlak M, Wilanowski T. Recent discoveries concerning the involvement of transcription factors from the Grainyhead-like family in cancer. Exp Biol Med (Maywood) 2015;240(11):1396–401. doi: 10.1177/1535370215588924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cieply B, Riley Pt, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, et al. Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer research. 2012;72(9):2440–53. doi: 10.1158/0008-5472.CAN-11-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang X, Deng Z, Zhuang X, Ju S, Mu J, Jiang H, et al. Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS One. 2012;7(12):e50781. doi: 10.1371/journal.pone.0050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen W, Yi JK, Shimane T, Mehrazarin S, Lin YL, Shin KH, et al. Grainyhead-like 2 regulates epithelial plasticity and stemness in oral cancer cells. Carcinogenesis. 2016;37(5):500–10. doi: 10.1093/carcin/bgw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das R, Gregory PA, Hollier BG, Tilley WD, Selth LA. Epithelial plasticity in prostate cancer: principles and clinical perspectives. Trends Mol Med. 2014;20(11):643–51. doi: 10.1016/j.molmed.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 47.Nieto CM, Rider LC, Cramer SD. Influence of stromal-epithelial interactions on androgen action. Endocrine-related cancer. 2014;21(4):T147–60. doi: 10.1530/ERC-14-0138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.