Abstract
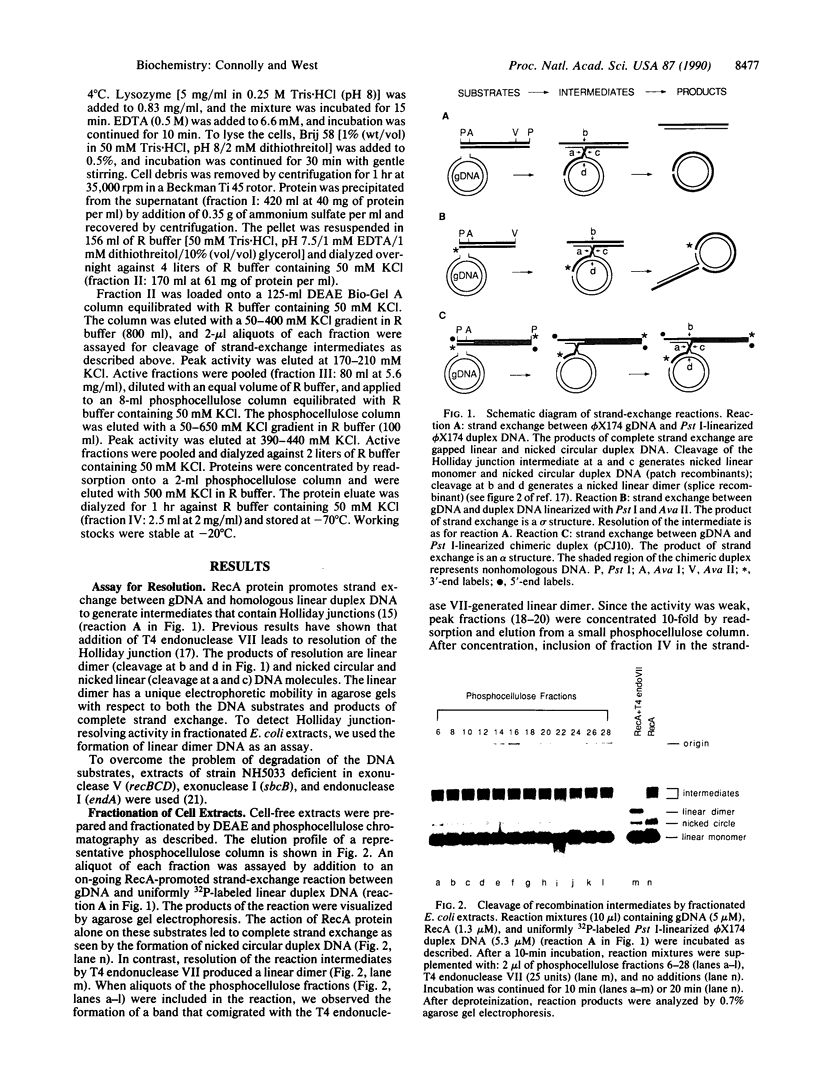
Escherichia coli RecA protein catalyzes reciprocal strand-exchange reactions between duplex DNA molecules, provided that one contains a single-stranded gap or tail, to form recombination intermediates containing Holliday junctions. Recombination reactions are thought to occur within helical RecA-nucleoprotein filaments in which DNA molecules are interwound. Structures generated in vitro by RecA protein have been used to detect an activity from fractionated E. coli extracts that resolves the intermediates into heteroduplex recombinant products. Resolution occurs by specific endonucleolytic cleavage at the Holliday junction. The products of cleavage are characteristic of patch and splice recombinants.
Full text
PDF

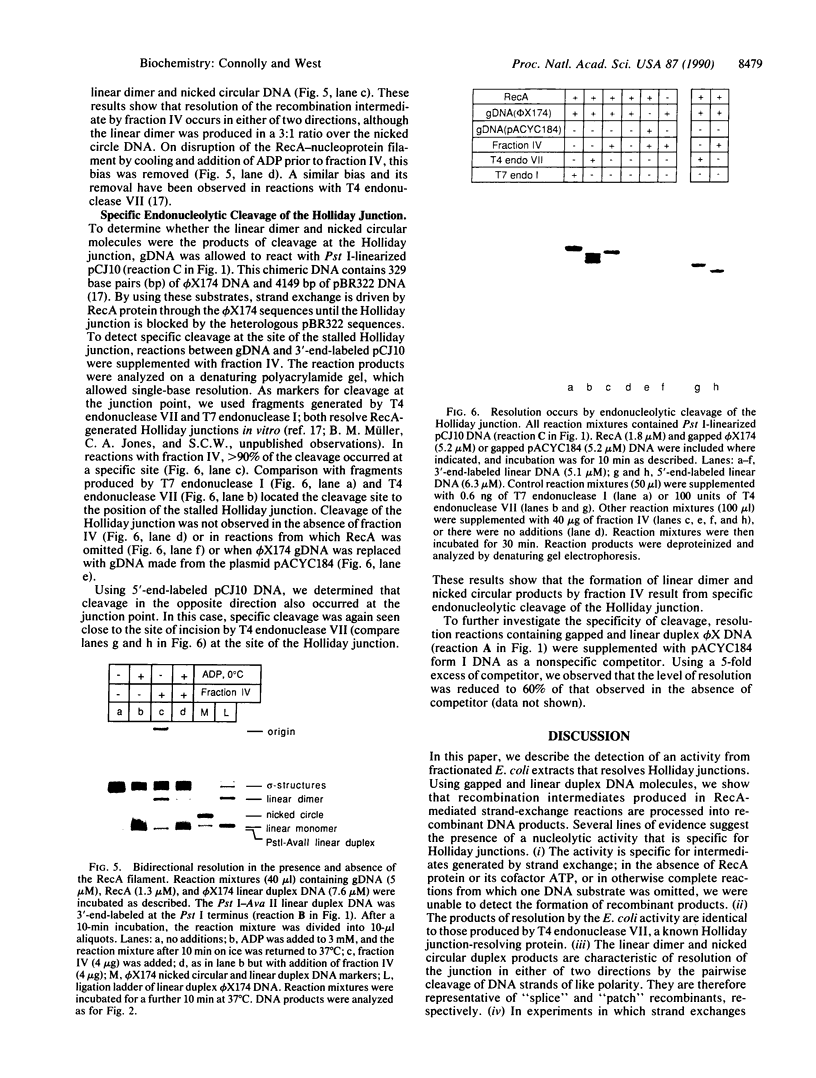

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- CLARK A. J., MARGULIES A. D. ISOLATION AND CHARACTERIZATION OF RECOMBINATION-DEFICIENT MUTANTS OF ESCHERICHIA COLI K12. Proc Natl Acad Sci U S A. 1965 Feb;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto E., Mursalim J., Howard-Flanders P. Homology-dependent cutting in trans of DNA in extracts of Escherichia coli: an approach to the enzymology of genetic recombination. Proc Natl Acad Sci U S A. 1978 Feb;75(2):620–624. doi: 10.1073/pnas.75.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley E. C., West S. C. Homologous pairing and the formation of nascent synaptic intermediates between regions of duplex DNA by RecA protein. Cell. 1989 Mar 24;56(6):987–995. doi: 10.1016/0092-8674(89)90632-6. [DOI] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. Enzymes of general recombination. Annu Rev Biochem. 1987;56:229–262. doi: 10.1146/annurev.bi.56.070187.001305. [DOI] [PubMed] [Google Scholar]
- DasGupta C., Wu A. M., Kahn R., Cunningham R. P., Radding C. M. Concerted strand exchange and formation of Holliday structures by E. coli RecA protein. Cell. 1981 Aug;25(2):507–516. doi: 10.1016/0092-8674(81)90069-6. [DOI] [PubMed] [Google Scholar]
- Elborough K. M., West S. C. Resolution of synthetic Holliday junctions in DNA by an endonuclease activity from calf thymus. EMBO J. 1990 Sep;9(9):2931–2936. doi: 10.1002/j.1460-2075.1990.tb07484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulds D., Dower N., Stahl M. M., Stahl F. W. Orientation-dependent recombination hotspot activity in bacteriophage lambda. J Mol Biol. 1979 Jul 15;131(4):681–695. doi: 10.1016/0022-2836(79)90197-9. [DOI] [PubMed] [Google Scholar]
- Howard-Flanders P., West S. C., Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984 May 17;309(5965):215–219. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- Jensch F., Kosak H., Seeman N. C., Kemper B. Cruciform cutting endonucleases from Saccharomyces cerevisiae and phage T4 show conserved reactions with branched DNAs. EMBO J. 1989 Dec 20;8(13):4325–4334. doi: 10.1002/j.1460-2075.1989.tb08619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Jensch F., von Depka-Prondzynski M., Fritz H. J., Borgmeyer U., Mizuuchi K. Resolution of Holliday structures by endonuclease VII as observed in interactions with cruciform DNA. Cold Spring Harb Symp Quant Biol. 1984;49:815–825. doi: 10.1101/sqb.1984.049.01.092. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Buckman C. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J Bacteriol. 1985 Nov;164(2):836–844. doi: 10.1128/jb.164.2.836-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi-DeLuca C., Lovett S. T., Kolodner R. D. Genetic and physical analysis of plasmid recombination in recB recC sbcB and recB recC sbcA Escherichia coli K-12 mutants. Genetics. 1989 Jun;122(2):269–278. doi: 10.1093/genetics/122.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Müller B., Jones C., Kemper B., West S. C. Enzymatic formation and resolution of Holliday junctions in vitro. Cell. 1990 Jan 26;60(2):329–336. doi: 10.1016/0092-8674(90)90747-3. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Rao B. J., Jwang B., Radding C. M. RecA protein reinitiates strand exchange on isolated protein-free DNA intermediates. An ADP-resistant process. J Mol Biol. 1990 Jun 20;213(4):789–809. doi: 10.1016/S0022-2836(05)80264-5. [DOI] [PubMed] [Google Scholar]
- Rosenberg S. M. Chi-stimulated patches are heteroduplex, with recombinant information on the phage lambda r chain. Cell. 1987 Mar 13;48(5):855–865. doi: 10.1016/0092-8674(87)90082-1. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. F., Smith G. R. Action of RecBCD enzyme on cruciform DNA. J Mol Biol. 1990 Jan 5;211(1):117–134. doi: 10.1016/0022-2836(90)90015-E. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Ogawa H. Structural genes of ATP-dependent deoxyribonuclease of Escherichia coli. Nat New Biol. 1972 Sep 6;239(88):14–16. doi: 10.1038/newbio239014a0. [DOI] [PubMed] [Google Scholar]
- West S. C., Cassuto E., Howard-Flanders P. Postreplication repair in E. coli: strand exchange reactions of gapped DNA by RecA protein. Mol Gen Genet. 1982;187(2):209–217. doi: 10.1007/BF00331119. [DOI] [PubMed] [Google Scholar]
- West S. C., Cassuto E., Howard-Flanders P. Role of SSB protein in RecA promoted branch migration reactions. Mol Gen Genet. 1982;186(3):333–338. doi: 10.1007/BF00729451. [DOI] [PubMed] [Google Scholar]
- West S. C., Countryman J. K., Howard-Flanders P. Enzymatic formation of biparental figure-eight molecules from plasmid DNA and their resolution in E. coli. Cell. 1983 Mar;32(3):817–829. doi: 10.1016/0092-8674(83)90068-5. [DOI] [PubMed] [Google Scholar]
- West S. C., Körner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M. Mutants of Escherichia coli lacking endonuclease I, ribonuclease I, or ribonuclease II. J Bacteriol. 1971 Jul;107(1):87–94. doi: 10.1128/jb.107.1.87-94.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., Weisberg R. A., Studier F. W. Gene 3 endonuclease of bacteriophage T7 resolves conformationally branched structures in double-stranded DNA. J Mol Biol. 1987 Jan 20;193(2):359–376. doi: 10.1016/0022-2836(87)90224-5. [DOI] [PubMed] [Google Scholar]