Abstract
The human large intestine is populated by an extremely high density of microorganisms, collectively termed the colonic microbiota1, which plays an important role in human health and nutrition2. The survival of microbiota members from the dominant Gram-negative phylum Bacteroidetes depends on their ability to degrade dietary glycans that cannot be metabolised by the host3. The genes encoding proteins involved in the degradation of specific glycans are organised into co-regulated polysaccharide utilisation loci (PULs)4–8. The archetypal PUL has been named Sus, for starch utilisation system, and consists of 7 proteins named SusA-G8–10. Glycan degradation occurs mainly intracellularly and depends critically on the import of oligosaccharides by an outer membrane (OM) protein complex composed of an extracellular SusD-like lipoprotein and an integral membrane SusC-like TonB-dependent transporter (TBDT)4–7,11–13. The presence of the partner SusD-like lipoprotein is the major difference that distinguishes SusC-like proteins from previously characterised TBDTs. Many sequenced gut Bacteroides spp encode over 100 C/D pairs, with the large majority of them having unknown functions and substrate specificities3,8,14,15. The central, unresolved question is how extracellular substrate binding by SusD proteins is coupled to OM passage via their cognate SusC transporter. Here we present X-ray crystal structures of two functionally distinct SusCD complexes purified from Bacteroides thetaiotaomicron and derive a general model for substrate translocation. The SusC transporters form homodimers, with each β-barrel protomer tightly capped by the SusD. Ligands are bound at the SusCD interface in a large solvent-excluded cavity. Molecular dynamics simulations and single channel electrophysiology reveal a "pedal bin" mechanism in which SusD moves away from SusC in a hinge-like fashion in the absence of ligand to expose the substrate binding site to the extracellular milieu. Our results provide mechanistic insights into OM nutrient import by members of the microbiota, which is of major significance for understanding the human-microbiota symbiosis.
To elucidate the mechanism of substrate acquisition by SusCD complexes via X-ray crystallography we initially tried to overexpress several B. thetaiotaomicron (B. theta) SusC homologues in the OM of E. coli, but were not successful. However, we discovered that a number of SusCD complexes are expressed at high levels (~ 1 mg/l) in B. theta (Extended Data Fig. 1). The remarkable abundance of TBDTs and the paucity of OM diffusion channels such as porins in B. theta (Extended Data Fig. 1a) shows that nutrients are acquired in a fundamentally different way in Bacteroidetes compared to e.g. E. coli. A four-component complex produced by B. theta in minimal medium could be purified to homogeneity via ion exchange chromatography (Extended Data Fig. 1b) and was identified as BT2261-2264 by mass spectrometry. The substrate of BT2261-2264 is not known, but the presence of homologues of both SusD (BT2263) and SusC (BT2264) support a function in nutrient acquisition. BT2261 and BT2262 are small OM lipoproteins (16 and 24 kDa) located on the periphery of the complex and that in B. theta are unique to the BT2261-64 system (Extended data Fig. 4). The BT2261-64 locus on the B. theta genome is not in an operon with any other genes.
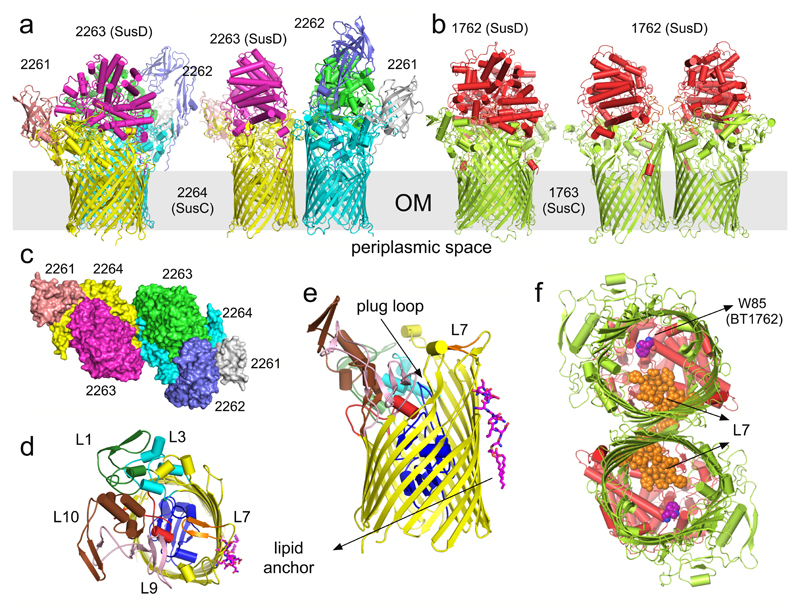
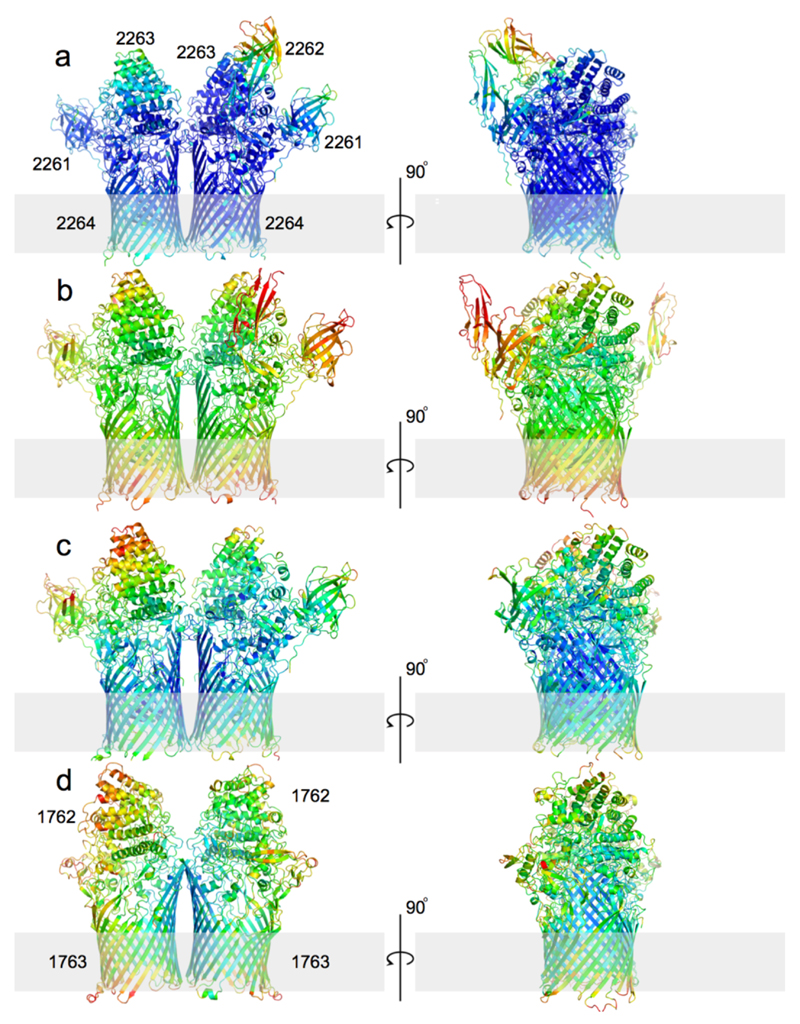
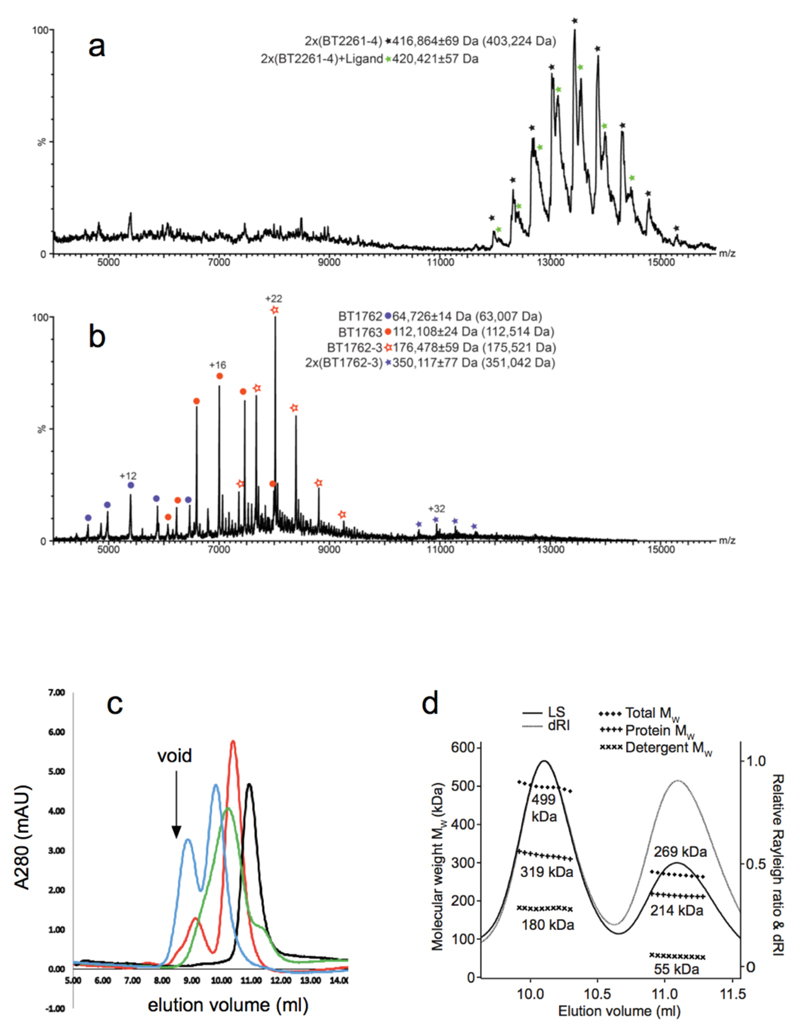
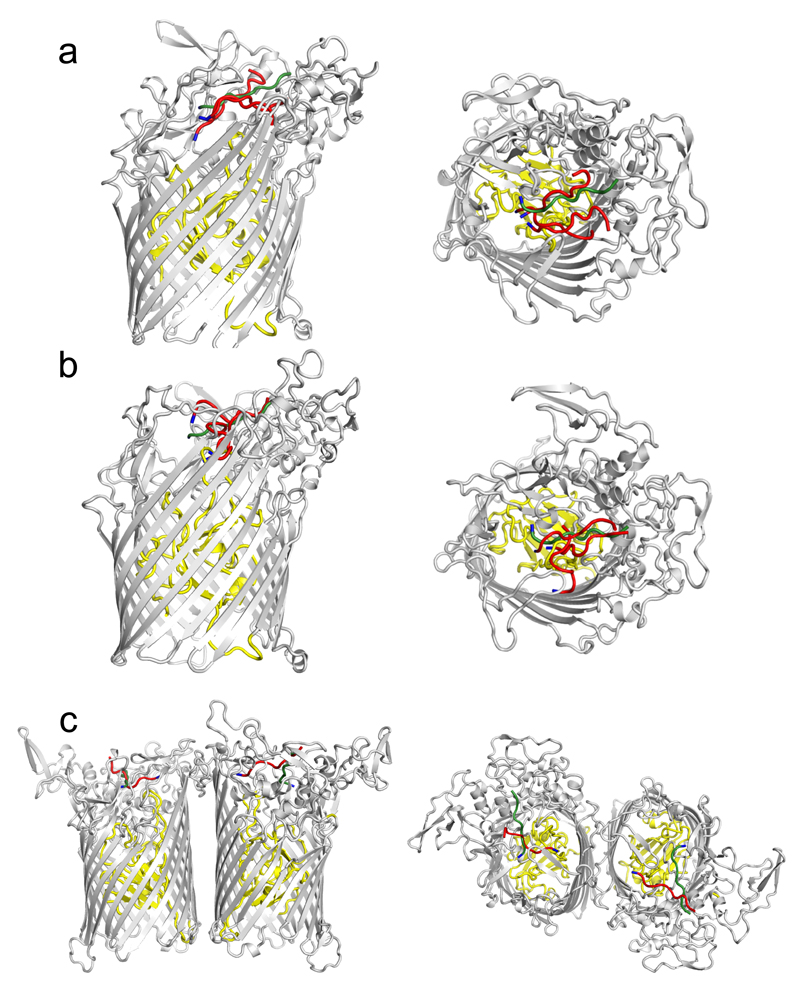
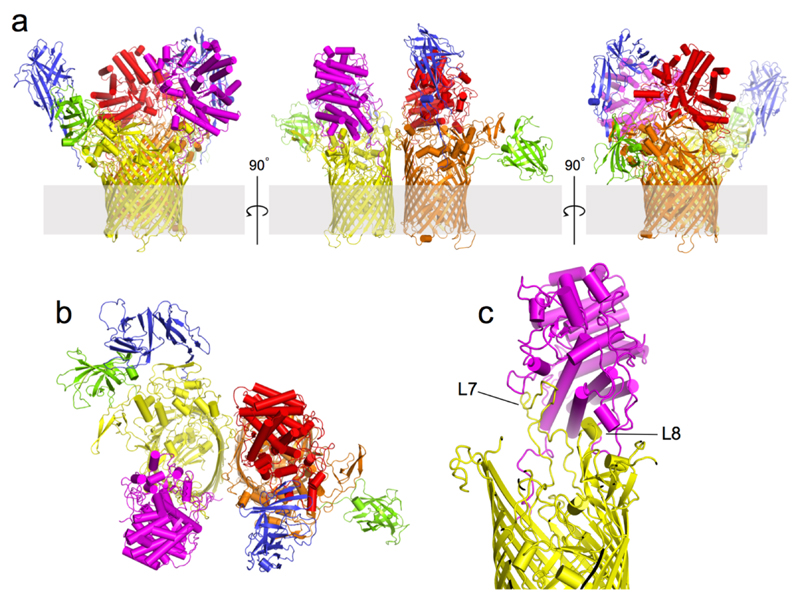
The structure of BT2261-64 was solved by molecular replacement using data to 2.75 Å resolution (Figs. 1a and c and Supplementary Tables 1-3) (Methods). The structure reveals that the central BT2264 (SusC) component of the complex is a typical, albeit large TBDT (984 residues), with an N-terminal plug domain inserted into a 22-stranded β-barrel (Figs. 1d and e)13. Strikingly, SusC forms homodimers burying ~2900 Å2 of surface area (Figs. 1a and c). Identical interfaces are observed for two other crystal forms (Extended Data Fig. 2), suggesting the dimer is not a crystal packing artefact. Native mass spectrometry confirms that BT2261-64 forms an octameric complex of ~400 kDa in size (Extended Data Fig. 3). To our knowledge, the dimeric organisation of SusC is unique for TBDTs.
Figure 1. Overall architecture of SusCD complexes.
a, b, Cartoon views from the side for BT2261-64 (a) and BT1762-63 (b) rotated by 90º, with individual subunits labelled. OM, outer membrane. c, Surface representation of BT2261-64 from the outside of the cell. d, e, Architecture of BT2264 (SusC-like) viewed from the extracellular side (d) and from the plane of the membrane (e) with several extracellular loops indicated (L). The plug domain in the 22-stranded β-barrel is dark blue, and the N-terminal ten residues of BT2263 (SusD-like) including the lipid anchor are shown as stick models in magenta. Loop L7 is shown in orange. f, View of the BT1762-63 dimer from the periplasmic side. The L7 loops (orange) of BT1763 (SusC-like) and the functionally important residue Trp85 in BT1762 (SusD-like) are shown as space-filling models.
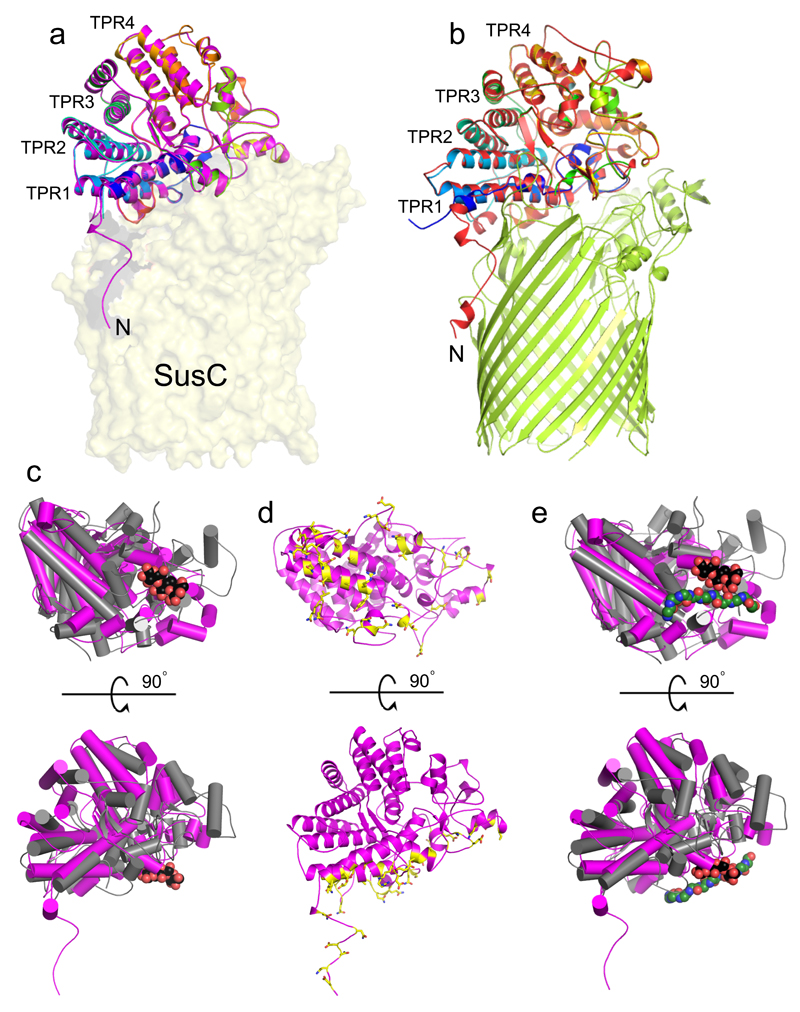
Surprisingly, the SusD-like subunit BT2263 (SusD) is located on top of the SusC forming an extracellular lid on the transporter, generating a large interface area of ~3800 Å2 (Figs. 1a and c). The SusCD interaction is stabilised by ~50 hydrogen bonds and three salt bridges, explaining the high stability of the complex in SDS-PAGE (Extended Data Fig. 1). SusC-bound SusD has an identical structure compared to SusD in isolation (Fig. 2a), demonstrating that no conformational changes occur upon binding to SusC. The face of SusD that interacts with SusC is distant from the conserved four tetratricopeptide (TPR) repeats (Fig. 2a), contrasting with earlier predictions16. Instead the interaction surface includes the highly variable region that forms the substrate binding site of SusD-like proteins (Fig. 2c-e)11,16.
Figure 2. The SusCD interaction involves the ligand binding face of SusD.
a Cartoon overlay of BT2263 SusD in isolation (rainbow colouring; N-terminus blue) and within the BT2261-2264 complex (magenta). The four TPR domains are labelled and BT2264 SusC is shown as a transparent surface. b, Cartoon overlay for BT1762 in isolation (rainbow colouring) and within BT1762-1763 (red). BT1763 SusC is shown in green. c, Superposition of BT2263 with the archetypal SusD BT3701 in grey (PDB ID: 3CKB), viewed from the bottom (top panel) and from the side as in a. Bound maltotriose in BT3701 is shown as a space-filling model (carbons, black; oxygens, red). d, BT2263 with residues forming hydrogen bonds and salt bridges with BT2264 coloured yellow. e, Superposition as in c, with the putative peptide ligand of BT2261-2264 included to show binding site overlap.
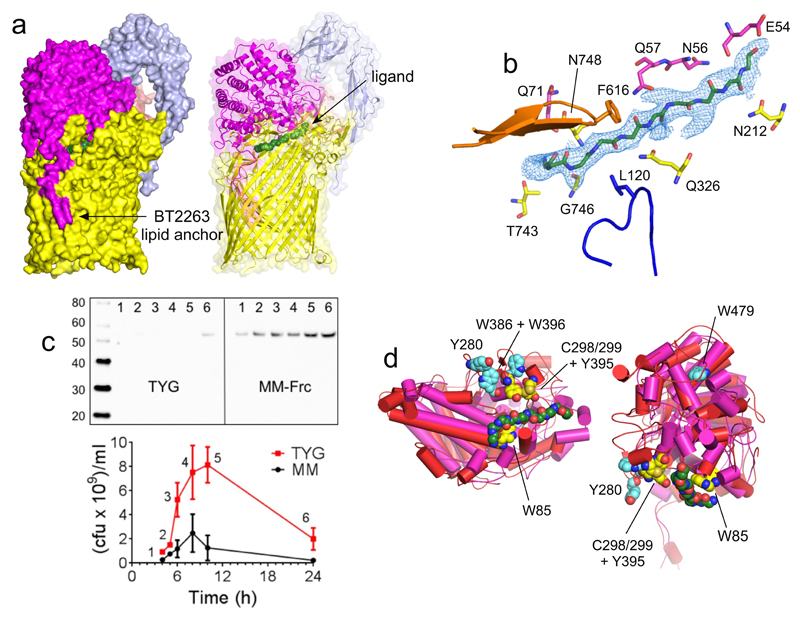
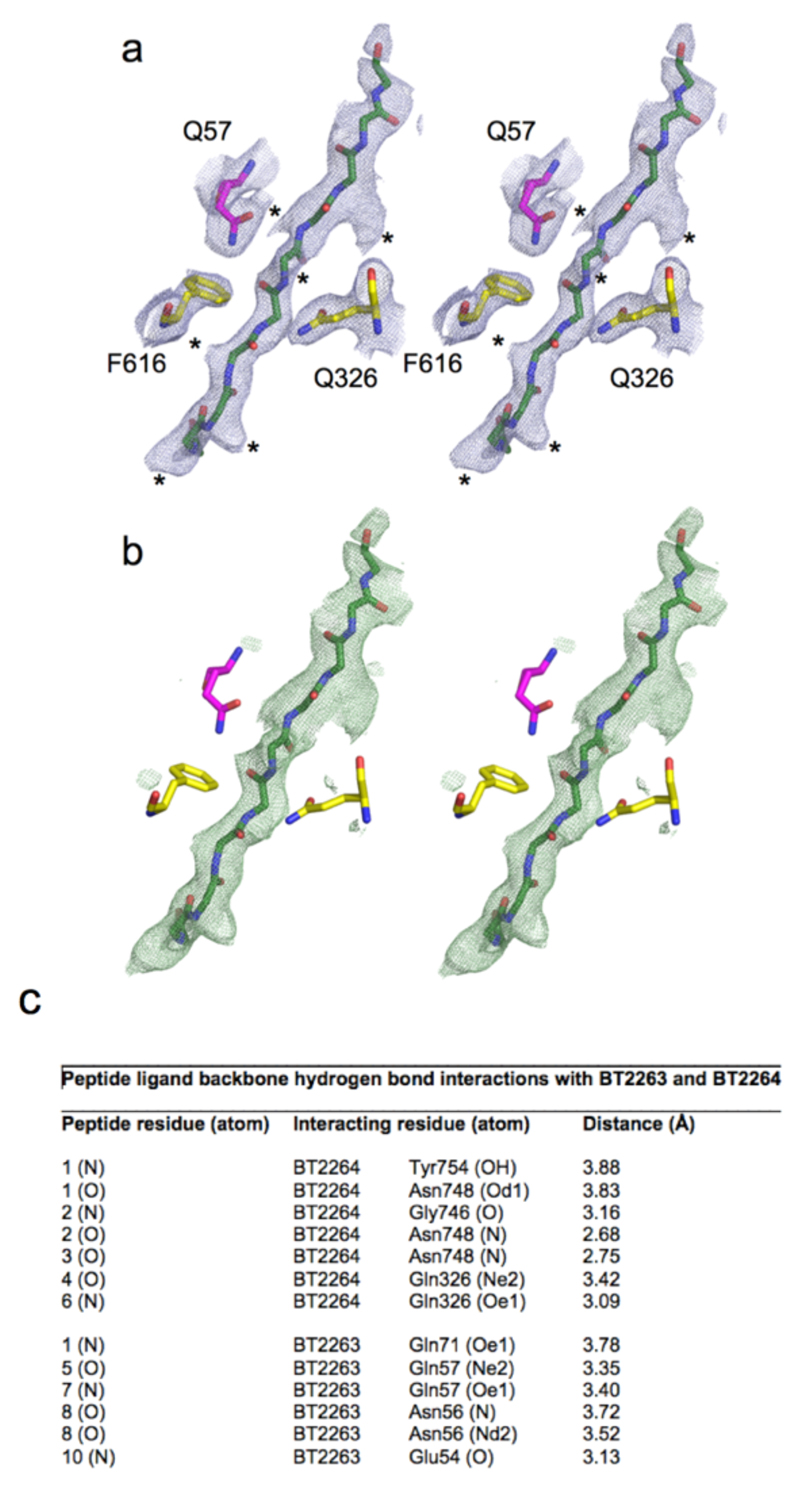
Inspection of the SusCD interface reveals the presence of a well-defined, ~30 Å long stretch of electron density that cannot be accounted for by anything added during bacterial growth and protein purification and crystallisation. The density is modelled well by a 10-residue peptide (Fig. 3a and b and Extended Data Fig. 5). At a resolution of 2.75 Å, the peptide sequence is ambiguous. Based on the relatively weak density for several side chains we favour the possibility that an ensemble of peptides is bound. An ensemble would explain better why we could not identify the ligand by native mass spectrometry (Extended Data Fig. 3), but we cannot exclude that a unique sequence is present, or even that the density might not correspond to a peptide. To account for the sequence ambiguity, we modelled the putative peptide ligand(s) as deca-glycine (Fig. 3b). The fact that the ligand survived a large number of purification steps indicates that it is tightly bound and originates from B. theta. The ligand is present in a large cavity between SusC and SusD that is not solvent accessible (Fig. 3a). Nine out of ten residues make backbone interactions with either SusC or SusD residues (Fig. 3b and Extended Data Fig. 5). A PISA17 analysis reveals that the ligand interacts more extensively with SusC compared to SusD (540 Å2 and 300 Å2 buried surface area respectively), suggesting that the complex is a transport intermediate in which a peptide ligand has been transferred from SusD to SusC and is now tightly bound as is typical for TBDT substrates13.
Figure 3. The SusCD ligand binding site is conserved.
a, Side views of BT2261-64 showing the putative peptide ligand in green. b, Close-up of the binding site, showing SusC (yellow) and SusD (magenta) residues forming hydrogen bonds with the peptide backbone. Loop L7 is orange and the plug loop dark blue. 2Fo-Fc density for deca-glycine is shown as a blue mesh at 1.5σ. c, Western blot analysis of BT2261-64 expression via FLAG-tagged BT2263 in rich medium (TYG) and fructose minimal medium (MM-Frc) (representative of three experiments). Bottom panel shows corresponding cfu values. d, Mutational analysis of levan binding by BT1762 (red). Views are from the periplasmic side (left panel) and from the front. Residues that abolish levan binding are shown in yellow with those that do not affect levan binding in cyan. BT2263 (magenta) with bound deca-glycine (green) is superposed to show the conservation of the binding sites.
To provide functional data for BT2261-64 we analysed expression of the complex using western blots. In contrast to known glycan uptake systems14, BT2261-64 is constitutively expressed during log phase growth in minimal medium and only upregulated on rich media during stationary phase when cell viability drops (Fig. 3c), suggesting a role for the complex during conditions of nutrient stress. These expression patterns are supported by previous transcriptomic data showing upregulation of BT2261-64 in minimal media18. A constructed ΔBT2264 (susC) knockout strain did not give a phenotype under various conditions (Methods), a result that is not surprising given that B. theta encodes over 100 SusCD systems with the majority having unknown functions3,8,14,15. We also did not observe binding to SusD (BT2263) by isothermal titration calorimetry (ITC) for two synthetic decapeptides with sequences consistent with the electron density (Methods). Despite this, the overall emerging picture is that BT2261-64 imports oligopeptides derived from other members of the microbiota. Given the extremely high microbial densities, bacterial turnover represents an underappreciated but potentially important source of nutrients in the gut. An alternative, intriguing possibility is that BT2261-64 serves as a receptor for signalling peptides, e.g in quorum sensing19.
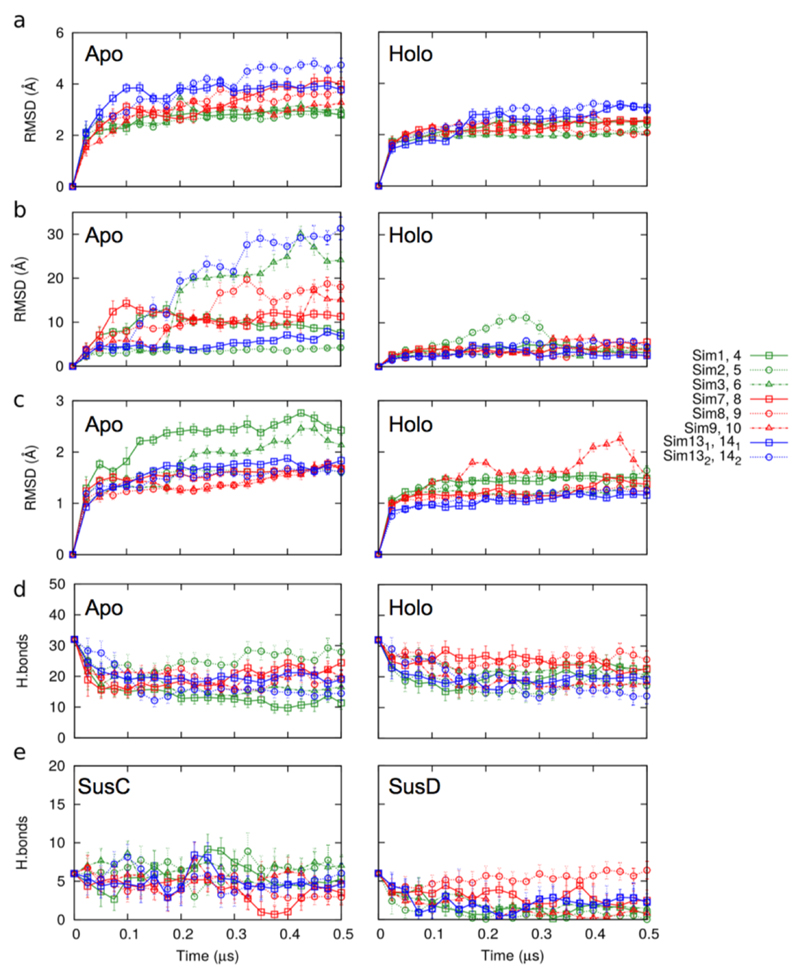
The ligand binding site in the complex is excluded from solvent (Fig. 3a) indicating that SusD must dissociate from SusC to capture substrate from the external environment. To answer the key question as to how the substrate is delivered by SusD to the transporter we performed unbiased molecular dynamics (MD) simulations on the central hub BT2263-64 (SusCD; dimer) and on BT2261-64 ("tetramer" and "octamer") in the presence and absence of a modelled peptide (holo- and apo-complex respectively; Methods). In the holo-simulations, all complexes are stable during the duration of the simulations (Figs. 4a-c, Extended Data Fig. 6 and Supplementary Videos 1-3). While remaining at the SusCD interface, the conformation of the peptide varies substantially and non-uniformly in all simulations (Fig. 4a, Extended Data Fig. 7 and Supplementary Videos 1-3). The peptide movement in the simulations appears hard to reconcile with the presence of well-defined electron density in the crystal structure. The most likely explanation for this discrepancy is that the peptide sequence used in the simulations is not stable in the binding site. Together with the peptide ITC data this suggests that the BT2261-64 system is at least to some extent sequence-specific. Despite the peptide movement, the total number of hydrogen bonds with SusC and SusD remains approximately constant (Extended Data Fig. 6).
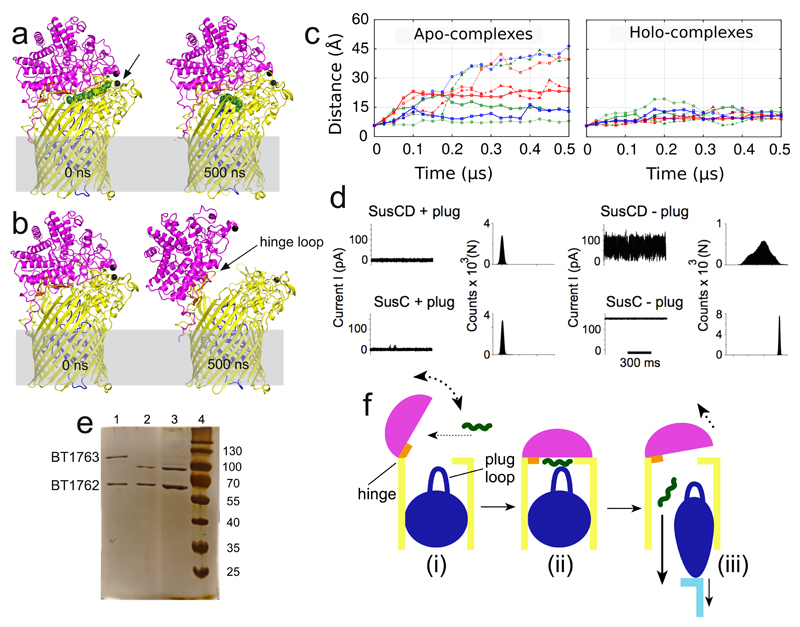
Figure 4. SusCD complexes transport their substrates via a pedal bin mechanism.
a and b, MD simulation snapshots of holo (a) and apo (b) BT2263-64 after 0 ns and 500 ns. SusC is coloured yellow with the plug domain in dark blue; SusD is shown in magenta. Cα atoms of Asn203 in SusC and Thr296 in SusD are shown as black spheres. c, Average distances vs. simulation time between SusC-Asn203 and SusD-Thr296 for SusCD dimers (green), BT2261-64 tetramers (red), and the (BT2261-64)x2 octamer (blue). d, Single channel electrophysiology traces and corresponding all-point histograms for BT1762-63 complexes (SusCD +/- plug) and for BT1763 (SusC +/- plug). Traces and histograms are representatives of 10 experiments. e, SDS-PAGE of BT1762-63 before (lane 1) and after (lane 2) incubation for 2 weeks at room temperature, and from crystal drops (lane 3). f, General mechanism for nutrient uptake via SusCD transport complexes, colour-coded as in panels a and b. Ligand is shown as a green wavy line and the C-terminal domain of TonB that interacts with the SusC plug is cyan.
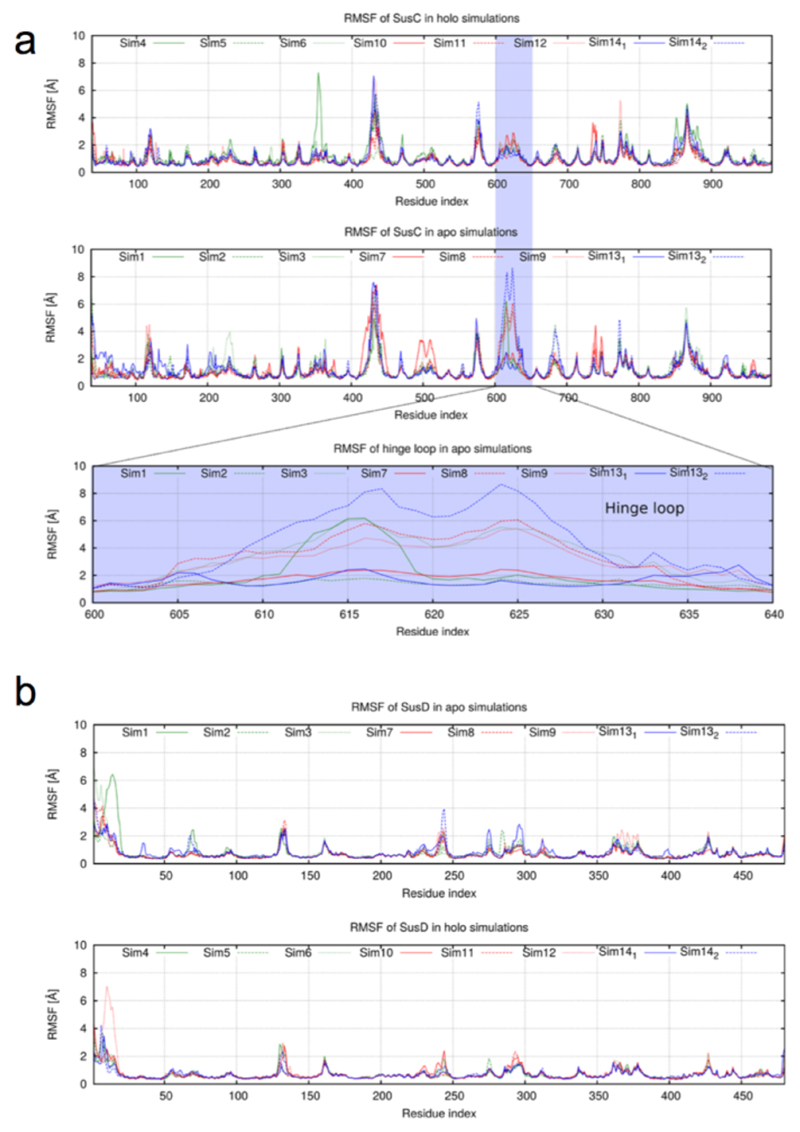
Remarkably, in the majority of apo simulations SusD undergoes a hinge-like motion to expose the ligand binding site to the external environment. The extent of opening between different simulations varies due to thermal fluctuations, but in the most-open states SusD has rotated ~40-45° and has completely lost its contacts with loops L1, L3-5, and L9-L11 of SusC (Fig. 4b; Supplementary Videos 1-3). Analysed in a different way, the Cα-Cα distance between residues at the front of the complex (Thr296 in SusD and Asn203 in SusC) increases from ~6 Å in the closed, ligand-bound complexes to ~40 Å in open, ligand-free states (Fig. 4c). The conformational changes are very similar in all simulations where lid opening is observed and correspond to a rigid-body movement of SusD (Extended Data Fig. 8). Loop L7 of SusC moves together with SusD and is responsible for the majority of the remaining SusCD interactions in the open state. We have named L7 the hinge loop, and it is the region in SusC that exhibits the largest conformational differences between the holo- and apo-simulations (Extended Data Fig. 8). The hinge point at the base of L7 is located at the back of the complex, near the SusD lipid anchor (Fig. 4b). The octamer simulation demonstrates that SusC dimerisation does not impede opening of the SusD lid and moreover suggests that both SusCD hubs function independently (Fig. 4c, Extended Data Fig. 9 and Supplementary Video 3). A final implication of the computational results is that since removal of the ligand produces a mechanistically sensible response (i.e. SusD lid opening), the MD simulations support our modelling of the BT2261-64 ligand as an oligopeptide. Even if the ligand is not a peptide, its location at the CD interface would stabilise the closed state of the complex, i.e. the results of the MD simulations of the holo complex would most likely not depend on the assigned identity of the ligand. This notion is supported by the fact that the SusD lid remains closed, despite the fact that the peptide assumes many different conformations during the simulations.
The large number and sequence diversity of SusCD gene pairs in Bacteroidetes and especially Bacteroides spp3,8,14,15 leads to the important question whether the BT2261-64 structure and transport mechanism are representative for those of other SusCD complexes. To address this issue we determined the crystal structure of BT1762-1763, which is part of a 7-gene PUL with an established function in levan (β2,6-fructan) degradation4. No other proteins co-purify with BT1762-63 (Extended Data Fig. 1d), suggesting that the accessory PUL OM lipoproteins are not an integral part of this complex. Native mass spectrometry and SEC-MALS show that BT1763 (SusC) also forms homodimers (Extended Data Fig. 3). The crystal structures confirm this, and show that the relative orientations of BT1762 (SusD) and BT1763 are the same as in the BT2263-64 complex, with SusD capping SusC (Figs. 1b and 2b). As is the case for BT2263-64, binding to SusC does not cause significant conformational changes in the SusD (Fig. 2b). Importantly, the hinge loop (L7) observed in BT2264 is also present in BT1763 (Fig. 1f). No substrate is bound in the BT1762-63 structure, in accordance with the known high specificity of the complex4 and the fact that no substrate was added during cell growth. The reason that the SusD lid is closed is likely related to the fact that crystallisation favours stable, compact states. Unexpectedly, no density is observed for the plug domain of BT1763 (Fig. 1f), and SDS-PAGE confirms the ~20 kDa lower molecular weight of BT1763 from crystal-containing drops (Fig. 4e). Removal of the plug and the preceding DUF4480 domain also occurs after prolonged incubation of freshly isolated complex, presumably via proteolytic cleavage by a co-purified contaminant (Fig. 4e). We took advantage of these different BT1762-63 complexes to obtain experimental evidence for opening of the SusD lid by single channel electrophysiology. Reconstitution of freshly purified, full-length SusCD complex in the planar lipid bilayer generates very small and relatively stable ion currents, consistent with the absence of a large channel in intact TBDTs. By contrast, plug-less complex (SusCD -plug) produces large channels with average conductance values of ~1.5 nS. The traces are extremely noisy and a wide range of conductance values are observed including zero, indicating dynamic opening and closing events (Fig. 4d). To exclude the possibility that gating by extracellular SusC loops was responsible for the observed dynamics in the plug-less BT1762-63 complex we isolated truncated BT1763 from SDS-PAGE gels, followed by refolding and reconstitution (SusC - plug). This produces large channels with similar conductance values (~3 nS) as measured for a genetically created plug-less TBDT, FhuA20. In contrast to those of plug-less SusCD, the plug-less SusC channels are stable, indicating that the extracellular loops of SusC are not responsible for the dynamics observed in plug-less SusCD (Fig. 4d). Lastly, full-length SusC (SusC + plug) behaves similarly to SusCD + plug and does not form channels, demonstrating that refolding and reconstitution generates correctly folded transporters. The electrophysiology data for the levan importer therefore provide experimental evidence that fully supports the dynamic opening and closing of the SusD lid in ligand-free complexes observed by MD simulations of the putative peptide importer.
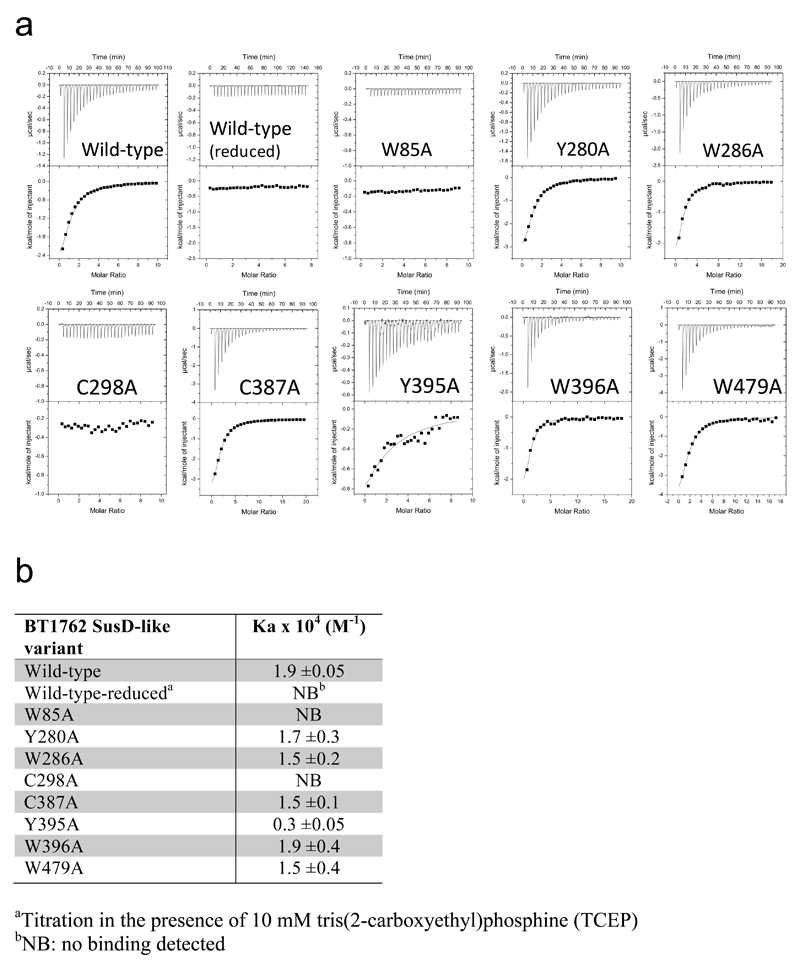
To provide functional support for the structures and transport mechanism we focused on BT1762-63 because robust growth of B. theta on levan is dependent on an intact copy of BT1762 (ref. 4). We predicted the aromatic residues Trp85, Trp286, Tyr395 and Trp396 as well as the disulphide-bonded residues Cys298 and Cys299 to be in or close to the putative levan binding site on BT1762 (SusD) based on sequence alignment and comparison with the ligand-bound structures of the archetypal SusD (BT3071; Fig. 2) and BT1043 (PDB 3EHN)21. Individual alanine mutants were produced in E. coli and levan binding to purified BT1762 variants was assessed by ITC. Mutation of Trp85 and Cys298 resulted in complete loss of levan binding, while the Tyr395 mutant displayed ~6-fold loss of affinity (Extended Data Fig. 10). Mutation of other aromatic residues, including ones far away from the binding site had no effect (e.g. Tyr280, Trp479). The locations of the residues abolishing levan binding to SusD are in excellent agreement with the positioning of SusD within the SusCD complex and also with the location of the ligand binding site in BT2261-64 (Fig. 3d).
Combining our structural and functional data and previous mechanistic insights for TBDTs13 we propose a "pedal bin" mechanism for nutrient acquisition by SusCD-like systems (Fig. 4f). (i) In the absence of substrate, the SusD lid of the empty transporter is mobile and samples a range of conformational states. (ii) Upon substrate binding, the closed state represented by the crystal structures is stabilised by ligand interactions with both SusC and SusD (Fig. 3b). (iii) The final step in transport involves TonB binding to the TonB box of the transporter22,23 to generate conformational changes in the plug and possibly extracellular loops of SusC that lead to substrate release and the formation of a transport channel into the periplasmic space13. The TonB box of BT2264 (residues 23-29) is not visible in the structures, consistent with the view that this segment is mobile and accessible in ligand-bound TBDTs to allow binding/recognition by TonB13. Residue L120 of the plug loop in BT2264 contacts the peptide via van der Waals interactions (Fig. 3b), providing a direct link between the ligand and the SusC plug and a potential way to signal TonB that the binding site in BT2264 is occupied13. The TonB-induced substrate dissociation reverts the transporter back to the dynamic open state, and the energy input from the ExbBD-TonB24,25 system is therefore equivalent to pressing the pedal to open the SusCD bin (Fig. 4e). The proposed transport mechanism also provides a rationale for the essential requirement of a surface located endo-acting enzyme in many PULs4–6,10since formation of the closed ‘transport competent’ complex appears to involve envelopment of the whole ligand molecule (Fig. 3a), precluding import of undigested, high molecular weight glycans.
Methods
Growth of B. theta, OM isolation and purification of SusCD complexes
Native BT2261-2264 complex
Wild-type B. theta was grown on minimal media containing fructose (0.5% w/v) as a carbon source at 37°C under anaerobic conditions overnight. At A600 of 1.5-2.0, cells were harvested by centrifugation at 11,305 x g for 30 minutes. Cells were resuspended in TSB buffer (20 mM Tris, 300 mM NaCl pH 8) and lysed at a pressure of 23 kpsi using a cell disrupter (Constant Systems 0.75 kW). The cells were centrifuged at 205,000 x g for 1 hour to collect total membranes. The pellets were resuspended and homogenised in 20 mM HEPES pH 7.5 containing 0.5 % (w/v) sodium lauroyl sarcosine. Typically, 100 ml buffer was used for 2 l of cells. The sample was incubated while stirring at 20 °C for 30 minutes and centrifuged at 205,000 x g for 30 minutes. The supernatant was discarded and the process was repeated. The clear, translucent pellets containing the OM proteins were resuspended and homogenised in 1.5 % (w/v) lauryldimethylamine-N-oxide (LDAO) buffer (10 mM Hepes, 50 mM NaCl pH 7.5) and incubated while stirring at 4°C overnight. The sample was centrifuged at 205,000 x g for 45 minutes and the supernatant containing OM proteins was collected. The sample was passed through a 0.45 μm filter, loaded on a 6 ml Resource Q anion exchange column and eluted with a linear gradient of NaCl (Extended Data Fig. 1). The BT2261-2264 peak was collected and further purified by gel filtration chromatography in 10 mM Hepes, 100 mM NaCl, 0.12% n-decyl-β-D-maltoside (DM) pH 7.5 using a HiLoad 16/60 Superdex 200 column. Peak fractions were pooled and the buffer was exchanged to 10 mM Hepes, 100 mM NaCl, 0.4% Tetraethylene Glycol Monooctyl Ether (C8E4) pH 7.5 using a 100 kD MWCO ultrafiltration device (Millipore). The protein was concentrated to 7-10 mg/ml, aliquoted and flash-frozen in liquid nitrogen.
Selenomethionine-substituted BT2261-2264 complex
10 litres of wild type B. theta were grown in rich TYG-haematin media at 37°C anaerobically for 20 hours to A600 of 1.2, and harvested by centrifugation at 11,305 x g for 30 minutes. The cells were resuspended and used to inoculate 10 litres of minimal media containing 0.5 % (w/v) fructose to induce expression of BT2261-2264. Seleno-methionine (SeMet) enrichment was performed based on the well-established methionine biosynthesis inhibition protocol for E. coli 26. Cells were incubated at 37°C anaerobically for 30 minutes before the addition of amino acids (Lys, Phe, Thr, Leu, Ile, Val) and SeMet (60 mg/l). The cells were incubated for a further 17 hours to a final A600 of ~1.7 and harvested. Bacteria were processed and BT2261-64 was purified as described above. BT1762-1763 complex. A 4xAla linker and 6xHis-tag were added to the C-terminus of genomic BT1762 (susD) using pExchange-tdk11. The mutated B. theta cells were grown on minimal media with 0.5% fructose under anaerobic conditions as fructose is known to activate expression of the levan PUL (BT1757-1763) (ref. 4). Cells were harvested and processed as outlined above. Following solubilisation with LDAO, the His-tagged complex was purified by cobalt-affinity chromatography (Talon) followed by gel filtration with a HiLoad 16/60 Superdex 200 column in 10 mM Hepes, 100 mM NaCl, 0.12% DM pH 7.5. For crystallisation the detergent was exchanged to C8E4 via a second gel filtration step in 10 mM Hepes, 100 mM NaCl, 0.4% C8E4 pH 7.5.
Overexpression and purification of BT2262 and BT2263
The mature parts of BT2262 and BT2263 lacking the N-terminal cysteine were amplified using PCR for ligation independent cloning into a pET9 vector containing a kanamycin resistance gene for selection, a PelB signal sequence for targeting proteins to the periplasm and a C-terminal TEV protease-cleavable 10xHis-tag. After TEV cleavage, the sequence "SM" is added to the first residue of the cloned sequence. The vector was digested using BsaI and annealed with the insert using T4 DNA polymerase. Proteins were expressed in BL21 (DE3) E. coli cells grown in LB media at 37°C to A600 of ~0.6 and induced with 0.2 mM IPTG then incubated at 18 °C overnight. Cells were harvested at 2,826 x g for 30 minutes, resuspended in TSB buffer and lysed at 23 kpsi using a cell disrupter. The lysed cells were centrifuged at 205,000 x g for 45 minutes and the supernatant was collected. The proteins were purified using nickel affinity chromatography (Chelating Sepharose; GE Healthcare) and eluted with TSB buffer containing 250 mM imidazole. The buffer was exchanged to TEV cleavage buffer (50 mM Tris, 50 mM NaCl, 0.5 mM EDTA, 0.5 mM TCEP pH 8). TEV protease was added to a ~ 5-10:1 w/w ratio (protein:TEV) and the His-tag was cleaved overnight at room temperature with gentle rocking. The cleaved proteins were subjected to a second round of nickel affinity chromatography. Tag-less proteins were further purified by gel filtration in 10 mM Hepes, 100 mM NaCl pH 7.5 using a HiLoad 16/60 Superdex 200 column, and concentrated to ~10-15 mg/ml for crystallisation.
Crystallisation of BT2262 and BT2263
Sitting drop crystallisation trials were set up using a Mosquito crystallisation robot (TTP Labtech) using commercial screens (Molecular Dimensions Structure, Index, PACT and JCSG+ screens). Initial hits were obtained in Structure, #37 for BT2263 (0.2 M sodium acetate trihydrate, 0.1 M Tris pH 8.5, 30% w/v PEG4000) and Structure, #30 for BT2262 (2 M ammonium sulfate, 0.1 M Hepes pH 7.5, 2% w/v PEG400).
Crystallisation and structure determination of BT1762
The mature part of BT1762 was overexpressed in the cytoplasmic space of E. coli BL21(DE3) and purified by cobalt affinity chromatography. The protein was crystalized using the sitting drop method using a Mosquito robot (TTP Labtech). Rod-like crystals grew in 2.0 M ammonium sulfate, 100 mM sodium acetate pH 4.6 and 8% PEG 4000. Both native and SeMet data were collected on beamline I03 at Diamond light source (Didcot, UK). Data were processed and integrated to 1.8 Å with XDS27 and scaled with Aimless28,29. Space group determination was tested with Pointless29. The phase problem was solved by SeMet single-wavelength anomalous dispersion (SAD) using HKL2MAP30 and Shelx C/D/E pipeline31. An initial model was produced by Buccaneer32 using the SAD data and used as a search model for molecular replacement using Molrep33 on the native data. The model was completed using iterative cycles of refinement with Refmac5 (ref. 34) and manual model-building using COOT35. The Rfree set of reflections was 5% of the unique reflections, randomly selected. The model was validated using Molprobity36.
Crystallisation and structure determination of OM complexes
BT2261-2264
Sitting drop crystallisation trials were set up using a Mosquito crystallisation robot (TTP Labtech) using commercial screens (Molecular Dimensions MemGold 1 and 2) and in-house screens. The triclinic and orthorhombic crystals for native BT2261-64 as well as the crystals for SeMet-substituted complex were produced by the same condition (MemGold 2 #44; 0.2 M magnesium formate dihydrate, 0.05 M Tris pH 8, 18-22 % w/v PEG 3350), although the triclinic crystals occurred very infrequently. Diffraction data were collected at 100 K at the Diamond Light Source (Didcot, UK), on beamlines I03 (BT2263), I04 (BT2261-2264 and BT1762-1763) and I24 (BT2261-2264). All diffraction data were processed with XDS and Pointless was used for space group determination. Data were scaled using Aimless.
After obtaining well-diffracting crystals for native BT2261-64 we performed molecular replacement (MR) trials in Phaser37 with both crystal forms, using components available within the PDB at that time. These consisted of FepA38 (PDB ID 1FEP; 14% identity to BT2264), BT2259 (PDB ID 4Q69; 26% identity to BT2263) and BT2261 (PDB ID 3H3I). However, none of these trials produced a plausible solution, including searches that were performed with Sculptor-modified39 search models. We then obtained crystals of soluble BT2263 and solved its structure via MR using BT2259 as a search model (26% sequence identity). Subsequently a definite MR solution for the complex was found by Phaser for a P1 crystal using BT2263 and Sculptor-modified FepA as search models (RFZ=14.7 TFZ=* PAK=0 LLG=503 RFZ=10.4 TFZ=29.9 PAK=0 LLG=1195 RFZ=5.5 TFZ=10.3 PAK=3 LLG=1263 RFZ=5.7 TFZ=9.0 PAK=7 LLG=1220 LLG=1789; 2 copies each of BT2263 and BT2264). Subsequent Autobuilding within Phenix39 yielded a model with Rfree of ~44%. Placement of two copies of BT2261 via MR was straightforward at this stage. Subsequent rounds of manual building of BT2264 and the N-terminal domain of BT2262 within COOT35 depended critically on using density-modified maps produced by RESOLVE40, in particular after application of 2-fold non-crystallographic symmetry (NCS). During the later stages of model building and Phenix refinement, we solved the structure of BT2262 by molecular replacement using the N-terminal domain (residues 4-110) from the complex as a search model. When the Rfree of the model for the complex reached ~28%, BT2262 was placed in the density, guided by its N-terminal domain. Refinement of B-factors was done isotropically including TLS, with each protein chain taken as a separate group. NCS was not used. For all structures, protein geometry was validated with MolProbity36. Figures were produced with PyMOL41 (Schrödinger LLC). In the final model of BT2261-64, only one copy of BT2262 is visible within the complex (residues 4-212; numbering from the first residue of the mature sequences), with poor density for the C-terminal domain. Besides BT2262, the final model contains two copies of BT2264 (residues 37-984), two copies of BT2263 (residues 1-480) and two copies of BT2261 (residues 3-148). Of the two complexes in the asymmetric unit, the best density is observed for the complex consisting of chains ABFG, and the electron density maps for the peptide reported here were based on this complex. For the other complex in the P1 asymmetric unit (chains CDE) as well as for some of the complexes in the other space groups, the peptide density is slightly different. It is not clear at this point whether the differences in density for the peptide are a consequence of the possible presence of a bound ensemble.
During the later stages of structure refinement of BT2261-64 we also obtained diffracting crystals for SeMet-substituted complex in space group P21. Both SAD and MAD datasets were collected for these crystals but none of these provided phases of sufficient quality to solve the structure de novo. Given that the anomalous signal was weak, the failure of phasing was most likely due to a low level of SeMet incorporation by B. theta. However, anomalous difference maps could be produced that provided a useful independent registry check via the 51 methionine residues present in the tetrameric BT2261-64 complex.
The structures of the BT2261-2264 complex in space groups P212121 and P21 were solved by MR in Phaser using the individual components from the P1 crystals as search models, and refined with Phenix. The orthorhombic complex contains 2 molecules of each of the subunits, with the C-terminal domains of both BT2262 subunits missing. In the monoclinic, SeMet complex, no density is visible for either of the BT2262 subunits, confirming the dynamic nature of this subunit (Extended Data Fig. 2). Extended Data Fig. 2 also shows that the triclinic structure has the lowest B-factors and consequently yielded the best-quality maps. The data collection and refinement statistics are summarised in Supplementary Table 1 (BT2261-64) and 2 (BT2263, BT2262 and BT1762).
BT1762-1763
Two different crystal forms were obtained for BT1762-63. First, a single diamond-shaped plate was obtained from MemGold 2, condition 18 (crystal form 1; 0.1 M magnesium formate dihydrate, 0.1 M MOPS pH 7, 17 % w/v PEG 3350), and cryoprotected by the addition of ~20% glycerol. A dataset to 3.1 Å resolution collected on this crystal at DLS beamline I04 generated a definite MR solution with Phaser (RFZ=6.8 TFZ=14.4 PAK=0 LLG=173 TFZ==14.7 RFZ=7.0 TFZ=7.6 PAK=9 LLG=627 TFZ==10.4 LLG=807 TFZ==10.0; space group P21212), using the BT1762 structure and a Sculptor-generated model of BT2264 (21% sequence identity) as search models. In this crystal, there is only one SusC protomer within the asymmetric unit, with the SusC homodimer generated by crystallographic symmetry. Autobuilding within Phenix resulted in a model with Rfree ~43-44% (R ~39%), comparable to BT2261-64 at a similar stage of refinement. Due to the lack of non-crystallographic symmetry (NCS) the maps were of insufficient quality to build a more complete model for BT1763. We then obtained another crystal form (crystal form 2; P212121) from a different preparation with 2 molecules in the AU from 18-20% w/v PEG 3350, 50 mM Hepes pH 7.0, 0.2 M NH4NO3. The best crystal diffracted X-rays to ~3.0 Å resolution. Running Phaser with BT1762 and the partial, Autobuilt model of BT1763 gave a single, definite solution (RFZ=11.7 TFZ=24.0 PAK=0 LLG=610 TFZ==27.7 RF++ TFZ=45.1 PAK=0 LLG=1771 TFZ==42.7 RFZ=6.1 TFZ=36.4 PAK=0 LLG=2747 TFZ==37.0 (& TFZ==31.6) LLG+=(2747 & 3697) LLG=3740 TFZ==38.1 PAK=0 LLG=3740 TFZ==41.5). Following Autobuilding within Phenix, a model was built via iterative cycles of manual building and refinement within Phenix. During later stages of refinement, NCS was omitted and TLS refinement of B-factors was applied (one domain per chain). The structure of crystal form 1 was then solved via MR, followed by refinement in Phenix as described above. The data collection and refinement statistics for BT1762-63 are summarised in Supplementary Table 3.
Knockout construction of BT2264 and growth experiments
Two rounds of PCR were used to create a product with the desired sequence, including 1000 bp upstream and downstream of the knockout region. This was ligated with pExchange-tdk vector11 which was then used for genetic recombination. The BT2264 knockout and WT strains were grown on TYG rich media and minimal media containing 0.5% fructose or glucose. In addition, growth on MM-Fructose plus tryptone (1%) was tested. The cells were incubated for 72 hours at 37 °C under anaerobic conditions and the A600 was taken every 15 minutes using a plate reader. The BT2264 knockout strain showed no growth phenotype under any of the conditions tested.
Western blot analysis
pExchange-tdk11 was used to add a 4xAla linker and FLAG-tag (DYDDDDK) to the C-terminus of BT2263 (SusD) in WT B. theta. The resulting mutant strain was grown in TYG rich media and minimal media containing 0.5% fructose, and samples were collected over 24 hrs. The whole cells were diluted to relative CFU (colony forming units) using Bugbuster detergent and solubilised. The solubilised material was analysed by western blot using Sigma F7425 Anti-FLAG (1 in 2000 dilution) as the primary antibody and Goat anti-rabbit IgG-HRP: sc-2004, Santa Cruz Biotechnology (1 in 5000 dilution) as the secondary antibody.
Isothermal Titration Calorimetry
BT1762
Site directed mutagenesis of the mature form of BT1762 cloned into pRSETA (Invitrogen) was performed using the Quikchange kit (Agilent) according to the manufacturers protocol. The affinity of wild type and mutants (numbered from the mature form of the protein) for levan was quantified by isothermal titration calorimetry (ITC) using a Microcal VP-ITC. Briefly, the protein sample (50–60 μM), stirred at 300 rpm in a 1.4 ml reaction cell, was injected with 27-29 x 10 μl aliquots of levan (0.5-2% w/v; Montana polysaccharides, USA). Titrations were carried out in 20mM Na-HEPES buffer, pH 7.5 at 25 °C. Integrated binding heats, minus dilution heat controls, were fit to a single set of sites binding model to derive Ka using Microcal Origin v7.0. At least two independent titrations were performed for each protein tested. The molar concentration of binding sites present in levan was determined by altering the concentration of ligand used for regression of the isotherm until the fit yielded a value of 1 for n (number of binding sites on each molecule of protein) for the wild-type and using this value for all titrations. The assumption that n = 1 was deemed valid based on the structure and mutant binding data.
BT2263
We probed the possible interaction between two synthetic oligopeptides (Biomatik Corporation, USA) and BT2263 SusD-like. The first peptide was a hypothetical, negatively charged sequence consistent with the density (GSSGGQQEGG), whereas the second contained several positively charged residues (GDSGSKQKKG), again based on a possible assignment. In addition, the binding to poly-DL-alanine (Sigma P9003; Mw 1000-5000) was investigated. Peptides at 10 mM in buffer were titrated to 40 μM BT2263 as described above.
Native mass spectrometry
The protein sample was buffer exchanged to 1 M ammonium acetate containing 0.5% C8E4 utilizing a biospin 6 column (Bio-Rad). Typically, 2-3 μl of the protein samples were loaded onto a gold-coated borosilicate capillary (1 mm outer diameter). Proteins were transmitted into a Synapt HDMS mass spectrometer (Waters) modified to facilitate the transmission of high mass species via a nano-ESI apparatus. Instrument parameters were set as follows: backing pressure- 6 mBar, capillary voltage- 1.6 kV, cone voltage-170 V, trap and transfer collision energy were 200 V for the BT2261-2264 complex and 100 V for BT1762-1763. Data analysis was carried out using Masslynx 4.1 (Waters).
Size exclusion chromatography multi-angle light scattering (SEC-MALS)
SEC-MALS was performed using an ÄKTA Pure with Superdex 200 10/300 GL Increase (GE Healthcare), with in-line DAWN HELIOS II MALS detector (eight fixed angles) and Optilab T-rEX differential refractometer (Wyatt Technology). BT1762-63 (100 μl of 7 mg/ml) was analysed in 10 mM Hepes pH 7.5, 150 mM NaCl, 0.05% dodecyl-β-D-maltoside. Data were collected and analysed using ASTRA 6 (Wyatt Technology). Molecular weights for the protein-detergent complex and its protein and detergent constituents were determined through protein-conjugate analysis with Zimm plot extrapolation using dn/dc values of 0.1850 ml/g and 0.1435 ml/g for protein and detergent respectively, and a protein UV extinction coefficient of 1.86 ml/mg/cm.
Refolding of Sus proteins from SDS-PAGE
The proteins were refolded from gels using a simple refolding protocol. Briefly, the samples were mixed with loading buffer (final concentrations: 75 mM Tris pH 6.8, 15 % glycerol, 0.6 % SDS, 5 % β-Mercaptoethanol, 0.01 mg/ml bromophenol blue) and heated for 15 min at 90° C. Each sample was loaded onto multiple wells for a higher yield on a 12% SDS gel along with the PageRuler™ Prestained Protein ladder (Thermo Scientific, Product no. 26617). Part of the gel was silver stained. Thin slices of the unstained gel were excised along the expected size of the band by comparing with its stained counterpart and were transferred to a vial containing 1% Genapol. Gel pieces were crushed gently for about half an hour using a micro pestle to make the solution homogenous. The suspension was kept for overnight incubation at 8°C with 550 rpm shaking on a Thermomixer. Following overnight incubation, the mixture was centrifuged at 3500 x g for 30 min and the supernatant was collected and checked for channel activity by electrophysiology.
Single Channel Electrophysiology
Electrophysiology measurements were carried out using a single channel reconstitution protocol as described elsewhere42. In short, a teflon cuvette was used in which a 25 μm thick Teflon film with an aperture size of around 50 -100 μm was inserted between the two chambers. 1-2 μl of a solution containing 1 % Hexadecane in Hexane was used to impregnate the area around the aperture to make it more hydrophobic (lipophilic) and left to dry. The two chambers were filled with 1 M KCl and 10 mM HEPES pH 7. Around 2 μl solution of 5 mg DPhPC (diphytanoylphosphatidylcholine, Avanti polar lipids, Alabaster, AL) dissolved in 1 ml n-pentane was used to form the membrane using the Montal and Muller technique43. A pair of Ag/AgCl electrodes (World Precision Instruments, Sarasota, FL) was used to measure the ionic current across the membrane where one of the electrodes is connected to the cis (ground) side of the membrane and the other was connected to the trans headstage of the Axopatch 200B amplifier (Axon Instruments). Protein dissolved in 1 % Genapol was always added to the cis side of the membrane. The ion current trace was filtered using a low pass Bessel filter at 10 kHz and was recorded using a sampling frequency of 50 kHz. Data were analysed using the Clampfit program. The data thus obtained were filtered at 5 kHz for obtaining a clear view of the traces. The data shown in Fig. 4d were obtained at +50 mV.
Molecular Dynamics simulations
Molecular dynamics (MD) simulations were carried out with GROMACS 4.6.5 (ref. 44) using the all atom CHARMM36 force field for protein, lipids and ions45,46 using standard procedures for membrane proteins47. Long-range Coulomb interactions were calculated using the partice-mesh Ewald (PME) summation method with a short-range cutoff of 1.2 nm and a Fourier grid spacing of 0.12 nm. At the same time, Lennard-Jones interactions were considered up to a distance of 1 nm, and a switch function was used to smoothly switch off the interactions to reach zero at 1.2 nm. The unbiased simulations were performed under constant pressure (1 bar) and constant temperature (300 K) with a time step of 2 fs by applying constraints to the bonds of hydrogen atoms using the LINCS algorithm48. The MD simulations were performed for different starting structures (dimer, tetramer and octamer) of the BT2261-2264 SusCD complex. The missing residues in the periplasmic turn (L570-F578) of SusC were modeled using Modeler 9.12 (ref. 49). In case of the octamer, the missing second copy of BT2261 was added manually. The starting structures of the SusCD complex in the peptide-bound conformations were embedded in a symmetric bilayer consisting of POPE (palmitoyloleoyl-phosphatidylethanolamine) lipids using the membrane builder in the CHARMM-GUI50. Subsequently, the lipid bilayer and the protein were solvated using a TIP3P water box and further neutralized by adding the appropriate number of potassium ions. The holo systems were equilibrated using the well-established protocol developed in the CHARMM-GUI Membrane Builder. We used the sequence GSSGGENQGG for the bound peptide in the holo simulations. This sequence is consistent with the observed density in the best-defined SusCD complex of the P1 structure (PDB ID 5QF8). For the apo simulations, the bound peptides were removed from the respective equilibrated structures and 20 ns of equilibration was carried out. Finally, a total of 7 μs unbiased simulations were performed with no constraints on the system for the holo-complexes (dimer and tetramer: 3 x 0.5 μs each and octamer: 1 x 0.5 μs) and apo-complexes (dimer and tetramer: 3 x 0.5 μs each and octamer: 1 x 0.5 μs).
Data Availability
The datasets generated (coordinate files and structure factors) for BT2261-2264 have been deposited in the Protein Data Bank with accession codes 5FQ8, 5FQ7 and 5FQ6 for BT2261-2264 (space groups P1, P212121 and P21 respectively), 5FQ3 for BT2262 and 5FQ4 for BT2263. For BT1762-1763 the accession numbers are 5LX8 for BT1762 and 5T3R and 5T4Y for BT1762-63 (space groups P21212 and P212121 respectively).
Extended Data
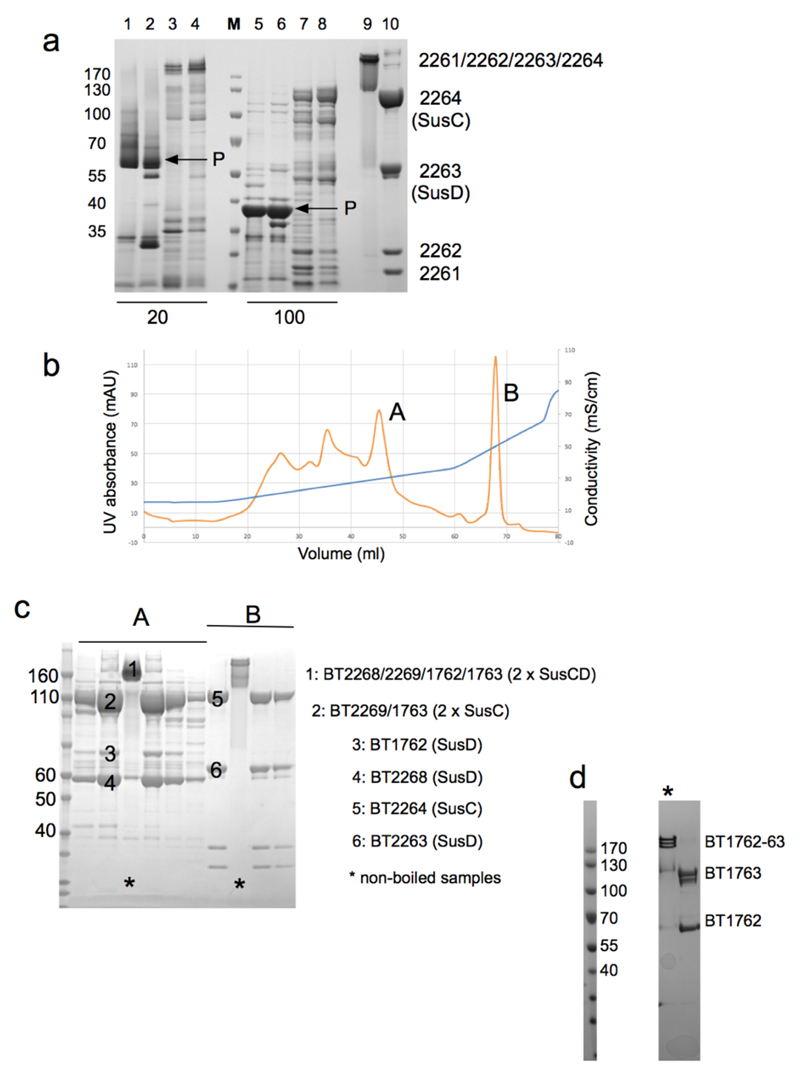
Extended Data Fig. 1. High abundance of SusCD complexes in B. theta OM.
a, SDS-PAGE gel of total outer membranes from E. coli (lanes 1,2,5,6) and B. theta (lanes 3,4,7,8). Each lane contains ~10 μg protein. Samples 5-8 were boiled (100). "P" denotes E. coli trimeric porins OmpF/C, which migrate at their monomeric molecular weights (~35 kDa) only after boiling. Note the relative lack of small-molecule OM diffusion channels (~30-50 kDa) in B. theta (lanes 7,8) and the low levels of large OM proteins including TBDTs (70-120 kDa) in E. coli (lanes 5,6). Purified BT2261-2264 complex is shown in lanes 9 (non-boiled) and 10 (boiled). b, Representative ion-exchange chromatogram from three separate experiments of B. theta total OM proteins separated on Resource-Q (6 ml, pH 7.5) after extraction in LDAO (Methods). Peaks A and B were further purified by gel filtration. c, SDS-PAGE gel of purified SusCD complexes from peaks A and B. Numbered gel bands were excised and subjected to identification by mass spectrometry. d, SDS-PAGE gel of purified BT1762-63 complex before (*) and after boiling. The SusCD complexes are highly stable and remain intact in 2% SDS.
Extended Data Fig. 2. The oligomeric nature of SusCD complexes is not a consequence of crystal packing.
Cartoon side views of BT2261-2264 complexes rotated by 90° for space groups P1 (a), P212121 (b) and SeMet P21 (c). d, Cartoon side view of BT1762-1763 (P212121). The protein backbones are coloured on the same scale by their B-factors (blue; 20 Å2, red; 130 Å2). The grey bars indicate the hydrophobic phase of the OM. Structures were determined using data obtained from a single crystal in each case.
Extended Data Fig. 3. BT2261-2264 and BT1762-1763 form oligomeric complexes.
a, Mass spectrum of BT2261-2264 shows two prominent masses corresponding to an octamer and a ligand bound octamer in m/Z=12000-15000. b, Mass spectrum of BT1762-1763 indicates that these two proteins form dimers and tetramers. The numbers in parentheses on the right are the theoretical masses. c, Analytical gel filtration chromatography of BT2261-64 (blue) and BT1762-63 (green). For comparison, samples were run for soluble horse spleen ferritin (440 kDa; red) and for the membrane protein ammonium transporter Mep2 from Candida albicans (160 kDa; black). Buffer: 10 mM Hepes/100mM NaCl/0.12% DM pH 7.5. Column: Superdex-200 Increase 10/300 GL. d, Size-exclusion chromatography multi-angle light scattering (SEC-MALS) analysis of BT1762-63. Light scattering (LS) and differential refractive index (dRI) are plotted alongside the fitted total protein-detergent complex molecular weight (diamonds), and constituent protein (pluses) and detergent (crosses) molecular weights, across each peak. BT1762-63 eluted as two species of 499 kDa (protein component 319 kDa, corresponding to a SusCD dimer) and 269 kDa (protein component 214 kDa). Chromatograms shown are from single experiments.
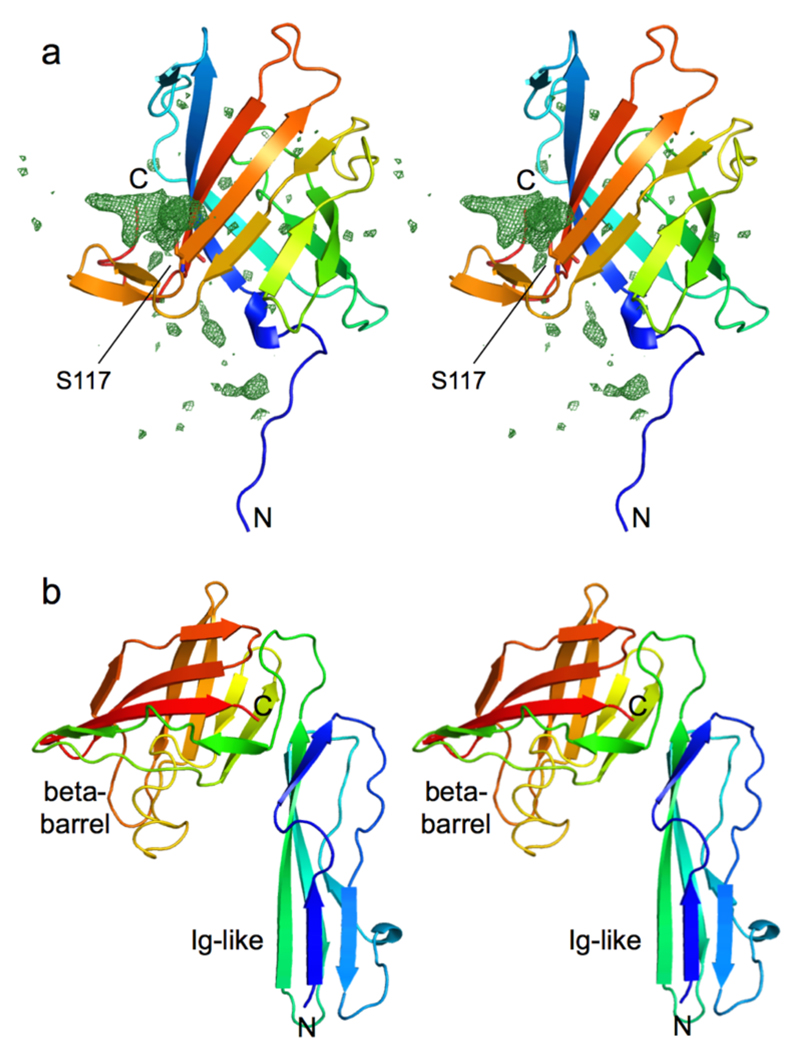
Extended Data Fig. 4. X-ray crystal structures of the small lipoproteins BT2261 and BT2262.
a, Stereo cartoon of BT2261 within the BT2261-2264 complex with rainbow colouring (blue; N-terminus). BT2261 is O-glycosylated on Ser117, consistent with the presence of the Bacteroidetes glycosylation motif D-S/T-A/L/V/I/M/T 51. Ser117 is shown as a stick model. Fo-Fc density within 20 Å of Ser117 is shown as a green mesh contoured at 3.0 σ. Three to four sugar moieties can be observed bound to Ser117. b, Stereo cartoon of soluble BT2262 with rainbow colouring. The protein consists of an N-terminal Ig-like domain and a C-terminal 8-stranded β-barrel. The functions of BT2261 and BT2262 are not clear, but both contain a small C-terminal 8-stranded β-barrel that displays structural similarity to lipid binding domains as judged by DALI 52. For BT2262, only one copy with a poorly ordered C-terminal domain is visible in the triclinic structure. Analogous to BT2263, the N-terminal segments of BT2261/62 that lead to the lipid anchors on the N-terminal cysteine residues are visible in the electron density; they are closely associated with SusC and do not appear to be flexible. Structures were determined using data obtained from a single crystal in each case.
Extended Data Fig. 5. Unbiased electron density for the bound ligand in BT2261-64.
Stereo views of simulated annealing omit maps using a starting temperature of 1000 K. a, 2Fo-Fc maps contoured at 1.5 σ, carve=2. b, Fo-Fc map contoured at 3.0 σ, carve=2. Selected residues contacting ligand are shown (yellow; BT2264/SusC, magenta, BT2263/SusD). Density for at least six amino acid side chains is present (denoted with * in the 2Fo-Fc map). c, Interaction Table showing hydrogen bond distances between the putative peptide ligand backbone and residues in BT2263 and BT2264.
Extended Data Fig. 6. MD simulations for BT2261-64.
a, Plots of BT2264 (SusC) Cα RMSD vs. simulation time for holo and apo complexes. b and c, Plots of BT2263 (SusD) Cα RMSD vs. simulation time for holo and apo simulations, relative to the starting conformation (b) and after SusD superposition (c). d,e Plots showing the number of hydrogen bonds between SusC and SusD vs. simulation time (d) and between holo SusCD and the modeled peptide (e). Simulations are numbered as follows: sim1-3, apo BT2263-64 (dimer); sim7-9, apo BT2261-64 (tetramer); sim13, apo (BT2261-64)x2 (octamer); sim4-6, holo BT2263-64; sim10-12, holo BT2261-64; sim14, holo (BT2261-64)x2. With the exception of those of the octamer owing to its very large size, the simulations were repeated three times with different initial atomic velocities to allow sampling in order to obtain a measure of the possible spread in results.
Extended Data Fig. 7. Dynamics of the bound peptide during MD simulations.
Side views (left panels) and top views showing the bound peptide in the BT2263-64 dimers (a), BT2261-64 tetramers (b) and the (BT2261-64)x2 octamer. For clarity, only one final conformation for BT2264 (SusC) is shown together with the starting conformation of the peptide (green) and the final peptide conformations after 500 ns of simulation (red). For orientation purposes the assigned N-termini of the peptides are coloured blue.
Extended Data Fig. 8. MD simulation root-mean-square-fluctuation (RMSF) analyses.
(a) Cα RMSF values for SusC in holo and apo complexes with the conformational fluctuations of the hinge loop L7 highlighted separately. (b) Cα RMSF values for SusD in apo and holo simulations. Simulations are numbered as in Extended Data Fig. 6: sim1-3, apo BT2263-64 (dimer); sim7-9, apo BT2261-64 (tetramer); sim13, apo (BT2261-64)x2 (octamer); sim4-6, holo BT2263-64; sim10-12, holo BT2261-64; sim14, holo (BT2261-64)x2.
Extended Data Fig. 9. Structure of the BT2261-64 apo-octamer after 500 ns of MD simulation (sim13), demonstrating the independent bin opening of the two SusCD hubs.
a, Views from the plane of the membrane rotated by 90º. b, View from the extracellular side. For clarity, the SusC and SusD subunits are shown in different colours (yellow and orange for SusC/BT2264, magenta and red for SusD/BT2263). BT2261 and BT2262 are shown in green and blue, respectively. c, Side view of the opened SusCD monomer highlighting the remaining interactions between SusC (yellow) and SusD (magenta) mediated by the hinge loop L7 and loop L8.
Extended Data Fig. 10. ITC analysis of levan binding for recombinant BT1762 SusD-like wild-type and mutants.
a, Titration curves from single experiments. Upper panels show raw injection heats of ligand (levan) into protein, lower panels show the integrated binding heats fit to a single set of sites binding model to determine Ka for all proteins except reduced wild-type (10 mM TCEP), W85A and C298A mutants that display no binding. Levan stock solution was between 0.5-2% (w/v) and protein ranged from 50-60 μM. b, Levan affinity of recombinant BT1762 SusD-like wild type and mutant proteins determined by ITC. Ka values shown are averages and standard deviations from at least two independent titrations. Residue numbering is that of the mature protein (first residue Cys1).
Supplementary Material
Acknowledgements
We would like to thank Joe Gray (Newcastle University) for B. theta OM protein identification, Richard Lewis (Newcastle University) for critical reading of the manuscript and Susan Buchanan (NIDDK; NIH) for the pET9 expression vector. We thank Owen Davies (Newcastle University) for help with SEC-MALS analysis. We are also indebted to the staff at beam lines I24, I04 and I03 of the Diamond Light Source UK for beam time (proposal mx9948) and assistance with data collection. AJG acknowledges support from the Barbour Foundation UK. SBP was funded by EU FP7-PEOPLE-2013-ITN Translocation network Nr. 607694. The research of KRP, UK, MW and BvdB has received support from the Innovative Medicines Initiatives Joint Undertaking under Grant Agreement No. 115525, resources which are composed of financial contributions from the European Union’s seventh framework programme (FP7/2007-2013) and European Federation of Pharmaceutical Industries and Associations companies in-kind contribution.
Footnotes
Author contributions AJG expressed, purified and crystallised proteins and carried out western blot analysis. KRP and UK performed molecular dynamics simulations. SPB and MW performed and supervised electrophysiology experiments, respectively. DC and CVR carried out mass-spectrometry analyses. AB collected crystallography data and managed the Newcastle Structural Laboratory. HZ expressed and purified BT1762 and performed ITC measurements. SJF and AB determined the structure of BT1762. DNB designed research and assisted in writing the paper. BvdB crystallised proteins and solved crystal structures, designed research and wrote the paper.
Author information The authors declare no financial interests.
References
- 1.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Marchesi JR, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2015 doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nature reviews Microbiology. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonnenburg ED, et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogowski A, et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nature communications. 2015;6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsbrink J, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuskin F, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. The Journal of biological chemistry. 2009;284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Elia JN, Salyers AA. Contribution of a neopullulanase, a pullulanase and an alpha-glucosidase to growth of Bacteroides thetaiotaomicron on starch. Journal of bacteriology. 1996;178:7173–7179. doi: 10.1128/jb.178.24.7173-7179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reeves AR, Wang GR, Salyers AA. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. Journal of bacteriology. 1997;179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shipman JA, Berleman JE, Salyers AA. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. Journal of bacteriology. 2000;182:5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annual review of microbiology. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martens EC, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS biology. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell host & microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolam DN, Koropatkin NM. Glycan recognition by the Bacteroidetes Sus-like systems. Current opinion in structural biology. 2012;22:563–569. doi: 10.1016/j.sbi.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 19.Kumar S, Engelberg-Kulka H. Quorum sensing peptides mediating interspecies bacterial cell death as a novel class of antimicrobial agents. Curr Opin Microbiol. 2014;21:22–27. doi: 10.1016/j.mib.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Mohammad MM, Howard KR, Movileanu L. Redesign of a plugged beta-barrel membrane protein. J Biol Chem. 2011;286:8000–8013. doi: 10.1074/jbc.M110.197723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koropatkin N, Martens EC, Gordon JI, Smith TJ. Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont. Biochemistry. 2009;48:1532–1542. doi: 10.1021/bi801942a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shultis DD, Purdy MD, Banchs CN, Wiener MC. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 2006;312:1396–9. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 23.Pawelek PD, et al. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 24.Jordan LD, et al. Energy-dependent motion of TonB in the Gram-negative bacterial inner membrane. Proc Natl Acad Sci U S A. 2013;110:11553–11558. doi: 10.1073/pnas.1304243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Celia H, et al. Structural insight into the role of the Ton complex in energy transduction. Nature. 2016;538:60–65. doi: 10.1038/nature19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. Journal of molecular biology. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 27.Kabsch W. XDS. Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans PR. Scaling and assessment of data quality. Acta Crystallogr D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 30.Pape T, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX programs. J Appl Cryst. 2004;37:843–844. [Google Scholar]
- 31.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Cryst. 2010;D66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowtan KD. The Buccaneer software for automated model building. Acta Crystallogr D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 33.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement'. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 34.Vagin AA, et al. REFMAC5 dictionary: organisation of prior chemical knowledge and guidelines for its use. Acta Crystallogr D. 2004;60:2284–2295. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchanan SK, et al. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Mol Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger TC. Maximum likelihood density modification. Acta Crystallogr D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The PyMOL Molecular Graphics System, Version 1.7.4. Schrödinger, LLC; [Google Scholar]
- 42.Schwarz G, Danelon C, Winterhalter M. On translocation through a membrane channel via an internal binding site: kinetics and voltage dependence. Biophys J. 2003;84:2990–2998. doi: 10.1016/S0006-3495(03)70025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montal M, Mueller P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc Natl Acad Sci U S A. 1972;69:3561–3566. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pronk S, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain x1 and x2 dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pothula KR, Solano CJ, Kleinekathöfer U. Simulations of Outer Membrane Channels and Their Permeability. Biochim et Biophys Acta. 2016;1858:1760–1771. doi: 10.1016/j.bbamem.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 48.Hess B, Bekker H, Berendsen HJC, Johannes GEMF. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 49.Eswar N, et al. Comparative Protein Structure Modeling Using Modeller. Current Protocols Bioinformatics. 2006;15:561–563. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 51.Coyne MJ, et al. Phylum-wide general protein O-glycosylation system of the Bacteroidetes. Molecular microbiology. 2013;88:772–783. doi: 10.1111/mmi.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucl Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated (coordinate files and structure factors) for BT2261-2264 have been deposited in the Protein Data Bank with accession codes 5FQ8, 5FQ7 and 5FQ6 for BT2261-2264 (space groups P1, P212121 and P21 respectively), 5FQ3 for BT2262 and 5FQ4 for BT2263. For BT1762-1763 the accession numbers are 5LX8 for BT1762 and 5T3R and 5T4Y for BT1762-63 (space groups P21212 and P212121 respectively).