Abstract
We showed before that long linear DNA molecules containing single-strand interruptions and undergoing pulsed-field gel electrophoresis (PFGE) tend to break into subfragments (electrophoretic nick instability). Here we show that circular chromosomal DNA with single-strand interruptions remains in the wells during PFGE. This means that the presence of nicks in immobile circular DNA is not enough to break this DNA during PFGE. In other words, under the conditions of our study, the artifactual conversion of nicks into double-strand breaks that we detect in linear DNA does not contribute to the overall level of chromosomal fragmentation, as measured by PFGE.
Keywords: double-strand breaks, single-strand breaks, ultraviolet light, nucleotide excision repair
Results
Pulsed-field gel electrophoresis (PFGE) extends the resolution range of agarose gel electrophoresis of nucleic acids from its upper limit of ~ 40 kbp [1] all the way to 10 Mbp [2]. PFGE is also a game-changer for detection of chromosomal fragmentation in prokaryotes [3], because intact prokaryotic chromosomes are mostly circular, and circular DNAs 30 kbp or longer fail to enter pulsed-field gels [4,5], while linear DNAs up to the full chromosome size enter the gel and resolve by size [2]. Development of methods for quantification of chromosomal fragmentation in E. coli facilitated mechanistic studies of this phenomenon in various mutants and/or under conditions of DNA damage [3,6–8]. In both types of perturbations of the DNA metabolism, the chromosomes suffer from multiple single-strand breaks (nicks), the most common type of DNA lesions. Some of these single strand interruptions would then be converted by chromosomal replication into double-strand breaks [9,10], and the resulting chromosomal fragmentation is detectable and quantifiable by PFGE.
Although circular DNA does not enter pulsed-field gels, and relaxation of the circular DNA should not change its (im)mobility, it was conceivable that some nicks could turn into double-strand breaks in PFGE conditions, and the resulting linear DNA would then enter the gel. For example, we have shown before [11] that long linear DNA fragments containing nicks are fragmented into smaller pieces during PFGE, displaying “electrophoretic nick instability”. At the same time, this artificial fragmentation during PFGE did not lead to increase in the fraction of the total DNA migrating in the gel as linear, suggesting that circular nicked species do not turn into linear species during PFGE and do not enter the gel [11]. To test directly whether nicked circular DNA in the agarose plug is broken artificially during electrophoresis, we did the following experiment.
Since a replicating chromosome, even if broken at a couple of places, may still have problem entering the gel due to its branched nature, we sought to minimize DNA replication in the chromosome that we nicked in vivo. For this, we grew E. coli cultures to early stationary phase to allow the replication activity to subside, irradiated these still metabolically active cells with UV doses (48 J/m2) producing ~3,000 pyrimidine dimers per genome equivalent [12], allowed short time for nucleotide excision repair (NER) to initiate excision of these lesions and isolated chromosomal DNA in agarose plugs for subsequent PFGE (Fig. 1A). Effectively, we have circular chromosomes with minimal replication activity, but at the same time with at least several single-strand interruptions due to initiated excision repair of multiple UV-lesions. If pulsed-field gel electrophoresis turns some of these single-strand breaks into double-strand breaks, we should have observed increased chromosomal fragmentation in the NER+ (Uvr+) UV-irradiated sample, and this increase would be mostly eliminated in the NER-negative (uvrA mutant) sample after UV-irradiation. Contrary to this expectation, we observed no statistically-significant increase in chromosomal fragmentation upon UV-irradiation, both in Uvr+ and uvrA mutant cultures (Fig. 1BC), suggesting no electrophoretic nick instability of circular DNA in PFGE.
Fig. 1. Multiply-nicked circles do not enter pulsed-field gel.
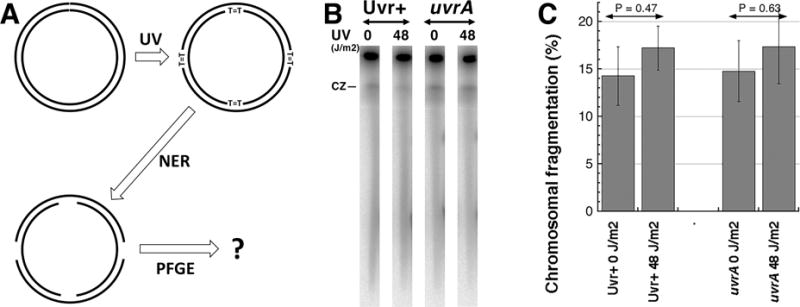
A. A scheme of the experiment: early-stationary cells (so that most chromosomes have no replication bubbles), grown with 32P orthophosphoric acid to label chromosomal DNA, are irradiated with high UV doses (introducing ~1,500 pyrimidine dimers per genome equivalent) as described [7,11]. Following UV exposures, the cells were diluted in spent LB to keep replication inhibited and, after giving 10 minutes at 37°C for nucleotide excision repair (NER) to initiate lesion removal, their chromosomal DNA is isolated in agarose plugs and subjected to pulsed-gel electrophoresis to reveal chromosomal fragmentation [7,8].
B. A representative pulsed-field gel of NER+ (AB1157) versus NER− (uvrA mutant, SRK303-1) cells. CZ, compression zone.
C. Quantification of several gels like in “B”. Chromosomal fragmentation is quantified as the percentage of the total DNA (well + lane) found in the lane. The data are averages ± SEM, with n = 4 or 5. P is a two-tail probability that two values are different (should be 0.05 or less for them to be considered different).
There was a possibility that NER loses its efficiency in the early stationary cells. To check whether there is enough nicks in these chromosomal DNA preparations, we used the demonstrated ability of PFGE to break long linear DNA molecules at nicks [11]. For this, we cut the same preparations of chromosomal DNA from the early stationary UV-irradiated cells with NotI restriction enzyme and followed the fate in PFGE of the longest NotI fragment (1.003 Mbp) (Fig. 2A). According to our previous findings, the presence of nicks in this fragment should destabilize it in PFGE conditions, reducing its representation in the overall digest signal. This is exactly what we have observed: UV-irradiation has reduced the 1 Mbp NotI fragment from the NER+ cells in half, but did not affect this same fragment from the NER− cells (Fig. 2BC). Since this fragment is ~4.5-times shorter than the overall E. coli chromosome, and yet is significantly degraded by the combined action of NER and PFGE, we conclude that circular chromosomes are not affected by the same electrophoretic nick instability.
Fig. 2. Nicked linear pieces are additionally fragmented during electrophoresis.
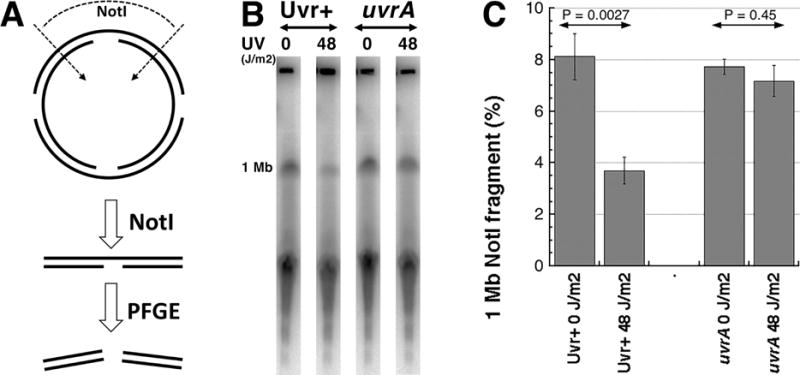
A. A scheme of the experiment: early-stationary cells were exposed to UV and made into agarose plugs as described in figure 1. The agarose plugs were washed in TE, digested with Not I and subjected to pulsed-gel electrophoresis to reveal any decrease in 1 Mbp band [11].
B. A representative pulsed-field gel of NER+ (AB1157) versus NER− (uvrA mutant, SRK303-1) cells. 1 Mb, the 1 Mbp Not I fragment, whose quantity is shown in “C” as the fraction of the total DNA signal in the gel.
C. Quantification of several gels like in “B”. The data are averages ± SEM, with n = 4 or 5. P is two-tail probability that the two values are different. The two values in the NER+ cells are (P<<0.05).
Our current results further support our previous conclusion that migration through the gel is required for breakage of long linear DNA at nicks. Besides circular DNA, chromosome-size replication intermediates are also unable to migrate into pulsed-field gel, because of their branched structure. Therefore, a small number of single-strand interruptions, even if converted into DSBs, may still be insufficient to force the branched DNA into the gel, making artificial inflation of chromosomal fragmentation after DNA damaging treatments of replicating cells even less likely. Another corollary of our finding is that, since double-strand breaks in DNA rarely happen without accompanying (and frequently more numerous) single-strand breaks, prokaryotic cells (with their circular chromosomes) offer a more reliable system to quantify the true density of double-strand breaks than eukaryotic cells (with their linear chromosomes) do.
In summary, the results of this study show that if, for any reason (in this case, the circular nature of the chromosomes) DNA molecules fail to enter the gel, they resist breakage during PFGE even if they harbor a few nicks.
Acknowledgments
This work was supported by grant # GM 073115 from the National Institutes of Health. The authors have no conflict of interest to declare.
Abbreviations
- UV
ultraviolet light
- NER
nucleotide excision repair
- CZ
compression zone
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maniatis T, Fritsch EF, Sambrook J. A laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1982. Molecular cloning; p. 545. [Google Scholar]
- 2.Cantor CR, Smith CL, Mathew MK. Pulsed-field gel electrophoresis of very large DNA molecules. Annu Rev Biophys Biophys Chem. 1988;17:287–304. doi: 10.1146/annurev.bb.17.060188.001443. [DOI] [PubMed] [Google Scholar]
- 3.Michel B, Ehrlich SD, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beverley SM. Characterization of the ‘unusual’ mobility of large circular DNAs in pulsed field-gradient electrophoresis. Nucleic Acids Res. 1988;16:925–939. doi: 10.1093/nar/16.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levene SD, Zimm BH. Separations of open-circular DNA using pulsed-field electrophoresis. Proc Natl Acad Sci U S A. 1987;84:4054–4057. doi: 10.1073/pnas.84.12.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarino E, Salguero I, Jiménez-Sánchez A, Guzmán EC. Double-strand break generation under deoxyribonucleotide starvation in Escherichia coli. J Bacteriol. 2007;189:5782–5786. doi: 10.1128/JB.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan SR, Kuzminov A. Replication forks stalled at ultraviolet lesions are rescued via RecA and RuvABC protein-catalyzed disintegration in Escherichia coli. J Biol Chem. 2012;287:6250–6265. doi: 10.1074/jbc.M111.322990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kouzminova EA, Rotman E, Macomber L, Zhang J, Kuzminov A. RecA-dependent mutants in E. coli reveal strategies to avoid replication fork failure. Proc Natl Acad Sci USA. 2004;101:16262–16267. doi: 10.1073/pnas.0405943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kouzminova EA, Kuzminov A. Fragmentation of replicating chromosomes triggered by uracil in DNA. J Mol Biol. 2006;355:20–33. doi: 10.1016/j.jmb.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 10.Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci USA. 2001;98:8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan SR, Kuzminov A. Trapping and breaking of in vivo nicked DNA during pulsed field gel electrophoresis. Anal Biochem. 2013;443:269–281. doi: 10.1016/j.ab.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]


